Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse
Abstract
1. Introduction
2. Results
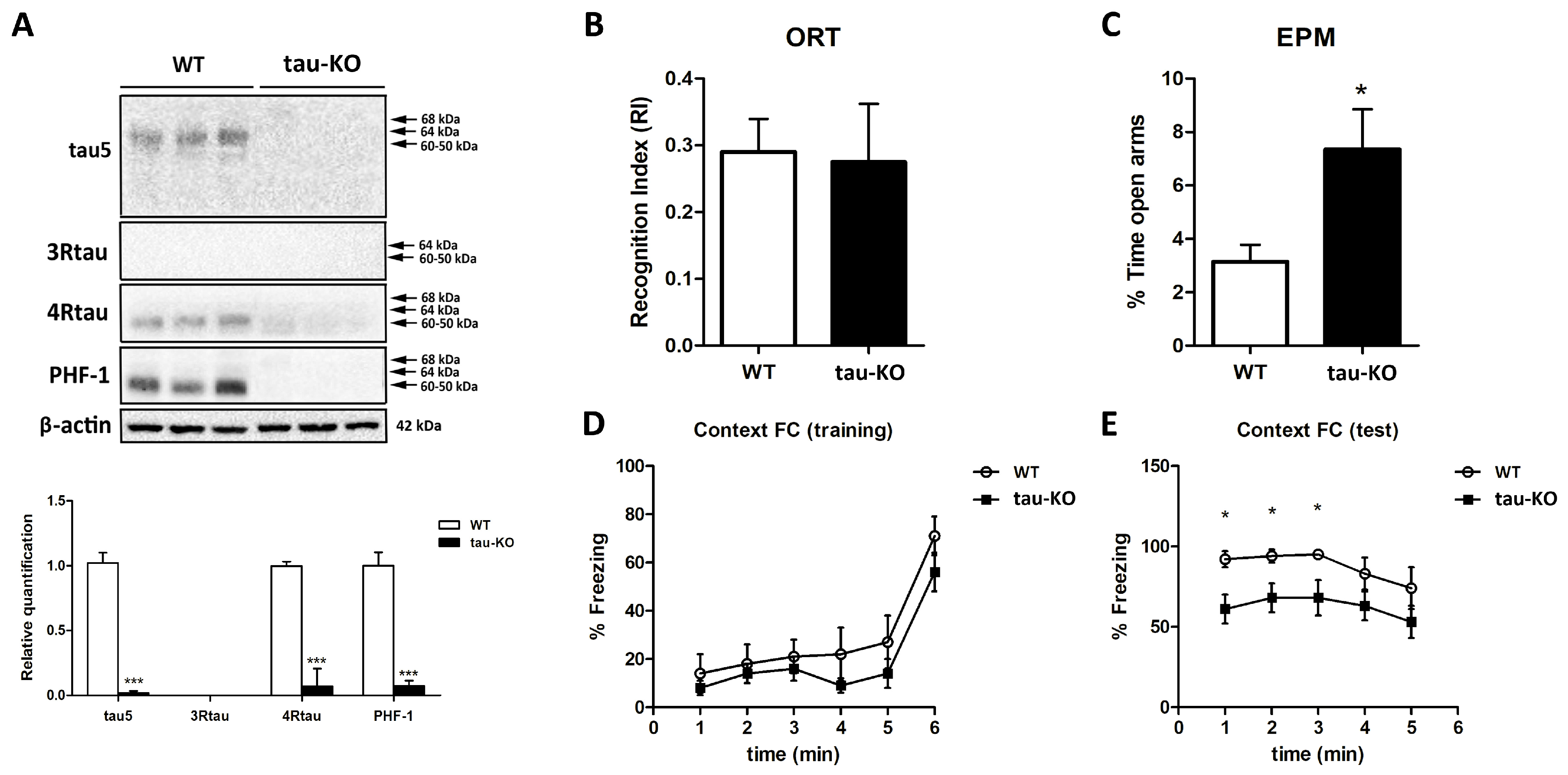
2.1. Molecular Characterization of Tau Protein and Behavioral Status of Tau-KO Mice
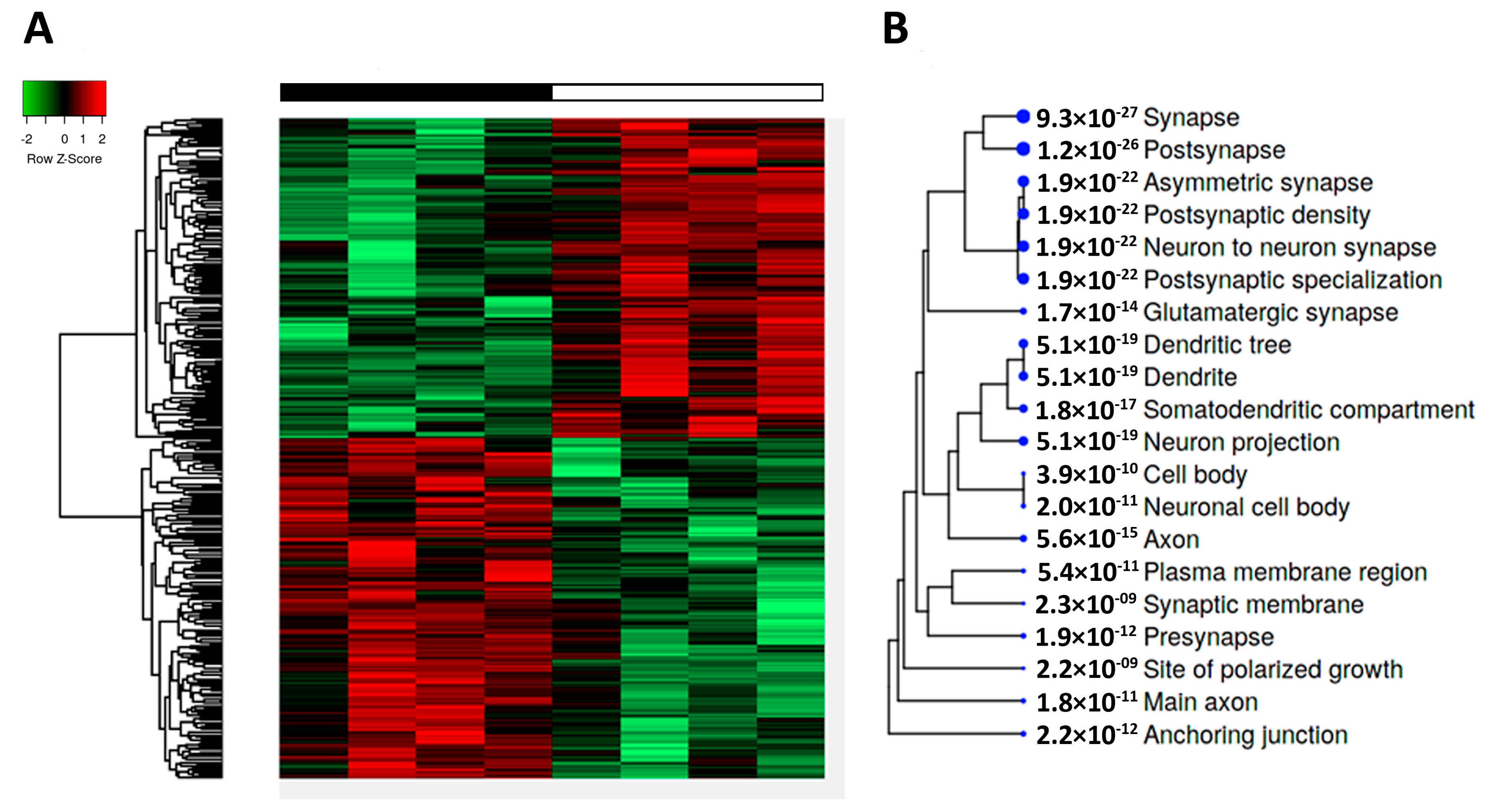
2.2. Transcriptomic Alterations in tau-KO Animals Using RNA-seq
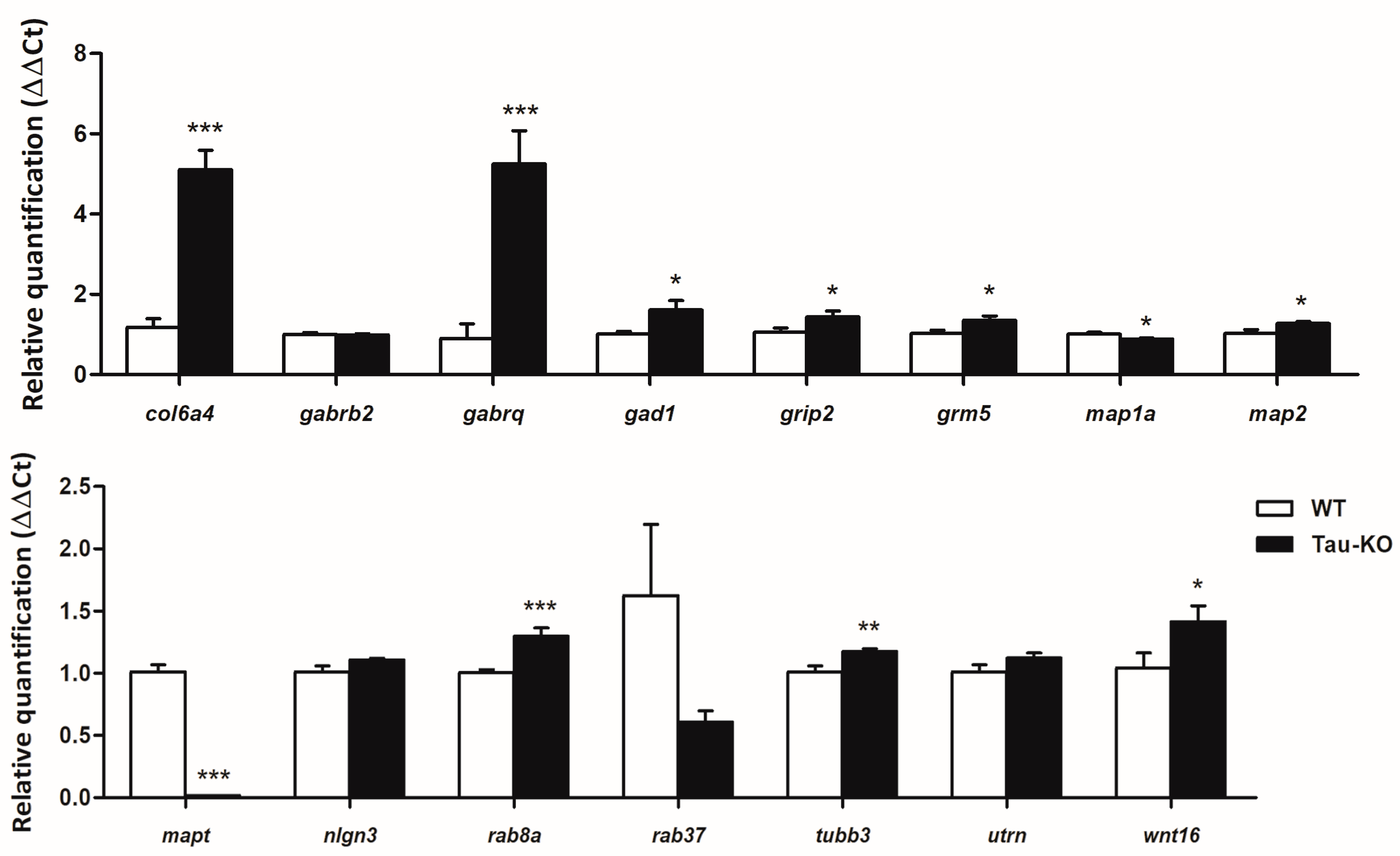
2.3. Gene Expression Validation
2.4. Phosphoproteome in Tau-KO Mice
2.5. Protein Validations Using Immunoblotting
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Mice Behavioral Evaluation Tests
- -
- Cognitive performance (two-object recognition test): This paradigm consists of placing the animals for 9 min in a V-maze containing two identical objects at the ends of the arms. Twenty-four hours after the training session, the animals are placed in the V-maze where one of the two familiar objects is replaced by a novel object. The time that the animals spend exploring the two objects is recorded in the training and test sessions; the object Recognition Index (RI) is calculated as the difference between the time spent exploring the novel (TN) and the familiar object (TF) during the test session, divided by the total time spent exploring the two objects [RI = (TN − TF)/(TN + TF)]. Animals exhibiting memory impairments revealed a lower object RI;
- -
