Potential Use of a Combined Bacteriophage–Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study
Abstract
1. Introduction
2. Results
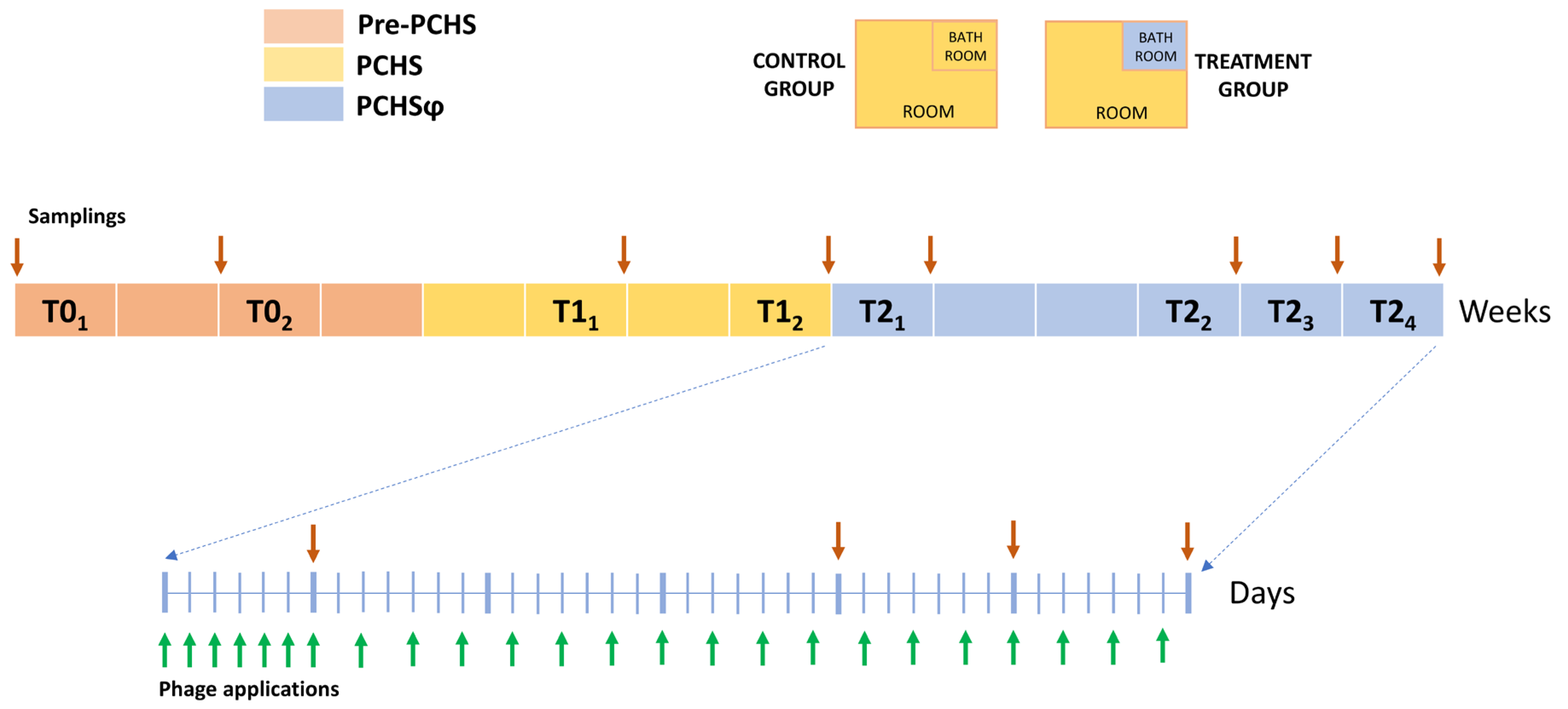
2.1. Study Set-Up
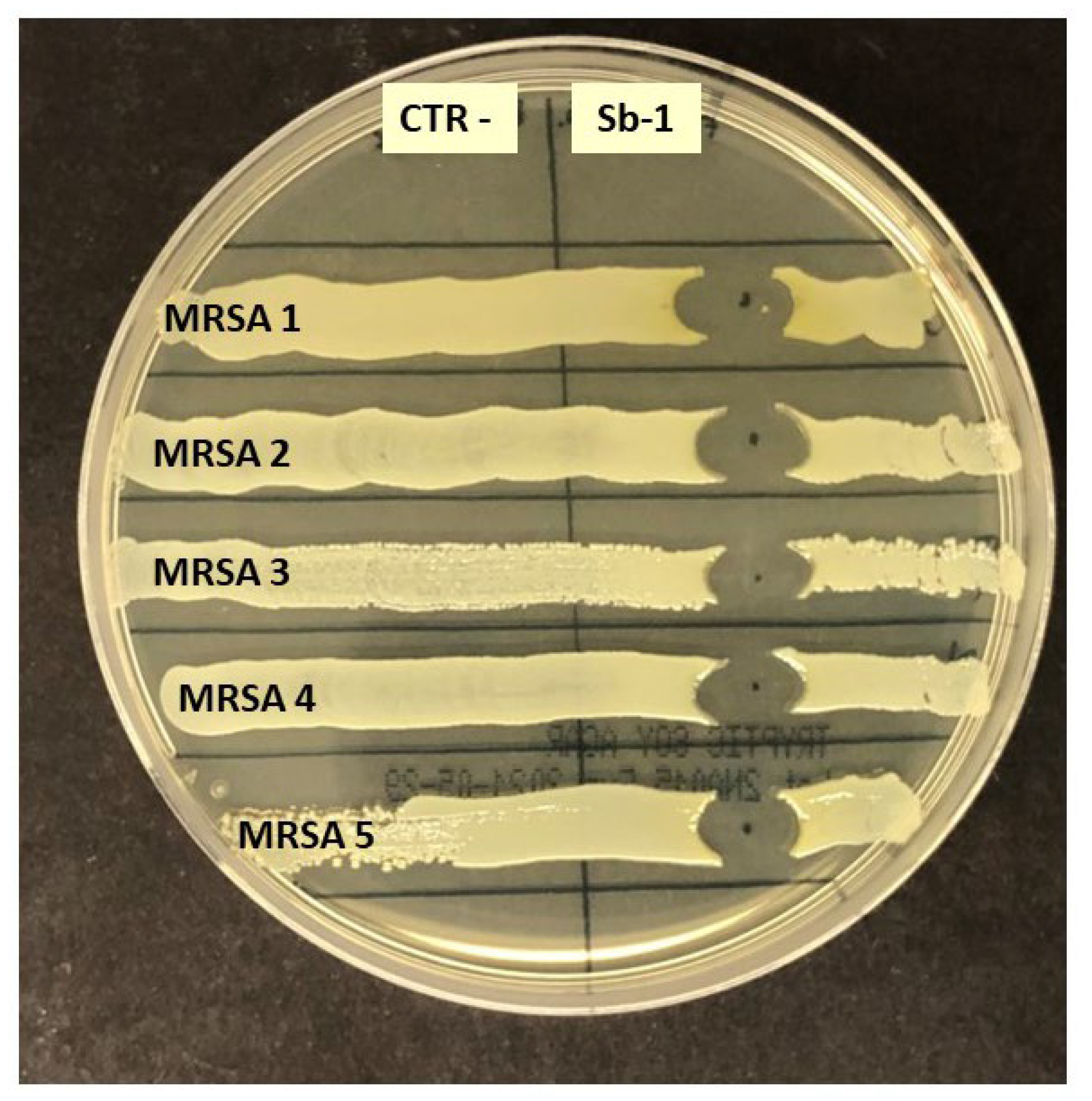
2.2. Phage Susceptibility of Staphylococcus spp. Hospital Isolates
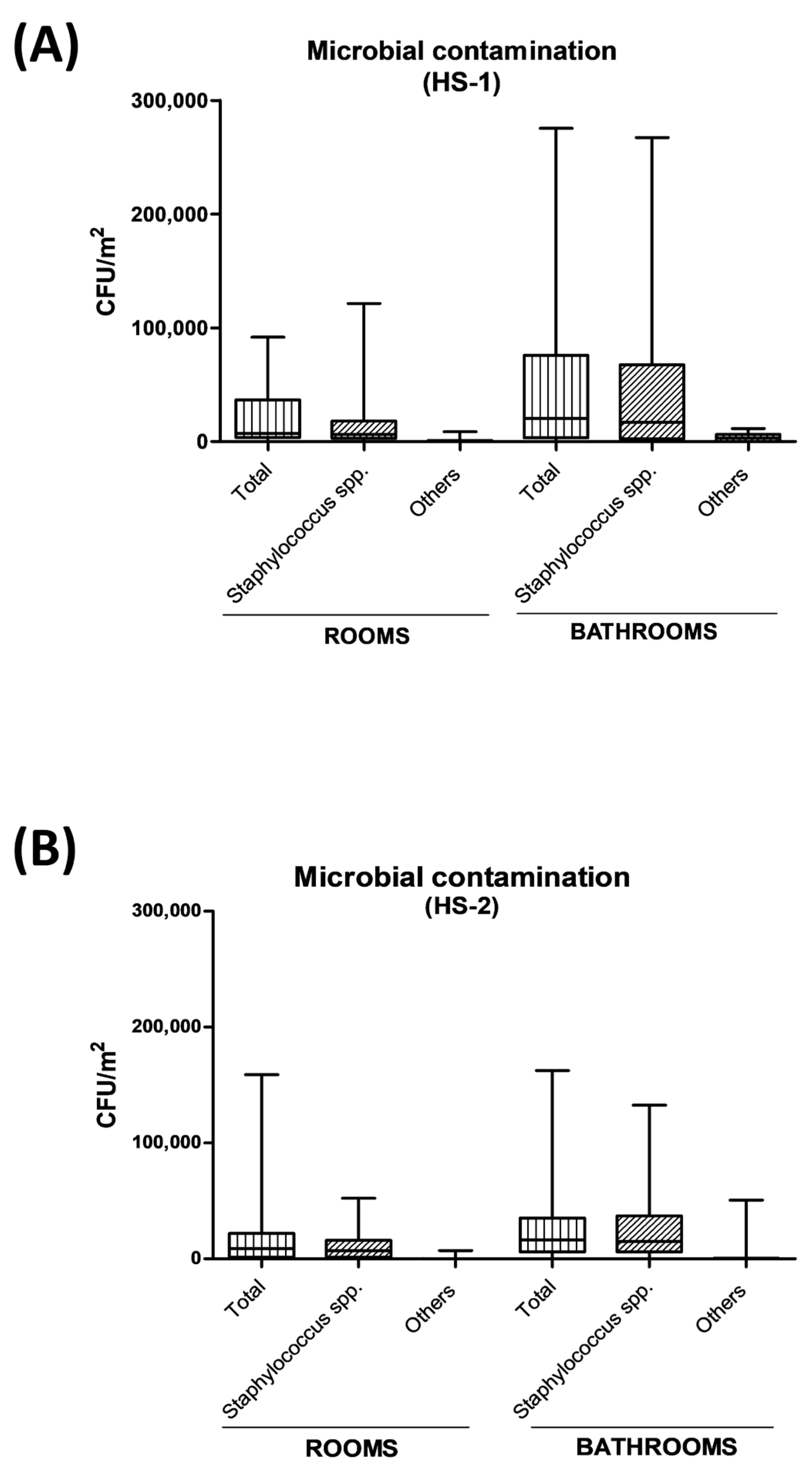
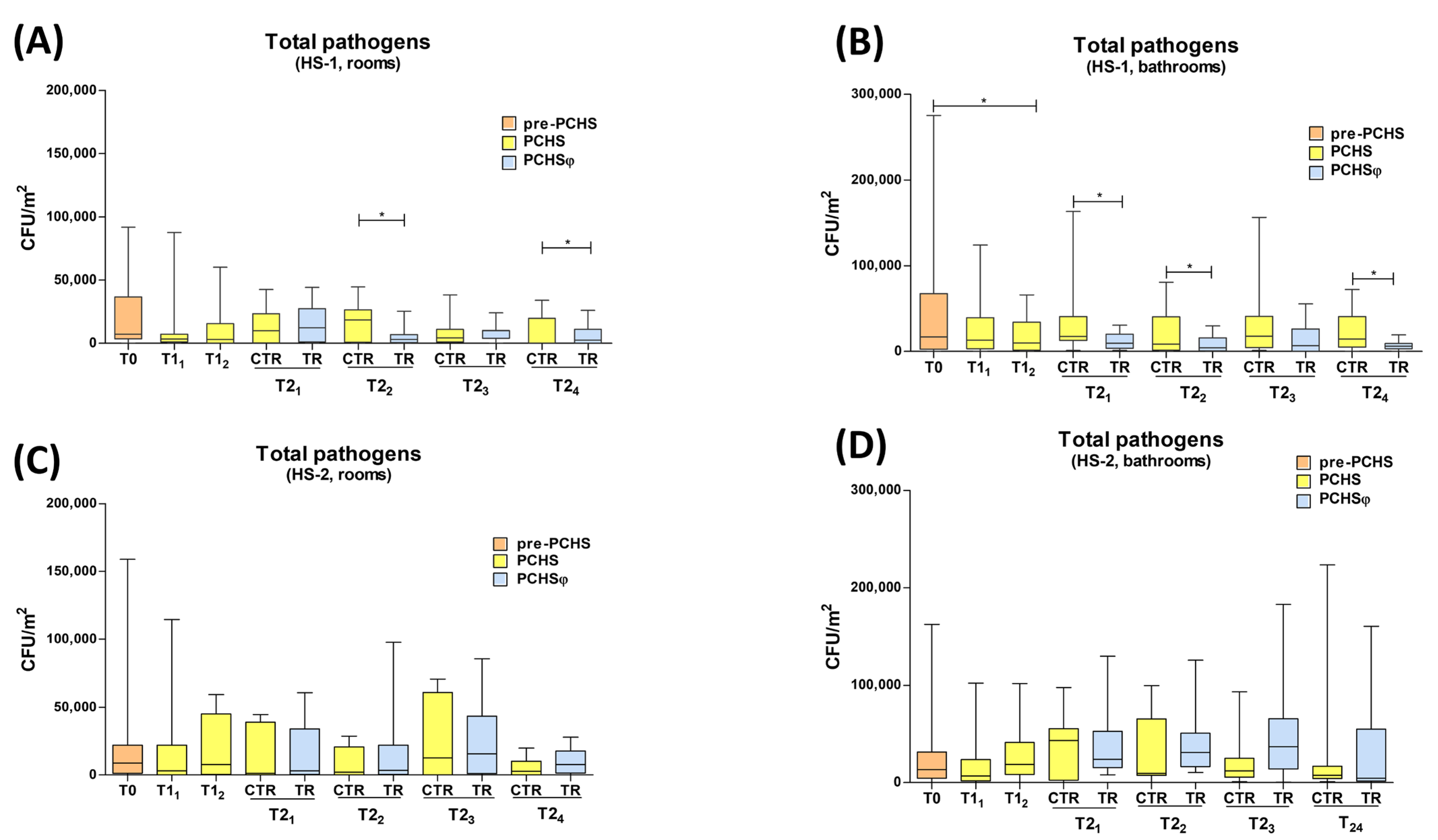
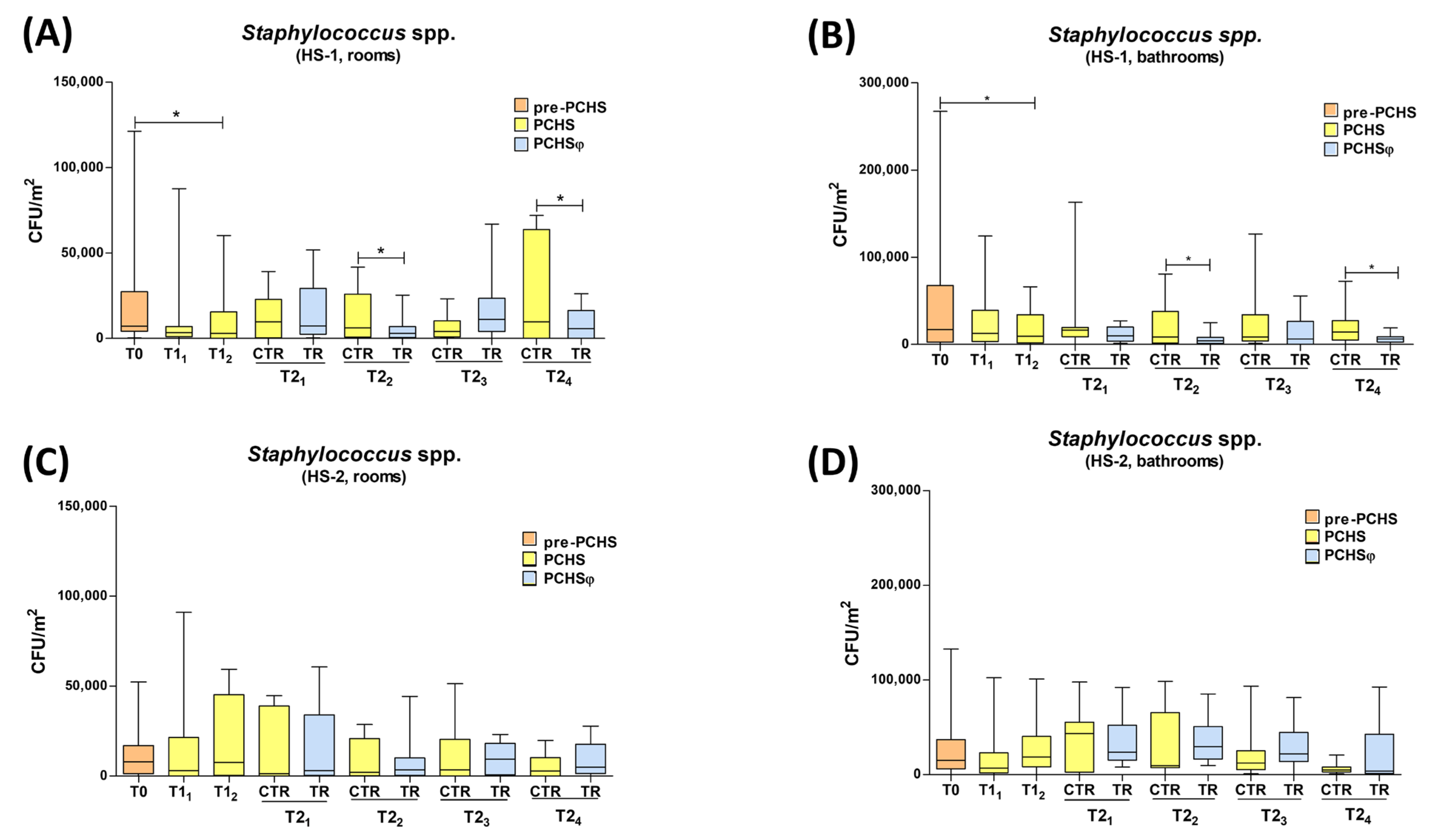
