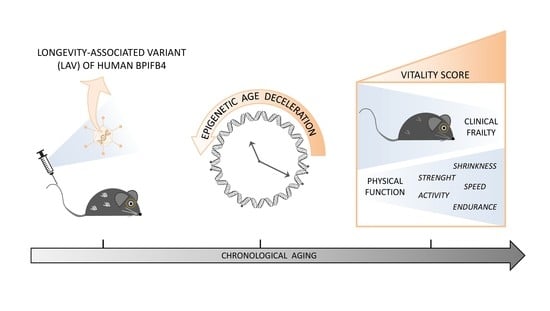
Effects of Human LAV-BPIFB4 Gene Therapy on the Epigenetic Clock and Health of Aged Mice
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
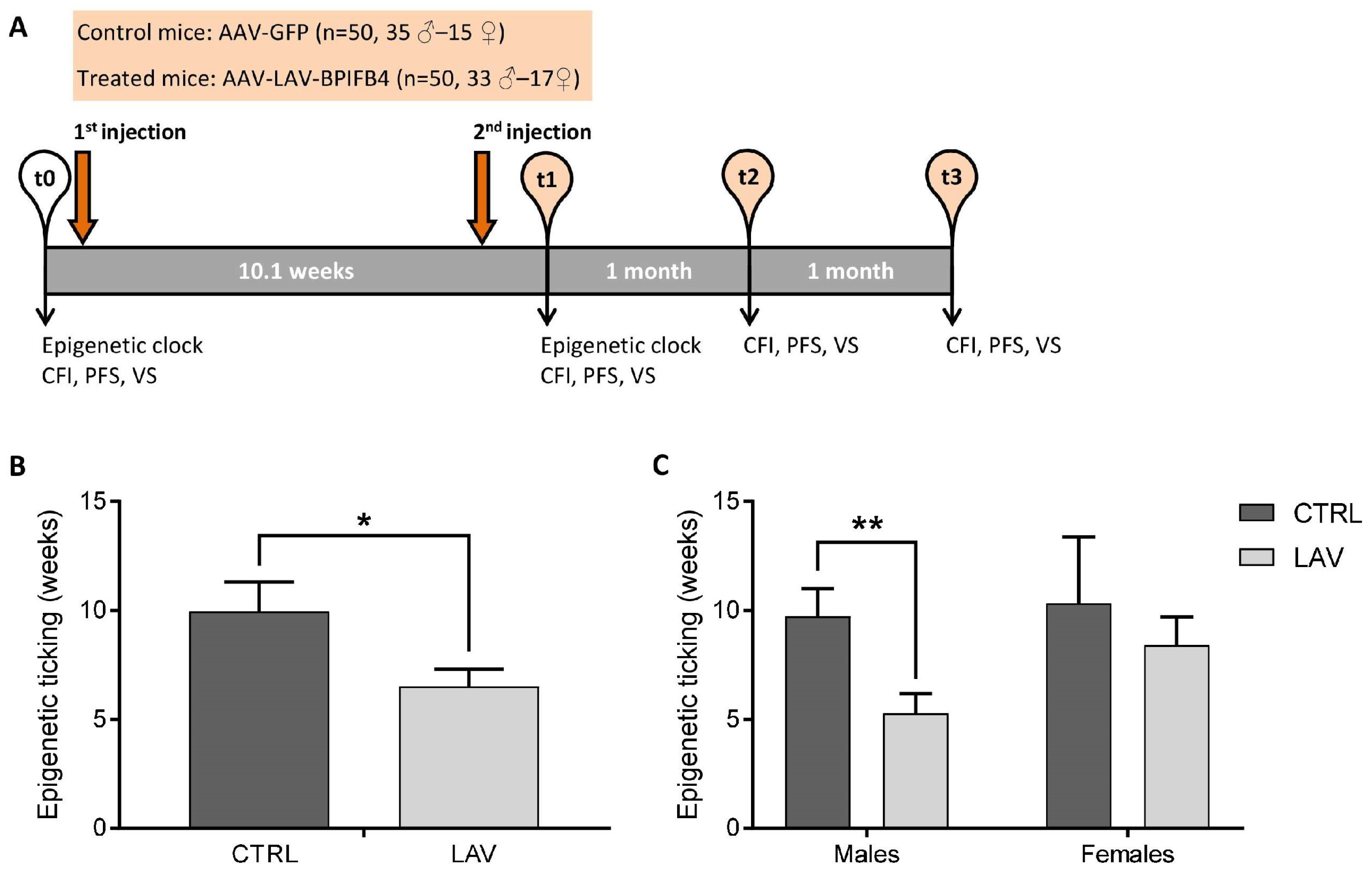
4.1. Experimental Design
4.2. Epigenetic Clock Analysis
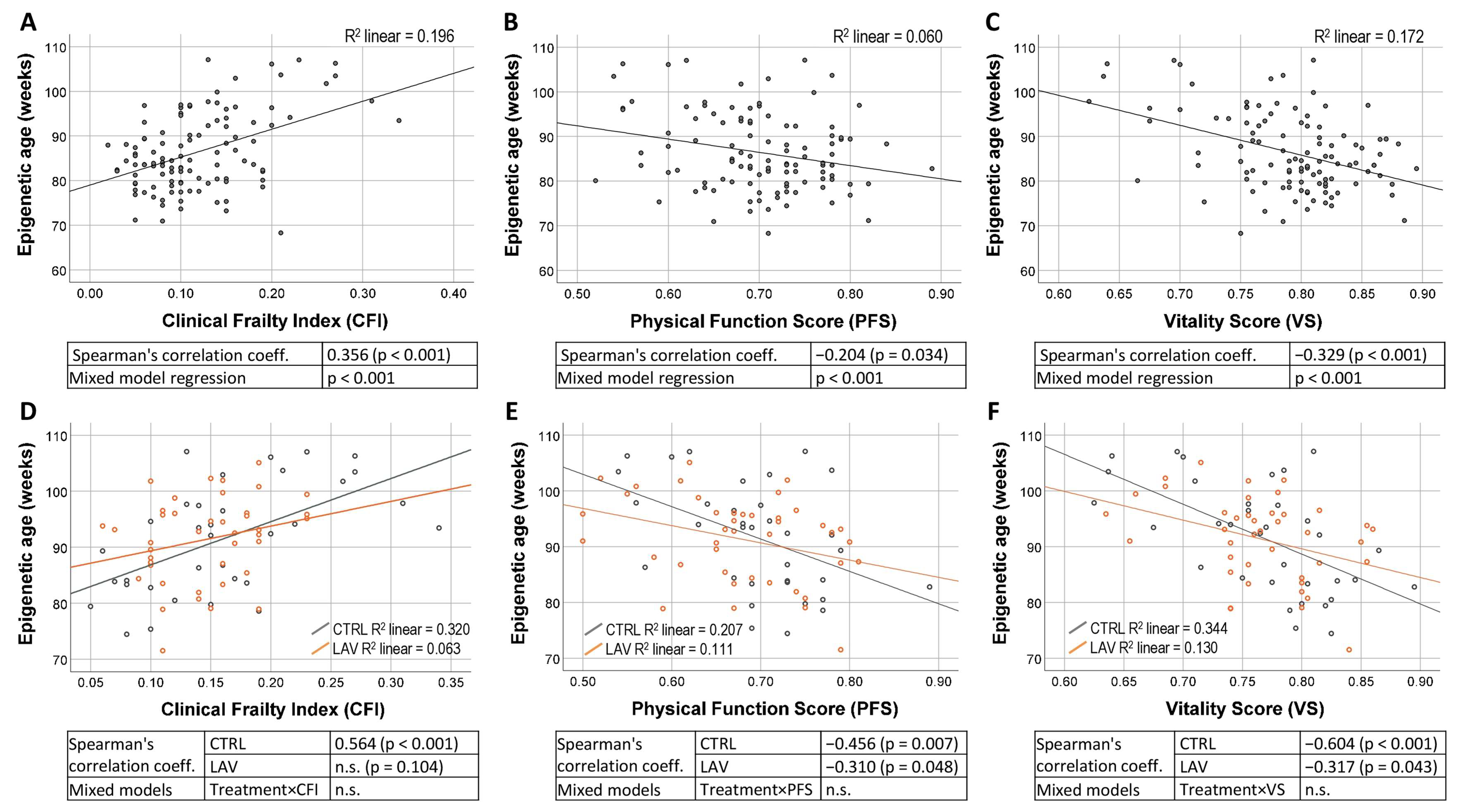
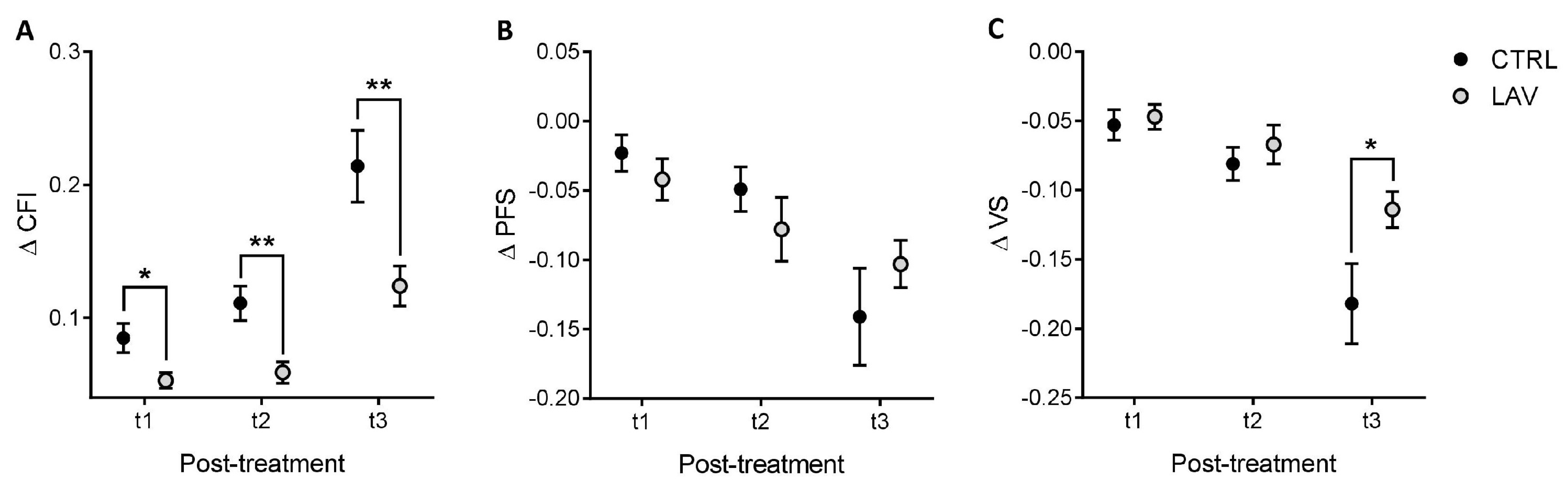
4.3. Measurement of Clinical Frailty Index, Physical Function Score and Vitality Score
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fox, J.G.; Barthold, S.W.; Davisson, M.T.; Newcomer, C.E.; Quimby, F.W.; Smith, A.L. The Mouse in Biomedical Research; Academic Press: New York, NY, USA, 2007; Volume 1–4. [Google Scholar]
- Marcozzi, S.; Bigossi, G.; Giuliani, M.E.; Giacconi, R.; Cardelli, M.; Piacenza, F.; Orlando, F.; Segala, A.; Valerio, A.; Nisoli, E.; et al. Comprehensive Longitudinal Non-Invasive Quantification of Healthspan and Frailty in a Large Cohort (n = 546) of Geriatric C57BL/6 J Mice. Geroscience 2023. [Google Scholar] [CrossRef]
- Ferrucci, L.; Levine, M.E.; Kuo, P.-L.; Simonsick, E.M. Time and the Metrics of Aging. Circ. Res. 2018, 123, 740–744. [Google Scholar] [CrossRef]
- Hartmann, A.; Hartmann, C.; Secci, R.; Hermann, A.; Fuellen, G.; Walter, M. Ranking Biomarkers of Aging by Citation Profiling and Effort Scoring. Front. Genet. 2021, 12, 686320. [Google Scholar] [CrossRef]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Ritchie, S.J.; Muniz-Terrera, G.; Harris, S.E.; Gibson, J.; Redmond, P.; Cox, S.R.; Pattie, A.; et al. The Epigenetic Clock Is Correlated with Physical and Cognitive Fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 2015, 44, 1388–1396. [Google Scholar] [CrossRef]