- Fear conditioning: A contextual fear conditioning procedure was carried out with a computerized StartFear system (Panlab-Harvard, Barcelona, Spain). Shocks were delivered and controlled using Freezing v1.3.04 software (Panlab-Harvard). During fear and conditioning testing, the fear chamber consisted of a black methacrylate box with a transparent front door (25 × 25 × 25 cm) inside a sound-attenuating chamber (67 × 53 × 55 cm). During fear conditioning, mice were placed in the fear chambers for 5 min and then received two-foot shocks (1 s, 0.3 mA) separated by a one minute resting period. After the last shock, mice were left in the chamber for another minute. The association between the context and the shock was assessed 24 h after the conditioning during the test session. Mice were placed in the fear chambers for 5 min, and freezing behavior, a rodent’s natural response to fear, was automatically scored. Freezing was defined as the absence of movement except for respiration, and freezing scoring was carried out by a high-sensitivity weight transducer system at the bottom of the experimental chambers that records and analyzes the signal generated by the mouse’s movement. Mice were considered to have frozen when remaining immobile for more than 2000 ms. Episodes were averaged in 60 s slots using Freezing 911 v1.3.04 software (Panlab-Harvard);
- -
- Emotional Evaluation: Elevated Plus Maze: The Elevated Plus Maze assesses animals’ anxiety-like behavior. It involves placing the mice in a cross-shaped maze elevated 40 cm above the ground. Two arms of the maze are open laterally so that the animal can perceive the elevation of the labyrinth. Animals presenting lower anxiety levels exhibit higher open-arms exploration scores.