2.3. Impact of PCHS and PCHSφ Sanitation on Microbial Contamination
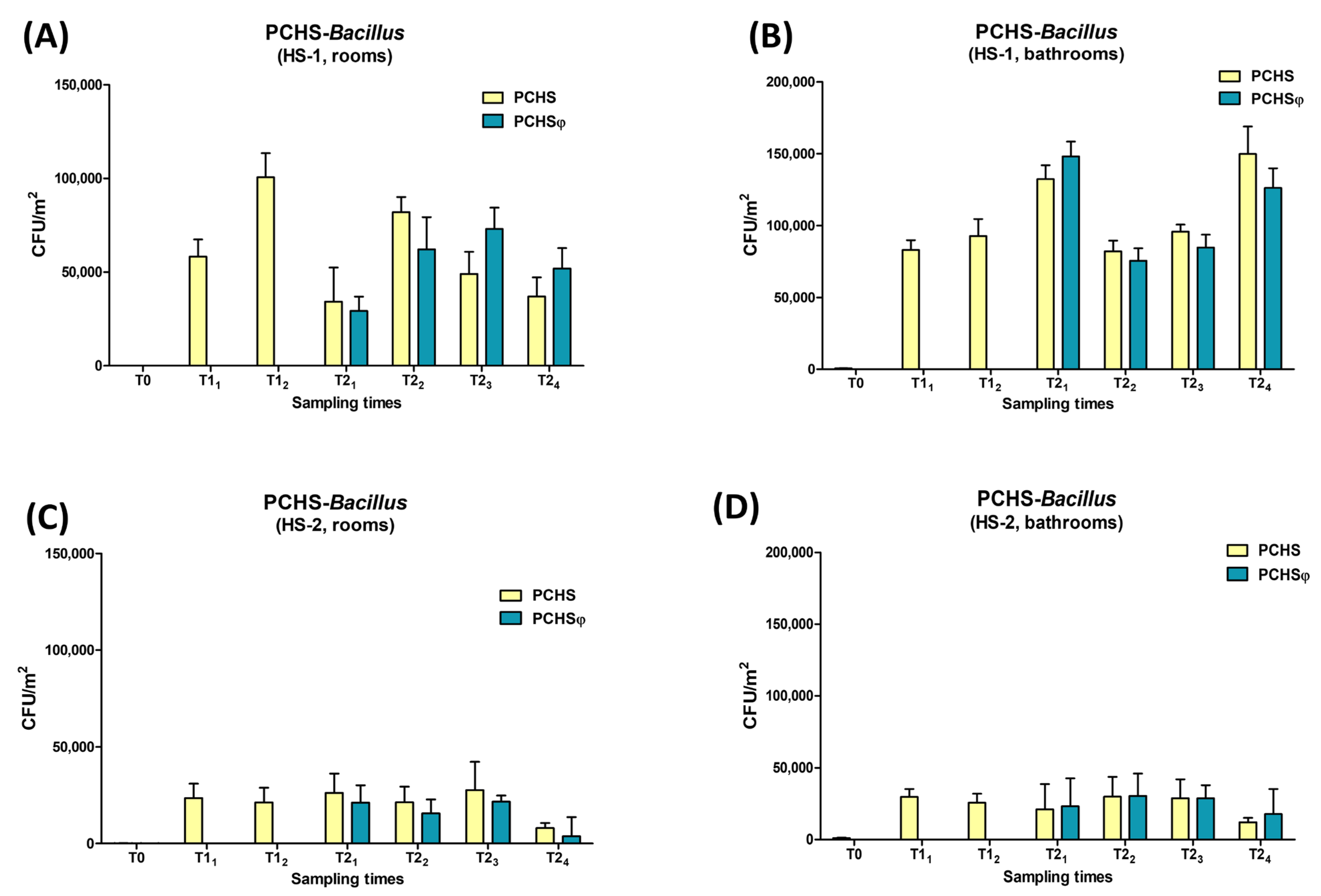
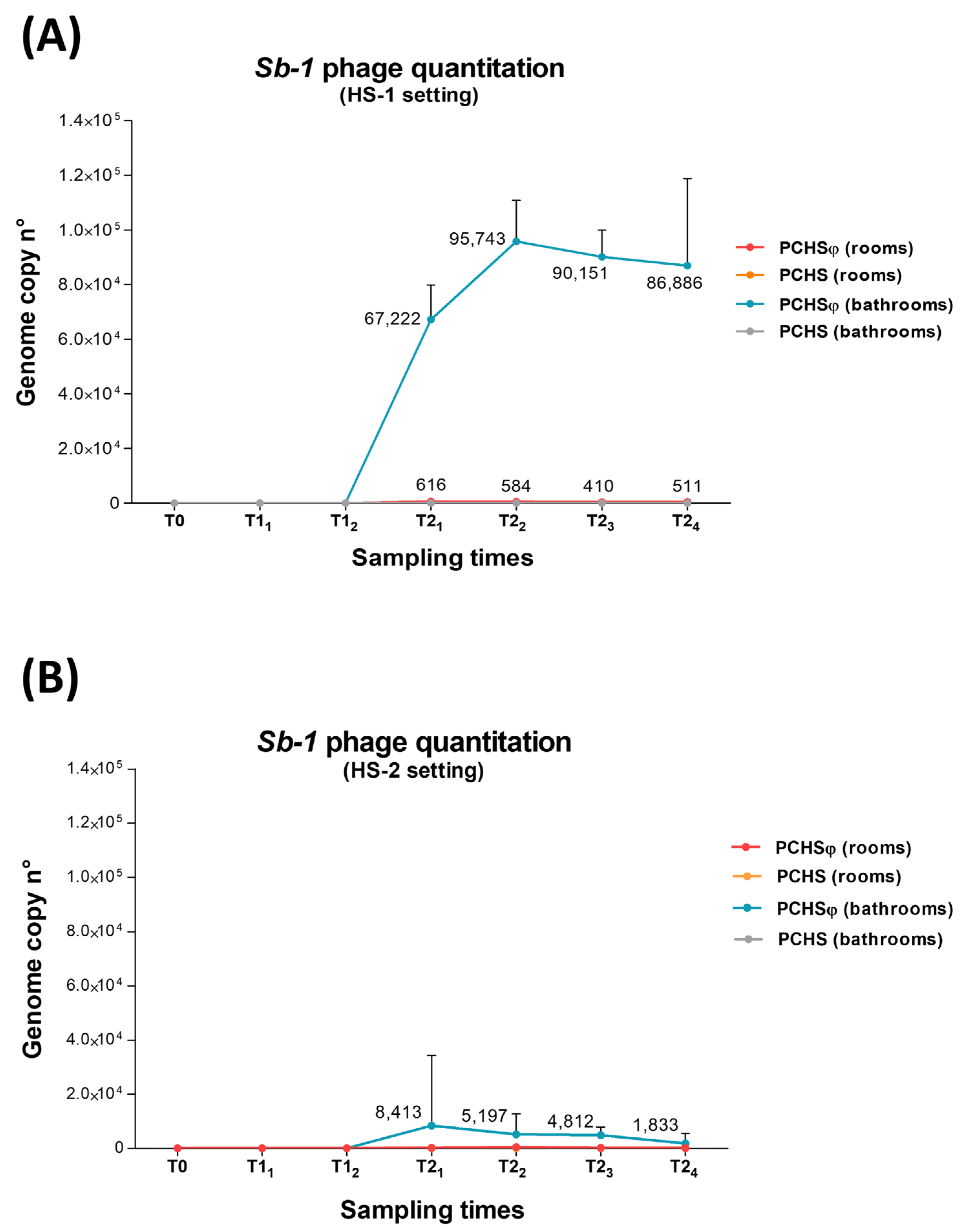
2.4. Assessment of PCHS-Bacillus and Sb-1 Phage on Treated Hospital Surfaces
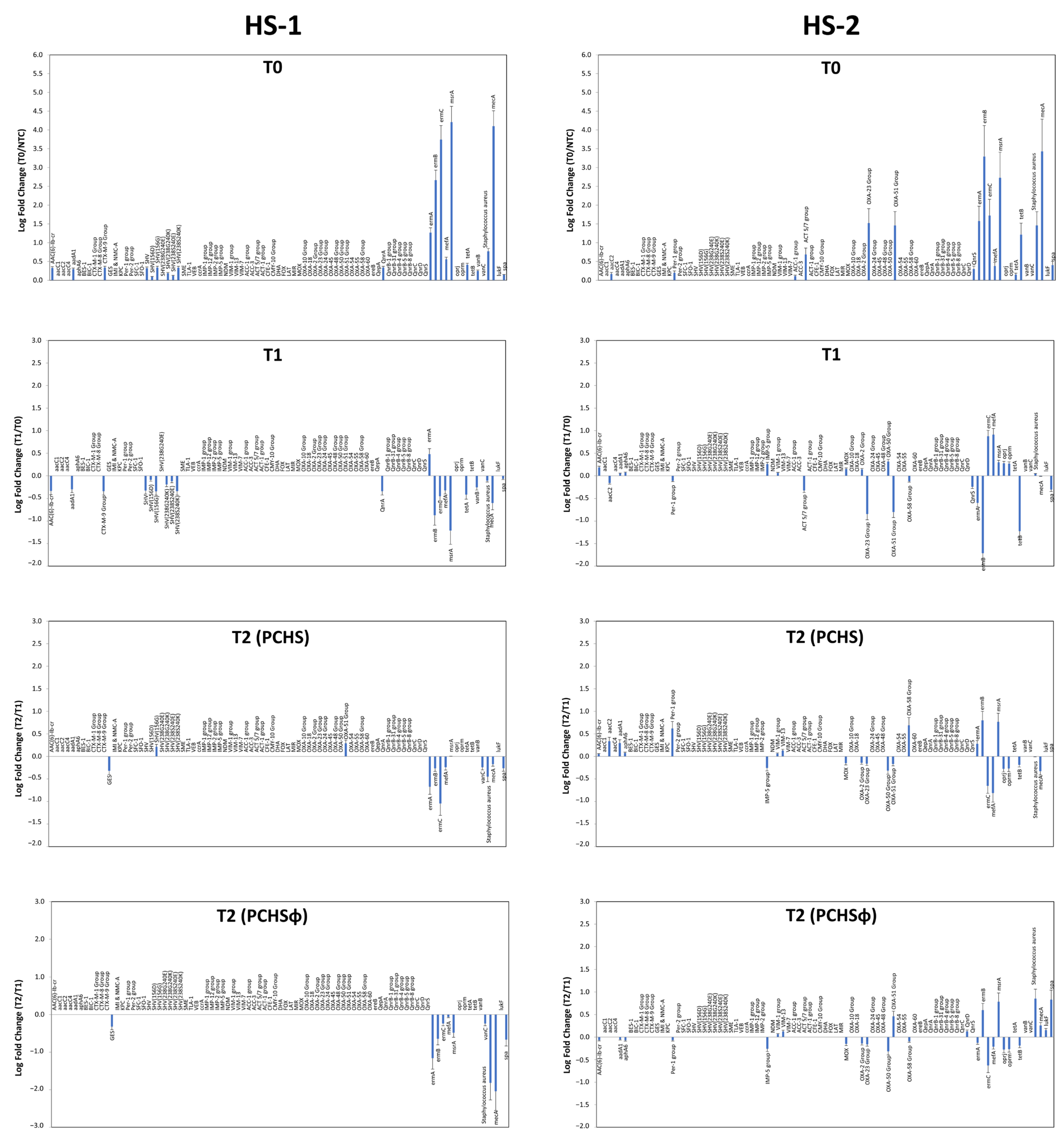
2.5. Impact of PCHS and PCHSφ Sanitation on AMR
3. Discussion
4. Materials and Methods
4.1. Study Settings
4.2. Study Design and Sanitation Procedures