- Bell, C.G.; Lowe, R.; Adams, P.D.; Baccarelli, A.A.; Beck, S.; Bell, J.T.; Christensen, B.C.; Gladyshev, V.N.; Heijmans, B.T.; Horvath, S.; et al. DNA Methylation Aging Clocks: Challenges and Recommendations. Genome Biol. 2019, 20, 249. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyö, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The Role of Retrotransposable Elements in Ageing and Age-Associated Diseases. Nature 2021, 596, 43–53. [Google Scholar] [CrossRef]
- Gabbianelli, R.; Malavolta, M. Epigenetics in Ageing and Development. Mech. Ageing Dev. 2018, 174, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Weidner, C.I.; Wagner, W. The Epigenetic Tracks of Aging. Biol. Chem. 2014, 395, 1307–1314. [Google Scholar] [CrossRef]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.-C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA Methylation-Based Measures of Biological Age: Meta-Analysis Predicting Time to Death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef]
- Bernabeu, E.; McCartney, D.L.; Gadd, D.A.; Hillary, R.F.; Lu, A.T.; Murphy, L.; Wrobel, N.; Campbell, A.; Harris, S.E.; Liewald, D.; et al. Refining Epigenetic Prediction of Chronological and Biological Age. Genome Med. 2023, 15, 12. [Google Scholar] [CrossRef]
- Han, Y.; Eipel, M.; Franzen, J.; Sakk, V.; Dethmers-Ausema, B.; Yndriago, L.; Izeta, A.; de Haan, G.; Geiger, H.; Wagner, W. Epigenetic Age-Predictor for Mice Based on Three CpG Sites. Elife 2018, 7, e37462. [Google Scholar] [CrossRef]
- Perez-Correa, J.-F.; Tharmapalan, V.; Geiger, H.; Wagner, W. Epigenetic Clocks for Mice Based on Age-Associated Regions That Are Conserved Between Mouse Strains and Human. Front. Cell. Dev. Biol. 2022, 10, 902857. [Google Scholar] [CrossRef]