4.3. RNA Extraction
4.4. Transcriptomic Analysis
4.5. Phosphoproteomic Analysis
- -
- Label-free phosphoproteomics: A quantity of 600 µg of protein was used to obtain phosphorylated fractions. The reduction was performed for protein digestion by adding dithiothreitol (DTT) to a final concentration of 10 mM and incubating at RT for 30 min. Subsequent alkylation with 30 mM (final concentration) iodoacetamide was performed for 30 min in the dark at room temperature. An additional reduction step was performed with 30 mM DTT (final concentration), allowing the reaction to stand at room temperature for 30 min. The mixture was diluted to 0.6 M urea using MilliQ water, and after the addition of trypsin (Promega, Madison, WI, USA) (enzyme:protein, 1:50 w/w), the sample was incubated at 37 °C for 16 h. Digestion was quenched by acidification (pH < 6) with acetic acid. After protein enzymatic cleavage, peptide cleaning was performed using Pierce™ Peptide Desalting Spin Columns (ThermoFisher Scientific, Waltham MA, USA). The High-Select™ TiO2 Phosphopeptide Enrichment Kit (ThermoFisher Scientific, Waltham, MA, USA) was used to obtain the phosphorylated peptide fractions according to the manufacturer’s instructions. Phosphopeptide mixtures were separated by reversed-phase chromatography using an Eksigent NanoLC ultra-2D pump fitted with an Acclaim™ PepMap™ 100 C18 column (0.075 × 250 mm, particle size 3 µm; ThermoFisher Scientific, Waltham, MA, USA). Samples were first loaded for concentration into an Acclaim™ PepMap™ 100 C18 trap column (0.1 × 20 mm, particle size 5 µm; ThermoFisher Scientific, Waltham, MA, USA). Mobile phases were 100% water, 0.1% formic acid (FA) (buffer A), and 100% acetonitrile with 0.1% FA (buffer B). A column gradient was developed from 2% B to 40% B in 120 min. The column was equilibrated at 95% B for 10 min and 2% B for 10 min. The pre-column was aligned with the column, and flow was maintained along the gradient at 300 nL/minute. Eluting peptides were analyzed using a 5600 Triple-TOF mass spectrometer (Sciex, Framingham, MA, USA). Informational data acquisition was obtained upon a survey scan performed in a mass range from 350 m/z up to 1250 m/z in a scan time of 25 ms. The top 15 peaks were selected for fragmentation. The minimum accumulation time for MS/MS was set to 200 ms, giving a total cycle time of 3.3 s. Product ions were scanned in a mass range from 100 m/z up to 1500 m/z and excluded for further fragmentation for 15 s. The raw MS/MS spectra searches were processed using the MaxQuant software (v 1.6.7.0) [80] and searched against the Uniprot proteome reference for Mus Musculus (Proteome ID: UP000000_10090, March 2021). The parameters used were as follows: initial maximum precursor (15 ppm); fragment mass deviations (20 ppm); fixed modification (Carbamidomethyl (C)); variable modification (Oxidation (M)); acetyl (protein N-terminal; phospho (STY)); enzyme (trypsin) with a maximum of 2 missed cleavages; minimum peptide length (7 amino acids); and false discovery rate (FDR) for PSM and protein identification (1%). Frequently observed laboratory contaminants were removed. The Perseus software (version 1.6.14.0) [81] was used for statistical analysis and data visualization;
- -
- SWATH-MS: For protein expression analysis, a pool containing the same amount of µg per sample was used as input for the generation of the SWATH-MS assay library. Twenty micrograms were diluted in Laemmli buffer and loaded into a 0.75 mm-thick polyacrylamide gel with a 4% stacking gel cast over a 12.5% resolving gel. The total gel was stained with Coomassie Brilliant Blue, and 12 equal slides from each pooled sample were excised from the gel and transferred into 1.5 mL Eppendorf LoBind tubes. Protein enzymatic cleavage was carried out with trypsin (1:20, w/w, Promega, Madison, WI, USA) at 37 °C for 16 h, as previously described [82]. Purification and concentration of peptides were performed using C18 Zip Tip Solid Phase Extraction (Millipore, Burlington, MA, USA). The peptides, recovered from in-gel and in-solution digestion processing, were reconstituted into a final concentration of 0.5 µg/µL of 2% ACN, 0.5% FA, and 97.5% MilliQ water before mass spectrometric analysis. MS/MS datasets for spectral library generation were acquired on a Triple TOF 5600+ mass spectrometer (Sciex) interfaced to the Eksigent nanoLC ultra-2D pump system (Sciex) as previously described. MS/MS data acquisition was performed using AnalystTF 1.7 (Sciex), and spectra files were processed through the ProteinPilot v5.0 search engine (Sciex) using the ParagonTM Algorithm (v.4.0.0.0) [83] for database search. The identified proteins were grouped based on MS/MS spectra by the Progroup™ Algorithm, regardless of the peptide