4.3. Environmental Sampling
4.4. Microbiological Analyses
4.5. Molecular Analyses
4.6. Antimicrobial and Phage Susceptibility Tests
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, J.A.; Stephens, B. Microbiology of the built environment. Nat. Rev. Microbiol. 2018, 16, 661–670. [Google Scholar] [CrossRef]
- Mahnert, A.; Moissl-Eichinger, C.; Zojer, M.; Bogumil, D.; Mizrahi, I.; Rattei, T.; Martinez, J.L.; Berg, G. Man-made microbial resistances in built environments. Nat. Commun. 2019, 10, 968. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering and Medicine; National Academy of Engineering. Microbiomes of the Built Environment: A research Agenda for Indoor Microbiology, Human Health, and Buildings; The National Academies Press: Washington, DC, USA, 2017; p. 317. [Google Scholar]
- Allegranzi, B.; Bagheri Nejad, S.; Combescure, C.; Graafmans, W.; Attar, H.; Donaldson, L.; Pittet, D. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet 2011, 377, 228–241. [Google Scholar] [CrossRef]
- ECDC. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals. 2013. Available online: https://www.ecdc.europa.eu/en/publications-data/point-prevalence-survey-healthcare-associated-infections-and-antimicrobial-use-0 (accessed on 15 December 2022).