- Fried, L.P.; Tangen, C.M.; Walston, J.; Newman, A.B.; Hirsch, C.; Gottdiener, J.; Seeman, T.; Tracy, R.; Kop, W.J.; Burke, G.; et al. Frailty in Older Adults: Evidence for a Phenotype. J. Gerontol.—Ser. A Biol. Sci. Med. Sci. 2001, 56, M146–M156. [Google Scholar] [CrossRef] [PubMed]
- Blodgett, J.M.; Theou, O.; Howlett, S.E.; Rockwood, K. A Frailty Index from Common Clinical and Laboratory Tests Predicts Increased Risk of Death across the Life Course. Geroscience 2017, 39, 447–455. [Google Scholar] [CrossRef]
- Kane, A.E.; Huizer-Pajkos, A.; MacH, J.; Mitchell, S.J.; De Cabo, R.; Le Couteur, D.G.; Howlett, S.E.; Hilmer, S.N. A Comparison of Two Mouse Frailty Assessment Tools. J. Gerontol.—Ser. A Biol. Sci. Med. Sci. 2017, 72, 904–910. [Google Scholar] [CrossRef]
- Malavolta, M.; Dato, S.; Villa, F.; De Rango, F.; Iannone, F.; Ferrario, A.; Maciag, A.; Ciaglia, E.; D’amato, A.; Carrizzo, A.; et al. LAV-BPIFB4 Associates with Reduced Frailty in Humans and Its Transfer Prevents Frailty Progression in Old Mice. Aging 2019, 11, 6555–6568. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Malovini, A.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Maciag, A.; Madonna, M.; Bellazzi, R.; Milanesi, L.; Vecchione, C.; et al. Serum BPIFB4 Levels Classify Health Status in Long-Living Individuals. Immun. Ageing 2015, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Malovini, A.; Maciąg, A.; Damato, A.; Auricchio, A.; Spinetti, G.; Sangalli, E.; et al. Genetic Analysis Reveals a Longevity-Associated Protein Modulating Endothelial Function and Angiogenesis. Circ. Res. 2015, 117, 333–345. [Google Scholar] [CrossRef]