sequence assigned. The false discovery rate (FDR) was determined using a non-linear fitting method [84], and the displayed results were those reporting a 1% Global FDR or better. Then, individual protein extracts from all sample sets (n = 10) were subjected to in-solution digestion, peptide purification, and reconstitution before mass spectrometric analysis. Protein extracts (20 g) from each sample were reduced by adding DTT to a final concentration of 10 mM and incubation at room temperature. The reduction was performed by adding DTT to a final concentration of 10 mM and incubating at room temperature for 30 min. Subsequent alkylation by 30 mM iodoacetamide was performed for 30 min in the dark. An additional reduction step was performed with 30 mM DTT, allowing the reaction to stand at room temperature for another 30 min. The mixture was then diluted to 0.6 M urea using MilliQ water, and after trypsin addition (Promega, Madison, WI, USA) (enzyme:protein, 1:50 w/w), the sample was incubated at 37 °C for 16 h. Digestion was quenched by acidification with acetic acid. The digestion mixture was dried in a SpeedVac. Purification and concentration of peptides were performed using C18 Zip Tip Solid Phase Extraction (Millipore, Burlington, MA, USA). The peptides recovered were reconstituted into a final concentration of 1 µg/µL of 2% ACN, 0.5% FA, and 97.5% MilliQ water before mass spectrometric analysis. For SWATH-MS-based experiments, the TripleTOF 5600+ instrument was configured as described in [85]. Using an isolation width of 16 Da (15 Da for optimal ion transmission efficiency and 1 Da for the window overlap), a set of 37 overlapping windows was constructed, covering the mass range 450–1000 Da. Using this method, 1 μL of each sample was loaded onto an Acclaim™ PepMap™ 100 C18 trap column (0.1 × 20 mm, particle size 5 µm; ThermoFisher Scientific, Waltham, MA, USA) and desalted with 100% water and 0.1% formic acid at 2 μL/minute for 10 min. The peptides were loaded onto an Acclaim™ PepMap™ 100 C18 column (0.075 × 250 mm, particle size 3 µm; ThermoFisher Scientific, Waltham, MA, USA) equilibrated in 2% acetonitrile and 0.1% FA. Peptide elution was carried out with a linear gradient of 2%–40% B for 120 min (mobile phases A: 100% water, 0.1% formic acid (FA), and B: 100% acetonitrile, 0.1% FA) at a flow rate of 300 nL/minute. Eluted peptides were infused into the mass spectrometer. The Triple TOF was operated in SWATH mode, in which a 0.050 s TOF MS scan from 350 to 1250 m/z was performed, followed by 0.080 s product ion scans from 230 to 1800 m/z on the 37 defined windows (3.05 s/cycle). The collision energy was set to optimum for a 2+ ion at the center of each SWATH block with a 15 eV collision energy spread. The resulting ProteinPilot group file from library generation was loaded into PeakView® (v2.1, Sciex), and peaks from SWATH runs were extracted with a peptide threshold of 99% confidence (Unused Score ≥ 1.3) and a FDR lower than 1%. For this, the MS/MS spectra of the assigned peptides were extracted by ProteinPilot, and only the proteins that fulfilled the following criteria were validated: (1) peptide mass tolerance lower than 10 ppm, (2) 99% confidence level in peptide identification, and (3) complete b/y ion series found in the MS/MS spectrum. Only proteins quantified with at least two unique peptides were considered. Then, quantitative data were analyzed using the Perseus software (version 1.6.14.0) for statistical analysis and data visualization. The two-sample Student’s t-test was applied to compare between groups. Then, only phosphopeptides with a p-value < 0.05 were considered differentially expressed. Proteomic experiments generate a large number of peptides or proteins that need to be independently evaluated using statistical tests and may yield type I errors [86]. However, it is important to note that, due to the often low power of proteomic experiments, the use of these corrections may fail to detect even true positives [87]. In this case, the use of only five samples per group, together with the low fold changes quantified in our data, were determinants for the statistical analysis, and as a consequence, using FDR corrections was not able to detect any significant phosphopeptide. MS data and search result files were deposited in the Proteome Xchange Consortium via the JPOST partner repository (https://repository.jpostdb.org, accessed on 1 March 2022) [88] with the identifiers PXD040416 for ProteomeXchange and JPST002064 for jPOST. (https://repository.jpostdb.org/preview/165924140563fcaaf142e99; Access key 9943);
- -
- Bioinformatics: The identification of significantly deregulated regulatory/metabolic pathways in proteomic datasets was performed using ShinyGO 0.77 [89].