- ECDC. European Surveillance of Healthcare Associated Infections in Intensive Care Units. 2015. Available online: http://ecdc.europa.eu/en/healthtopics/Healthcare-associated_infections (accessed on 15 December 2022).
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef]
- Pochtovyi, A.A.; Vasina, D.V.; Kustova, D.D.; Divisenko, E.V.; Kuznetsova, N.A.; Burgasova, O.A.; Kolobukhina, L.V.; Tkachuk, A.P.; Gushchin, V.A.; Gintsburg, A.L. Contamination of hospital surfaces with bacterial pathogens under the current COVID-19 outbreak. Int. J. Environ. Res. Public Health 2021, 18, 9042. [Google Scholar] [CrossRef]
- Avershina, E.; Shapovalova, V.; Shipulin, G. Fighting antibiotic resistance in hospital-acquired infections: Current state and emerging technologies in disease prevention, diagnostics and therapy. Front. Microbiol. 2021, 12, 707330. [Google Scholar] [CrossRef]
- Marimuthu, K.; Pittet, D.; Harbarth, S. The effect of improved hand hygiene on nosocomial mrsa control. Antimicrob. Resist. Infect. Control 2014, 3, 34. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, D.; Khan, Z.; Khan, A.; Ualiyeva, D.; Zaman, N. Excessive use of disinfectants against COVID-19 posing a potential threat to living beings. Curr. Res. Toxicol. 2021, 2, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Vandini, A.; Temmerman, R.; Frabetti, A.; Caselli, E.; Antonioli, P.; Balboni, P.G.; Platano, D.; Branchini, A.; Mazzacane, S. Hard surface biocontrol in hospitals using microbial-based cleaning products. PLoS ONE 2014, 9, e108598. [Google Scholar] [CrossRef] [PubMed]
- D’Accolti, M.; Soffritti, I.; Bonfante, F.; Ricciardi, W.; Mazzacane, S.; Caselli, E. Potential of an eco-sustainable probiotic-cleaning formulation in reducing infectivity of enveloped viruses. Viruses 2021, 13, 2227. [Google Scholar] [CrossRef]
- Kampf, G. Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Prussin, A.J., 2nd; Marr, L.C.; Vikesland, P.J.; Edwards, M.A.; Pruden, A. Factors shaping the human exposome in the built environment: Opportunities for engineering control. Environ. Sci. Technol. 2017, 51, 7759–7774. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, Z.; Hu, D.; Cao, L.; He, Q. Understanding building-occupant-microbiome interactions toward healthy built environments: A review. Front. Environ. Sci. Eng. 2021, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- D’Accolti, M.; Soffritti, I.; Bini, F.; Mazziga, E.; Mazzacane, S.; Caselli, E. Pathogen control in the built environment: A probiotic-based system as a remedy for the spread of antibiotic resistance. Microorganisms 2022, 10, 225. [Google Scholar] [CrossRef]
- Caselli, E.; Brusaferro, S.; Coccagna, M.; Arnoldo, L.; Berloco, F.; Antonioli, P.; Tarricone, R.; Pelissero, G.; Nola, S.; La Fauci, V.; et al. Reducing healthcare-associated infections incidence by a probiotic-based sanitation system: A multicentre, prospective, intervention study. PLoS ONE 2018, 13, e0199616. [Google Scholar] [CrossRef]
- Caselli, E.; Arnoldo, L.; Rognoni, C.; D’Accolti, M.; Soffritti, I.; Lanzoni, L.; Bisi, M.; Volta, A.; Tarricone, R.; Brusaferro, S.; et al. Impact of a probiotic-based hospital sanitation on antimicrobial resistance and hai-associated antimicrobial consumption and costs: A multicenter study. Infect. Drug Resist. 2019, 12, 501–510. [Google Scholar] [CrossRef]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Lanzoni, L.; Bisi, M.; Volta, A.; Berloco, F.; Mazzacane, S. An innovative strategy for the effective reduction of mdr pathogens from the nosocomial environment. Adv. Exp. Med. Biol. 2019, 1214, 79–91. [Google Scholar]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Mazzacane, S. Spread of mcr-1-driven colistin resistance on hospital surfaces, italy. Emerg. Infect. Dis. 2018, 24, 1752–1753. [Google Scholar] [CrossRef]
- Caselli, E.; D’Accolti, M.; Vandini, A.; Lanzoni, L.; Camerada, M.T.; Coccagna, M.; Branchini, A.; Antonioli, P.; Balboni, P.G.; Di Luca, D.; et al. Impact of a probiotic-based cleaning intervention on the microbiota ecosystem of the hospital surfaces: Focus on the resistome remodulation. PLoS ONE 2016, 11, e0148857. [Google Scholar] [CrossRef]
- D’Accolti, M.; Soffritti, I.; Mazzacane, S.; Caselli, E. Fighting amr in the healthcare environment: Microbiome-based sanitation approaches and monitoring tools. Int. J. Mol. Sci. 2019, 20, 1535. [Google Scholar] [CrossRef]