- Villa, F.; Carrizzo, A.; Ferrario, A.; Maciag, A.; Cattaneo, M.; Spinelli, C.C.; Montella, F.; Damato, A.; Ciaglia, E.; Puca, A.A. A Model of Evolutionary Selection: The Cardiovascular Protective Function of the Longevity Associated Variant of BPIFB4. Int. J. Mol. Sci. 2018, 19, 3229. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, M.; Beltrami, A.P.; Thomas, A.C.; Spinetti, G.; Alvino, V.; Avolio, E.; Veneziano, C.; Rolle, I.G.; Sponga, S.; Sangalli, E.; et al. The longevity-associated BPIFB4 411 gene supports cardiac function and vascularization in aging cardiomyopathy. Cardiovasc. Res. 2023, 13, 412. [Google Scholar] [CrossRef]
- Ciaglia, E.; Lopardo, V.; Montella, F.; Carrizzo, A.; Di Pietro, P.; Malavolta, M.; Giacconi, R.; Orlando, F.; Cattaneo, M.; Madeddu, P.; et al. Transfer of the Longevity-Associated Variant of BPIFB4 Gene Rejuvenates Immune System and Vasculature by a Reduction of CD38+ Macrophages and NAD+ Decline. Cell Death Dis. 2022, 13, 86. [Google Scholar] [CrossRef]
- Puca, A.A.; Lopardo, V.; Montella, F.; Di Pietro, P.; Cesselli, D.; Rolle, I.G.; Bulfoni, M.; Di Sarno, V.; Iaconetta, G.; Campiglia, P.; et al. The Longevity-Associated Variant of BPIFB4 Reduces Senescence in Glioma Cells and in Patients’ Lymphocytes Favoring Chemotherapy Efficacy. Cells 2022, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Puca, A.A.; Carrizzo, A.; Spinelli, C.; Damato, A.; Ambrosio, M.; Villa, F.; Ferrario, A.; Maciag, A.; Fornai, F.; Lenzi, P.; et al. Single Systemic Transfer of a Human Gene Associated with Exceptional Longevity Halts the Progression of Atherosclerosis and Inflammation in ApoE Knockout Mice through a CXCR4-Mediated Mechanism. Eur. Heart J. 2020, 41, 2487A–2497A. [Google Scholar] [CrossRef]