4.6. Retrotranscription Reaction-qPCR
4.7. Gel Electrophoresis and Western Blotting
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, D.R. Tauopathies: Classification and Clinical Update on Neurodegenerative Diseases Associated with Microtubule-Associated Protein Tau. Intern. Med. J. 2006, 36, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Gong, C.-X. Tau Exon 10 Alternative Splicing and Tauopathies. Mol. Neurodegener. 2008, 3, 8. [Google Scholar] [CrossRef]
- Hefti, M.M.; Farrell, K.; Kim, S.H.; Bowles, K.R.; Fowkes, M.E.; Raj, T.; Crary, J.F. High-Resolution Temporal and Regional Mapping of MAPT Expression and Splicing in Human Brain Development. PLoS ONE 2018, 13, e0195771. [Google Scholar] [CrossRef] [PubMed]
- McMillan, P.; Korvatska, E.; Poorkaj, P.; Evstafjeva, Z.; Robinson, L.; Greenup, L.; Leverenz, J.; Schellenberg, G.D.; D’Souza, I. Tau Isoform Regulation Is Region- and Cell-Specific in Mouse Brain. J. Comp. Neurol. 2008, 511, 788–803. [Google Scholar] [CrossRef]
- Lee, G.; Cowan, N.; Kirschner, M. The Primary Structure and Heterogeneity of Tau Protein from Mouse Brain. Science 1988, 239, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple Isoforms of Human Microtubule-Associated Protein Tau: Sequences and Localization in Neurofibrillary Tangles of Alzheimer’s Disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Goedert, M. Tau Pathology and Neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and Tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef]
- Rauch, J.N.; Olson, S.H.; Gestwicki, J.E. Interactions between Microtubule-Associated Protein Tau (MAPT) and Small Molecules. Cold Spring Harb. Perspect. Med. 2017, 7, a024034. [Google Scholar] [CrossRef]
- Brandt, R.; Trushina, N.I.; Bakota, L. Much More Than a Cytoskeletal Protein: Physiological and Pathological Functions of the Non-Microtubule Binding Region of Tau. Front. Neurol. 2020, 11, 590029. [Google Scholar] [CrossRef]
- Goedert, M.; Wischik, C.M.; Crowther, R.A.; Walker, J.E.; Klug, A. Cloning and Sequencing of the CDNA Encoding a Core Protein of the Paired Helical Filament of Alzheimer Disease: Identification as the Microtubule-Associated Protein Tau. Proc. Natl. Acad. Sci. USA 1988, 85, 4051–4055. [Google Scholar] [CrossRef]
- Kosik, K.S.; Orecchio, L.D.; Bakalis, S.; Neve, R.L. Developmentally Regulated Expression of Specific Tau Sequences. Neuron 1989, 2, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Goode, B.; Feinstein, S. Identification of a Novel Microtubule Binding and Assembly Domain in the Developmentally Regulated Inter-Repeat Region of Tau. J. Cell Biol. 1994, 124, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Gustke, N.; Trinczek, B.; Biernat, J.; Mandelkow, E.-M.; Mandelkow, E. Domains of Tau Protein and Interactions with Microtubules. Biochemistry 1994, 33, 9511–9522. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Beck, M.; Stahl, T.; Holzer, M.; Brauer, K.; Bigl, V.; Arendt, T. Phylogenetic Diversity of the Expression of the Microtubule-Associated Protein Tau: Implications for Neurodegenerative Disorders. Mol. Brain Res. 1999, 68, 119–128. [Google Scholar] [CrossRef]
- Sergeant, N.; Bretteville, A.; Hamdane, M.; Caillet-Boudin, M.-L.; Grognet, P.; Bombois, S.; Blum, D.; Delacourte, A.; Pasquier, F.; Vanmechelen, E.; et al. Biochemistry of Tau in Alzheimer’s Disease and Related Neurological Disorders. Expert Rev. Proteomics 2008, 5, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.; Avila, J. Intra and Extracellular Protein Interactions with Tau. Curr. Alzheimer Res. 2010, 7, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Townsend, K.; Goldberg, T.E.; Davies, P.; Conejero-Goldberg, C. MAPT Isoforms: Differential Transcriptional Profiles Related to 3R and 4R Splice Variants. J. Alzheimer’s Dis. 2011, 22, 1313–1329. [Google Scholar] [CrossRef]
- Mena, R.; Edward, P.C.; Harrington, C.R.; Mukaetova-Ladinska, E.B.; Wischik, C.M. 508 Staging the Pathological Assembly of Tau Protein into Alzheimer Paired Helical Filaments. Neurobiol. Aging 1996, 17, S127. [Google Scholar] [CrossRef]
- Mohandas, E.; Rajmohan, V.; Raghunath, B. Neurobiology of Alzheimer′s Disease. Indian J. Psychiatry 2009, 51, 55. [Google Scholar] [CrossRef]
- Novak, M.; Zilka, N.; Kovacech, B.; Barath, P.; Kontsekova, E. F3-02-03: Tau Truncation: The Most Productive Post-translational Modification. Alzheimer’s Dement. 2012, 8, 424. [Google Scholar] [CrossRef]
- Beharry, C.; Cohen, L.S.; Di, J.; Ibrahim, K.; Briffa-Mirabella, S.; Alonso, A.d.C. Tau-Induced Neurodegeneration: Mechanisms and Targets. Neurosci. Bull. 2014, 30, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, S.; von Bergen, M.; Brutlach, H.; Steinhoff, H.-J.; Mandelkow, E. Global Hairpin Folding of Tau in Solution. Biochemistry 2006, 45, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.d.C.; Zaidi, T.; Novak, M.; Grundke-Iqbal, I.; Iqbal, K. Hyperphosphorylation Induces Self-Assembly of τ into Tangles of Paired Helical Filaments/Straight Filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 6923–6928. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-L.; Wang, N.; Sun, F.-R.; Cao, X.-P.; Zhang, W.; Yu, J.-T. Tau in Neurodegenerative Disease. Ann. Transl. Med. 2018, 6, 175. [Google Scholar] [CrossRef] [PubMed]
- Carrell, R.W.; Gooptu, B. Conformational Changes and Disease—Serpins, Prions and Alzheimer’s. Curr. Opin. Struct. Biol. 1998, 8, 799–809. [Google Scholar] [CrossRef]