- Evran, S.; Tayyarcan, E.K.; Acar-Soykut, E.; Boyaci, I.H. Applications of bacteriophage cocktails to reduce salmonella contamination in poultry farms. Food Environ. Virol. 2022, 14, 1–9. [Google Scholar] [CrossRef]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, D.; Rodriguez-Rubio, L.; Martinez, B.; Rodriguez, A.; Garcia, P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zeng, Z.; Mai, K.; Yang, Y.; Feng, J.; Bai, Y.; Sun, B.; Xie, Q.; Tong, Y.; Ma, J. Preliminary treatment of bovine mastitis caused by staphylococcus aureus, with trx-sa1, recombinant endolysin of s. Aureus bacteriophage ime-sa1. Vet. Microbiol. 2016, 191, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Goode, D.; Allen, V.M.; Barrow, P.A. Reduction of experimental salmonella and campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl. Environ. Microbiol. 2003, 69, 5032–5036. [Google Scholar] [CrossRef]
- Gutierrez, D.; Fernandez, L.; Rodriguez, A.; Garcia, P. Role of bacteriophages in the implementation of a sustainable dairy chain. Front. Microbiol. 2019, 10, 12. [Google Scholar] [CrossRef]
- Jassim, S.A.; Limoges, R.G.; El-Cheikh, H. Bacteriophage biocontrol in wastewater treatment. World J. Microbiol. Biotechnol. 2016, 32, 70. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Nakai, T.; Park, S.C. Bacteriophage therapy of infectious diseases in aquaculture. Res. Microbiol. 2002, 153, 13–18. [Google Scholar] [CrossRef]
- O’Sullivan, L.; Bolton, D.; McAuliffe, O.; Coffey, A. Bacteriophages in food applications: From foe to friend. Annu. Rev. Food Sci. Technol. 2019, 10, 151–172. [Google Scholar] [CrossRef]
- Rahimzadeh, G.; Zazouli, M.A.; Rezai, M.S. Potential of lytic bacteriophages as disinfectant to control of pseudomonas aeruginosa on fomites. J. Environ. Health Sci. Eng. 2022, 20, 219–225. [Google Scholar] [CrossRef]
- Jensen, K.C.; Hair, B.B.; Wienclaw, T.M.; Murdock, M.H.; Hatch, J.B.; Trent, A.T.; White, T.D.; Haskell, K.J.; Berges, B.K. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant staphylococcus aureus and potential use as a fomite decontaminant. PLoS ONE 2015, 10, e0131714. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.K.; Chang, J.C.; Chu, H.T.; Chen, Y.T.; Jiang, H.L.; Wang, L.S.; Teh, S.H.; Yang, H.H.; Chen, D.S.; Li, Y.Z.; et al. Preoptimized phage cocktail for use in aerosols against nosocomial transmission of carbapenem-resistant acinetobacter baumannii: A 3-year prospective intervention study. Ecotoxicol. Environ. Saf. 2022, 236, 113476. [Google Scholar] [CrossRef]
- Chen, L.K.; Liu, Y.L.; Hu, A.; Chang, K.C.; Lin, N.T.; Lai, M.J.; Tseng, C.C. Potential of bacteriophage phiab2 as an environmental biocontrol agent for the control of multidrug-resistant acinetobacter baumannii. BMC Microbiol. 2013, 13, 154. [Google Scholar] [CrossRef]
- D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Bisi, M.; Mazzacane, S.; Caselli, E. Efficient removal of hospital pathogens from hard surfaces by a combined use of bacteriophages and probiotics: Potential as sanitizing agents. Infect. Drug Resist. 2018, 11, 1015–1026. [Google Scholar] [CrossRef]
- D’Accolti, M.; Soffritti, I.; Lanzoni, L.; Bisi, M.; Volta, A.; Mazzacane, S.; Caselli, E. Effective elimination of staphylococcal contamination from hospital surfaces by a bacteriophage-probiotic sanitation strategy: A monocentric study. Microb. Biotechnol. 2019, 12, 742–751. [Google Scholar] [CrossRef]
- Curran, E.T.; Wilkinson, M.; Bradley, T. Chemical disinfectants: Controversies regarding their use in low risk healthcare environments (part 1). J. Infect. Prev. 2019, 20, 76–82. [Google Scholar] [CrossRef]
- Nabi, G.; Wang, Y.; Hao, Y.; Khan, S.; Wu, Y.; Li, D. Massive use of disinfectants against COVID-19 poses potential risks to urban wildlife. Environ. Res. 2020, 188, 109916. [Google Scholar] [CrossRef] [PubMed]
- Zeshan, B.; Karobari, M.I.; Afzal, N.; Siddiq, A.; Basha, S.; Basheer, S.N.; Peeran, S.W.; Mustafa, M.; Daud, N.H.A.; Ahmed, N.; et al. The usage of antibiotics by COVID-19 patients with comorbidities: The risk of increased antimicrobial resistance. Antibiotics 2021, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Chen, S.Y.; Ko, W.C.; Hsueh, P.R. Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antimicrob. Agents 2021, 57, 106324. [Google Scholar] [CrossRef]
- Clancy, C.J.; Buehrle, D.J.; Nguyen, M.H. Pro: The COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC Antimicrob. Resist. 2020, 2, dlaa049. [Google Scholar] [CrossRef]
- WHO. Tackling Antimicrobial Resistance in the COVID-19 Pandemic. Available online: https://www.who.int/bulletin/volumes/98/7/20-268573.pdf (accessed on 15 December 2022).