- Fox, N.; Harvey, R.J.; Rossor, M.N. Protein Folding, Nucleation Phenomena and Delayed Neurodegeneration in Alzheimer’s Disease. Rev. Neurosci. 1996, 7, 21–28. [Google Scholar] [CrossRef]
- Hyman, B.T.; Augustinack, J.C.; Ingelsson, M. Transcriptional and Conformational Changes of the Tau Molecule in Alzheimer’s Disease. Biochim. Biophys. Acta-Mol. Basis Dis. 2005, 1739, 150–157. [Google Scholar] [CrossRef]
- Zilka, N.; Korenova, M.; Novak, M. Misfolded Tau Protein and Disease Modifying Pathways in Transgenic Rodent Models of Human Tauopathies. Acta Neuropathol. 2009, 118, 71–86. [Google Scholar] [CrossRef]
- Ferrer, I.; López-González, I.; Carmona, M.; Arregui, L.; Dalfó, E.; Torrejón-Escribano, B.; Diehl, R.; Kovacs, G.G. Glial and Neuronal Tau Pathology in Tauopathies. J. Neuropathol. Exp. Neurol. 2014, 73, 81–97. [Google Scholar] [CrossRef]
- Dujardin, S.; Colin, M.; Buée, L. Invited Review: Animal Models of Tauopathies and Their Implications for Research/Translation into the Clinic. Neuropathol. Appl. Neurobiol. 2015, 41, 59–80. [Google Scholar] [CrossRef] [PubMed]
- Götz, J.J.; Götz, J. Experimental Models of Tauopathy—From Mechanisms to Therapies. Adv. Exp. Med. Biol. 2019, 1184, 381–391. [Google Scholar]
- Harada, A.; Oguchi, K.; Okabe, S.; Kuno, J.; Terada, S.; Ohshima, T.; Sato-Yoshitake, R.; Takei, Y.; Noda, T.; Hirokawa, N. Altered Microtubule Organization in Small-Calibre Axons of Mice Lacking Tau Protein. Nature 1994, 369, 488–491. [Google Scholar] [CrossRef]
- Dawson, H.N.; Ferreira, A.; Eyster, M.V.; Ghoshal, N.; Binder, L.I.; Vitek, M.P. Inhibition of Neuronal Maturation in Primary Hippocampal Neurons from τ Deficient Mice. J. Cell Sci. 2001, 114, 1179–1187. [Google Scholar] [CrossRef]
- Tucker, K.L.; Meyer, M.; Barde, Y.-A. Neurotrophins Are Required for Nerve Growth during Development. Nat. Neurosci. 2001, 4, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Fujio, K.; Sato, M.; Uemura, T.; Sato, T.; Sato-Harada, R.; Harada, A. 14-3-3 Proteins and Protein Phosphatases Are Not Reduced in Tau-Deficient Mice. Neuroreport 2007, 18, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.C.S.; Yao, S.; Ittner, A.; Bertz, J.; Ke, Y.D.; Ittner, L.M.; Delerue, F. Generation of a New Tau Knockout (TauΔex1) Line Using CRISPR/Cas9 Genome Editing in Mice. J. Alzheimer’s Dis. 2018, 62, 571–578. [Google Scholar] [CrossRef]
- Ke, Y.D.; Suchowerska, A.K.; van der Hoven, J.; De Silva, D.M.; Wu, C.W.; van Eersel, J.; Ittner, A.; Ittner, L.M. Lessons from Tau-Deficient Mice. Int. J. Alzheimers. Dis. 2012, 2012, 873270. [Google Scholar] [CrossRef]
- Lopes, S.; Vaz-Silva, J.; Pinto, V.; Dalla, C.; Kokras, N.; Bedenk, B.; Mack, N.; Czisch, M.; Almeida, O.F.X.; Sousa, N.; et al. Tau Protein Is Essential for Stress-Induced Brain Pathology. Proc. Natl. Acad. Sci. USA 2016, 113, E3755–E3763. [Google Scholar] [CrossRef]
- Biundo, F.; Del Prete, D.; Zhang, H.; Arancio, O.; D’Adamio, L. A Role for Tau in Learning, Memory and Synaptic Plasticity. Sci. Rep. 2018, 8, 3184. [Google Scholar] [CrossRef]
- Tan, X.L.; Zheng, P.; Wright, D.K.; Sun, M.; Brady, R.D.; Liu, S.; McDonald, S.J.; Mychasiuk, R.; Cenap, S.; Jones, N.C.; et al. The Genetic Ablation of Tau Improves Long-Term, but Not Short-Term, Functional Outcomes after Experimental Traumatic Brain Injury in Mice. Brain Inj. 2020, 34, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Criado-Marrero, M.; Sabbagh, J.J.; Jones, M.R.; Chaput, D.; Dickey, C.A.; Blair, L.J. Hippocampal Neurogenesis Is Enhanced in Adult Tau Deficient Mice. Cells 2020, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hall, A.M.; Kelinske, M.; Roberson, E.D. Seizure Resistance without Parkinsonism in Aged Mice after Tau Reduction. Neurobiol. Aging 2014, 35, 2617–2624. [Google Scholar] [CrossRef]
- Liu, S.; Shen, Y.; Shultz, S.R.; Nguyen, A.; Hovens, C.; Adlard, P.A.; Bush, A.I.; Chan, J.; Kwan, P.; O’Brien, T.J.; et al. Accelerated Kindling Epileptogenesis in Tg4510 Tau Transgenic Mice, but Not in Tau Knockout Mice. Epilepsia 2017, 58, e136–e141. [Google Scholar] [CrossRef]
- Bi, M.; Gladbach, A.; van Eersel, J.; Ittner, A.; Przybyla, M.; van Hummel, A.; Chua, S.W.; van der Hoven, J.; Lee, W.S.; Müller, J.; et al. Tau Exacerbates Excitotoxic Brain Damage in an Animal Model of Stroke. Nat. Commun. 2017, 8, 473. [Google Scholar] [CrossRef] [PubMed]
- Putra, M.; Puttachary, S.; Liu, G.; Lee, G.; Thippeswamy, T. Fyn-Tau Ablation Modifies PTZ-Induced Seizures and Post-Seizure Hallmarks of Early Epileptogenesis. Front. Cell. Neurosci. 2020, 14, 592374. [Google Scholar] [CrossRef]
- Chang, C.-W.; Evans, M.D.; Yu, X.; Yu, G.-Q.; Mucke, L. Tau Reduction Affects Excitatory and Inhibitory Neurons Differently, Reduces Excitation/Inhibition Ratios, and Counteracts Network Hypersynchrony. Cell Rep. 2021, 37, 109855. [Google Scholar] [CrossRef]
- Cloyd, R.A.; Koren, J.; Abisambra, J.F.; Smith, B.N. Effects of Altered Tau Expression on Dentate Granule Cell Excitability in Mice. Exp. Neurol. 2021, 343, 113766. [Google Scholar] [CrossRef]
- Kimura, T.; Whitcomb, D.J.; Jo, J.; Regan, P.; Piers, T.; Heo, S.; Brown, C.; Hashikawa, T.; Murayama, M.; Seok, H.; et al. Microtubule-Associated Protein Tau Is Essential for Long-Term Depression in the Hippocampus. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130144. [Google Scholar] [CrossRef]