- Tarricone, R.; Rognoni, C.; Arnoldo, L.; Mazzacane, S.; Caselli, E. A probiotic-based sanitation system for the reduction of healthcare associated infections and antimicrobial resistances: A budget impact analysis. Pathogens 2020, 9, 502. [Google Scholar] [CrossRef]
- D’Accolti, M.; Soffritti, I.; Mazzacane, S.; Caselli, E. Bacteriophages as a potential 360-degree pathogen control strategy. Microorganisms 2021, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- D’Accolti, M.; Soffritti, I.; Bini, F.; Mazziga, E.; Cason, C.; Comar, M.; Volta, A.; Bisi, M.; Fumagalli, D.; Mazzacane, S.; et al. Shaping the subway microbiome through probiotic-based sanitation during the COVID-19 emergency: A pre–post case–control study. Microbiome 2023, 11, 64. [Google Scholar] [CrossRef]
- Soffritti, I.; D’Accolti, M.; Cason, C.; Lanzoni, L.; Bisi, M.; Volta, A.; Campisciano, G.; Mazzacane, S.; Bini, F.; Mazziga, E.; et al. Introduction of probiotic-based sanitation in the emergency ward of a children’s hospital during the COVID-19 pandemic. Infect. Drug Resist. 2022, 15, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Chai, S.; Guo, J.; Wu, W. Inactivation of lactobacillus virulent bacteriophage by thermal and chemical treatments. J. Food Prot. 2018, 81, 1673–1678. [Google Scholar] [CrossRef]
- Ge, Y.; Zhang, X.; Shu, L.; Yang, X. Kinetics and mechanisms of virus inactivation by chlorine dioxide in water treatment: A review. Bull. Environ. Contam. Toxicol. 2021, 106, 560–567. [Google Scholar] [CrossRef] [PubMed]
- ISS. Interim recommendations on cleaning and disinfection of non-healthcare settings during COVID-19 health emergency: Surfaces, indoor environments and clothing. In ISS COVID-19 Working Group on Biocides, ISS ed.; Istituto Superiore Sanità: Rome, Italy, 2020; Volume Rapporto ISS COVID-19 n. 25/2020. (In Italian) [Google Scholar]
| Healthcare Center | Study Period | ||
|---|---|---|---|
| T0 | T1 | T2 | |
| HS-1 | 2 | 2 | 3 |
| HS-2 | 2 | 6 | 17 |
| Healthcare Center | Sanitation Type | ||
|---|---|---|---|
| Chemical | PCHS | PCHSφ | |
| HS-1 | 17,053 CFU/m2 (100%) | 10,949 CFU/m2 (−35.8%) | 6473 CFU/m2 (−62.1%) |
| HS-2 | 14,948 CFU/m2 (100%) | 16,316 CFU/m2 (+9%) | 17,474 CFU/m2 (+16%) |
| R gene/Species | Resistance to | Bacteria | Settings | |
|---|---|---|---|---|
| HS-1 | HS-2 | |||
| AAC (6)-Ib-cr | Fluoroquinolones | Enterobacteriaceae | 0.33 | 0 |
| AadA1 | Aminoglycosides | Enterobacteriaceae, Staphylococcus, Streptococcus | 0.29 | 0 |
| AacC2 | Aminoglycosides | Enterobacteriaceae | 0 | 0.16 |
| CTX-M-9 Group | β-lactams (ESBL) | Enterobacteriaceae | 0.34 | 0 |
| Per-1 group | β-lactams | Pseudomonas | 0 | 0.18 |
| SHV | β-lactams (ESBL) | Enterobacteriaceae | 0.33 | 0 |
| SHV (156G) | β-lactams (ESBL) | Enterobacteriaceae | 0.34 | 0 |
| SHV (238S240K) | β-lactams (ESBL) | Enterobacteriaceae | 0.34 | 0 |
| ACT 5/7 group | β-lactams (ESBL) | Enterobacteriaceae | 0 | 0.68 |
| OXA-23 Group | β-lactams | Acinetobacter | 0 | 1.18 |
| OXA-51 Group | Carbapenems (carbapenemases) | Gram-negative | 0 | 1.12 |
| QnrA | Fluoroquinolones | Gram-negative | 0.34 | 0 |
| QnrS | Fluoroquinolones | Gram-negative | 0 | 0.24 |
| ermA | Macrolides (erythromycin) | Staphylococcus, Streptococcus | 1.27 | 1.58 |
| ermB | Macrolides (erythromycin) | Staphylococcus, Streptococcus | 2.66 | 3.29 |
| ermC | Macrolides (erythromycin) | Staphylococcus, Streptococcus | 3.74 | 1.83 |
| mefA | Macrolides (erythromycin) | Staphylococcus, Streptococcus, Enterococcus, Clostridium, Bacteroides | 0.56 | 0.38 |
| msrA | Macrolides (efflux pump) | Staphylococcus, Streptococcus, Enterococcus, Pseudomonas | 4.21 | 2.95 |
| tetA | Tetracyclin (efflux pump) | Enterobacteriaceae | 0.41 | 0.16 |
| tetB | Tetracyclin (efflux pump) | Gram-negative | 0 | 1.41 |
| vanB | Vancomycin | Enterococcus | 0.25 | 0 |
| mecA | Methicillin (β-lactamase) | Staphylococcus, Enterococcus | 4.10 | 3.40 |
| S. aureus | Bacterial species | S. aureus | 0.78 | 0.99 |
| spa | S. aureus virulence gene | S. aureus | 0.14 | 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Accolti, M.; Soffritti, I.; Bini, F.; Mazziga, E.; Arnoldo, L.; Volta, A.; Bisi, M.; Antonioli, P.; Laurenti, P.; Ricciardi, W.; et al. Potential Use of a Combined Bacteriophage–Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study. Int. J. Mol. Sci. 2023, 24, 6535. https://doi.org/10.3390/ijms24076535
D’Accolti M, Soffritti I, Bini F, Mazziga E, Arnoldo L, Volta A, Bisi M, Antonioli P, Laurenti P, Ricciardi W, et al. Potential Use of a Combined Bacteriophage–Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study. International Journal of Molecular Sciences. 2023; 24(7):6535. https://doi.org/10.3390/ijms24076535
Chicago/Turabian StyleD’Accolti, Maria, Irene Soffritti, Francesca Bini, Eleonora Mazziga, Luca Arnoldo, Antonella Volta, Matteo Bisi, Paola Antonioli, Patrizia Laurenti, Walter Ricciardi, and et al. 2023. "Potential Use of a Combined Bacteriophage–Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study" International Journal of Molecular Sciences 24, no. 7: 6535. https://doi.org/10.3390/ijms24076535
APA StyleD’Accolti, M., Soffritti, I., Bini, F., Mazziga, E., Arnoldo, L., Volta, A., Bisi, M., Antonioli, P., Laurenti, P., Ricciardi, W., Vincenti, S., Mazzacane, S., & Caselli, E. (2023). Potential Use of a Combined Bacteriophage–Probiotic Sanitation System to Control Microbial Contamination and AMR in Healthcare Settings: A Pre-Post Intervention Study. International Journal of Molecular Sciences, 24(7), 6535. https://doi.org/10.3390/ijms24076535