- Ahmed, T.; Van der Jeugd, A.; Blum, D.; Galas, M.-C.; D’Hooge, R.; Buee, L.; Balschun, D. Cognition and Hippocampal Synaptic Plasticity in Mice with a Homozygous Tau Deletion. Neurobiol. Aging 2014, 35, 2474–2478. [Google Scholar] [CrossRef]
- Regan, P.; Piers, T.; Yi, J.-H.; Kim, D.-H.; Huh, S.; Park, S.J.; Ryu, J.H.; Whitcomb, D.J.; Cho, K. Tau Phosphorylation at Serine 396 Residue Is Required for Hippocampal LTD. J. Neurosci. 2015, 35, 4804–4812. [Google Scholar] [CrossRef]
- Dioli, C.; Patrício, P.; Pinto, L.; Marie, C.; Morais, M.; Vyas, S.; Bessa, J.M.; Pinto, L.; Sotiropoulos, I. Adult Neurogenic Process in the Subventricular Zone-olfactory Bulb System Is Regulated by Tau Protein under Prolonged Stress. Cell Prolif. 2021, 54, e13027. [Google Scholar] [CrossRef] [PubMed]
- Dioli, C.; Patrício, P.; Trindade, R.; Pinto, L.G.; Silva, J.M.; Morais, M.; Ferreiro, E.; Borges, S.; Mateus-Pinheiro, A.; Rodrigues, A.J.; et al. Tau-Dependent Suppression of Adult Neurogenesis in the Stressed Hippocampus. Mol. Psychiatry 2017, 22, 1110–1118. [Google Scholar] [CrossRef]
- Lopes, S.; Teplytska, L.; Vaz-Silva, J.; Dioli, C.; Trindade, R.; Morais, M.; Webhofer, C.; Maccarrone, G.; Almeida, O.F.X.; Turck, C.W.; et al. Tau Deletion Prevents Stress-Induced Dendritic Atrophy in Prefrontal Cortex: Role of Synaptic Mitochondria. Cereb. Cortex 2016, 27, bhw057. [Google Scholar] [CrossRef] [PubMed]
- Oyama, F.; Kotliarova, S.; Harada, A.; Ito, M.; Miyazaki, H.; Ueyama, Y.; Hirokawa, N.; Nukina, N.; Ihara, Y. Gem GTPase and Tau. J. Biol. Chem. 2004, 279, 27272–27277. [Google Scholar] [CrossRef] [PubMed]
- Gómez de Barreda, E.; Dawson, H.N.; Vitek, M.P.; Avila, J. Tau Deficiency Leads to the Upregulation of BAF-57, a Protein Involved in Neuron-Specific Gene Repression. FEBS Lett. 2010, 584, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Benito, P.; Carmona, M.; Jordán, M.; Fernández-Irigoyen, J.; Santamaría, E.; del Rio, J.A.; Ferrer, I. Host Tau Genotype Specifically Designs and Regulates Tau Seeding and Spreading and Host Tau Transformation Following Intrahippocampal Injection of Identical Tau AD Inoculum. Int. J. Mol. Sci. 2022, 23, 718. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Yao, H.; Li, Y.; Ding, R.; Ren, X.; Tan, Y.; Ren, W.; Yu, H.; Zhan, X.; Wang, X.; et al. Regulation of Tau Protein on the Antidepressant Effects of Ketamine in the Chronic Unpredictable Mild Stress Model. Front. Psychiatry 2019, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-L.; Zuo, X.; Yang, F.; Ubeda, O.J.; Gant, D.J.; Alaverdyan, M.; Kiosea, N.C.; Nazari, S.; Chen, P.P.; Nothias, F.; et al. Loss of MAP Function Leads to Hippocampal Synapse Loss and Deficits in the Morris Water Maze with Aging. J. Neurosci. 2014, 34, 7124–7136. [Google Scholar] [CrossRef]
- Liu, G.; Thangavel, R.; Rysted, J.; Kim, Y.; Francis, M.B.; Adams, E.; Lin, Z.; Taugher, R.J.; Wemmie, J.A.; Usachev, Y.M.; et al. Loss of Tau and Fyn Reduces Compensatory Effects of MAP2 for Tau and Reveals a Fyn-independent Effect of Tau on Calcium. J. Neurosci. Res. 2019, 97, 1393–1413. [Google Scholar] [CrossRef]
- Cohen, P. The Regulation of Protein Function by Multisite Phosphorylation—A 25 Year Update. Trends Biochem. Sci. 2000, 25, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.N.; Lewis, R.J. Structural Basis for Control by Phosphorylation. Chem. Rev. 2001, 101, 2209–2242. [Google Scholar] [CrossRef]
- Yaffe, M.B. Phosphotyrosine-Binding Domains in Signal Transduction. Nat. Rev. Mol. Cell Biol. 2002, 3, 177–186. [Google Scholar] [CrossRef]
- Salazar, C.; Höfer, T. Multisite Protein Phosphorylation—From Molecular Mechanisms to Kinetic Models. FEBS J. 2009, 276, 3177–3198. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Barik, D.; Domingo Sananes, M.R.; Tyson, J.J.; Novák, B. Bistability by Multiple Phosphorylation of Regulatory Proteins. Prog. Biophys. Mol. Biol. 2009, 100, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Muzio, L. Lo The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID Interaction Database: 2019 Update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef]
- Bodakuntla, S.; Jijumon, A.S.; Villablanca, C.; Gonzalez-Billault, C.; Janke, C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends Cell Biol. 2019, 29, 804–819. [Google Scholar] [CrossRef]
- Bullmann, T.; Holzer, M.; Mori, H.; Arendt, T. Pattern of Tau Isoforms Expression during Development in Vivo. Int. J. Dev. Neurosci. 2009, 27, 591–597. [Google Scholar] [CrossRef]
- Bullmann, T.; De Silva, R.; Holzer, M.; Mori, H.; Arendt, T. Expression of Embryonic Tau Protein Isoforms Persist during Adult Neurogenesis in the Hippocampus. Hippocampus 2007, 17, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, R.A.; Wijesekara, N.; Fraser, P.E.; De Felice, F.G. Behavioral Abnormalities in Knockout and Humanized Tau Mice. Front. Endocrinol. 2020, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- Denk, F.; Wade-Martins, R. Knock-out and Transgenic Mouse Models of Tauopathies. Neurobiol. Aging 2009, 30, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Tabrizi, S.J. Gene Suppression Approaches to Neurodegeneration. Alzheimer’s Res. Ther. 2017, 9, 82. [Google Scholar] [CrossRef]
- Alzheimer’s Association International Conference—2021 Antisense Therapy Stifles CSF Tau in Mild Alzheimer’s Disease. Available online: https://www.alzforum.org/news/conference-coverage/antisense-therapy-stifles-csf-tau-mild-alzheimers-disease-0 (accessed on 23 March 2023).
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Montoya, A.; Beltran, L.; Casado, P.; Rodríguez-Prados, J.-C.; Cutillas, P.R. Characterization of a TiO2 Enrichment Method for Label-Free Quantitative Phosphoproteomics. Methods 2011, 54, 370–378. [Google Scholar] [CrossRef]
- Collins, B.C.; Hunter, C.L.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.L.; Chan, D.W.; Gibson, B.W.; Gingras, A.-C.; Held, J.M.; et al. Multi-Laboratory Assessment of Reproducibility, Qualitative and Quantitative Performance of SWATH-Mass Spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant Computational Platform for Mass Spectrometry-Based Shotgun Proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-Gel Digestion for Mass Spectrometric Characterization of Proteins and Proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The Paragon Algorithm, a Next Generation Search Engine That Uses Sequence Temperature Values and Feature Probabilities to Identify Peptides from Tandem Mass Spectra. Mol. Cell. Proteomics 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Shilov, I.V.; Seymour, S.L. Nonlinear Fitting Method for Determining Local False Discovery Rates from Decoy Database Searches. J. Proteome Res. 2008, 7, 3661–3667. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-Independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteomics 2012, 11, O111.016717. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, E.S.; Gritsenko, M.; Piehowski, P.D.; Gao, Y.; Orton, D.J.; Schepmoes, A.A.; Fillmore, T.L.; Frohnert, B.I.; Rewers, M.; Krischer, J.P.; et al. Tutorial: Best Practices and Considerations for Mass-Spectrometry-Based Protein Biomarker Discovery and Validation. Nat. Protoc. 2021, 16, 3737–3760. [Google Scholar] [CrossRef]
- Pascovici, D.; Handler, D.C.L.; Wu, J.X.; Haynes, P.A. Multiple Testing Corrections in Quantitative Proteomics: A Useful but Blunt Tool. Proteomics 2016, 16, 2448–2453. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. JPOSTrepo: An International Standard Data Repository for Proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]


| Gene | Gene Symbol | Reference |
|---|---|---|
| Collagen alpha-4(VI) chain | col6a4 | Mm01231949_m1 |
| Gamma-aminobutyric acid type A receptor subunit beta2 | gabrb2 | Mm00433467_m1 |
| Gamma-aminobutyric acid type A receptor subunit tyheta | gabrq | Mm00445057_m1 |
| Glutamate decarboxylase 1 | gad1 | Mm00725661_s1 |
| Glutamate metabotropic receptor 5 | grm5 | Mm00690332_m1 |
| Glutamate receptor-interacting protein 2 | grip2 | Mm01183453_m1 |
| Microtubule associated protein 1A | map1a | Mm01330378_m1 |
| Microtubule associated protein 2 | map2 | Mm00485231_m1 |
| Microtubule associated protein tau | mapt | Mm00521988_m1 |
| neuroligin-3 | nlgn3 | Mm01225951_m1 |
| RAB37, member RAS oncogene family | rab37 | Mm00445351_m1 |
| Ras-related protein Rab-8A | rab8a | Mm00445684_m1 |
| Tubulin beta 3 class III | tubb3 | Mm00727586_s1 |
| Utrophin | utrn | Mm01168866_m1 |
| Wnt family member 16 | wnt16 | Mm00446420_m1 |
| alanyl-tRNA synthetase | aars | Mm00507627_m1 |
| Hypoxanthine-guanine phosphoribosyltransferase | hprt-1 | Mm01545399_m1 |
| X-prolyl aminopepidase P1 | xpnpep1 | Mm00460040_m1 |
| β-glucuronidase | gusβ | Mm01197698_m1 |
| Antibody | Supplier | Reference | Host | Dilution WB |
|---|---|---|---|---|
| Tau 5 | ThermoFisher | MA5-12808 | Ms | 1/500 |
| 3R tau | Millipore | 05-803 | Ms | 1/1000 |
| 4R tau | Millipore | 05-804 | Ms | 1/1000 |
| PHF-1 | Dr. Peter Davies | - | Ms | 1/1000 |
| β-actin | Sigma | A5316 | Ms | 1/30,000 |
| map1a | Millipore | MAB362 | Ms | 1/500 |
| tubulin b-III | Signalway | 21617 | Rb | 1/1000 |
| pNFL (Ser473) | Millipore | MABN2431 | Ms | 1/500 |
| GAD1 | Cell Signaling | #5305 | Rb | 1/200 |
| vinculin | Santa Cruz | Sc-73614 | Ms | 1/1000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrés-Benito, P.; Flores, Á.; Busquet-Areny, S.; Carmona, M.; Ausín, K.; Cartas-Cejudo, P.; Lachén-Montes, M.; Del Rio, J.A.; Fernández-Irigoyen, J.; Santamaría, E.; et al. Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse. Int. J. Mol. Sci. 2023, 24, 6559. https://doi.org/10.3390/ijms24076559
Andrés-Benito P, Flores Á, Busquet-Areny S, Carmona M, Ausín K, Cartas-Cejudo P, Lachén-Montes M, Del Rio JA, Fernández-Irigoyen J, Santamaría E, et al. Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse. International Journal of Molecular Sciences. 2023; 24(7):6559. https://doi.org/10.3390/ijms24076559
Chicago/Turabian StyleAndrés-Benito, Pol, África Flores, Sara Busquet-Areny, Margarita Carmona, Karina Ausín, Paz Cartas-Cejudo, Mercedes Lachén-Montes, José Antonio Del Rio, Joaquín Fernández-Irigoyen, Enrique Santamaría, and et al. 2023. "Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse" International Journal of Molecular Sciences 24, no. 7: 6559. https://doi.org/10.3390/ijms24076559
APA StyleAndrés-Benito, P., Flores, Á., Busquet-Areny, S., Carmona, M., Ausín, K., Cartas-Cejudo, P., Lachén-Montes, M., Del Rio, J. A., Fernández-Irigoyen, J., Santamaría, E., & Ferrer, I. (2023). Deregulated Transcription and Proteostasis in Adult mapt Knockout Mouse. International Journal of Molecular Sciences, 24(7), 6559. https://doi.org/10.3390/ijms24076559