- Dang, Z.; Avolio, E.; Thomas, A.C.; Faulkner, A.; Beltrami, A.P.; Cervellin, C.; Carrizzo, A.; Maciag, A.; Gu, Y.; Ciaglia, E.; et al. Transfer of a Human Gene Variant Associated with Exceptional Longevity Improves Cardiac Function in Obese Type 2 Diabetic Mice through Induction of the SDF-1/CXCR4 Signalling Pathway. Eur. J. Heart Fail. 2020, 22, 1568–1581. [Google Scholar] [CrossRef]
- Cattaneo, M.; Maciag, A.; Milella, M.S.; Ciaglia, E.; Bruno, A.; Puca, A.A. Longevity-Associated Variant of BPIFB4 Confers Neuroprotection in the STHdh Cell Model of Huntington Disease. Int. J. Mol. Sci. 2022, 23, 15313. [Google Scholar] [CrossRef] [PubMed]
- Belsky, D.W.; Moffitt, T.E.; Cohen, A.A.; Corcoran, D.L.; Levine, M.E.; Prinz, J.A.; Schaefer, J.; Sugden, K.; Williams, B.; Poulton, R.; et al. Eleven Telomere, Epigenetic Clock, and Biomarker-Composite Quantifications of Biological Aging: Do They Measure the Same Thing? Am. J. Epidemiol. 2018, 187, 1220–1230. [Google Scholar] [CrossRef]
- Lopardo, V.; Conti, V.; Montella, F.; Iannaccone, T.; Esposito, R.M.; Sellitto, C.; Manzo, V.; Maciag, A.; Ricciardi, R.; Pagliano, P.; et al. Gender Differences Associated with the Prognostic Value of BPIFB4 in COVID-19 Patients: A Single-Center Preliminary Study. J. Pers. Med. 2022, 12, 1058. [Google Scholar] [CrossRef]
- Dane, A.P.; Wowro, S.J.; Cunningham, S.C.; Alexander, I.E. Comparison of Gene Transfer to the Murine Liver Following Intraperitoneal and Intraportal Delivery of Hepatotropic AAV Pseudo-Serotypes. Gene Ther. 2013, 20, 460–464. [Google Scholar] [CrossRef]
- Ciaglia, E.; Montella, F.; Maciag, A.; Scala, P.; Ferrario, A.; Banco, C.; Carrizzo, A.; Spinelli, C.C.; Cattaneo, M.; De Candia, P.; et al. Longevity-Associated Variant of BPIFB4 Mitigates Monocyte-Mediated Acquired Immune Response. J. Gerontol.—Ser. A Biol. Sci. Med. Sci. 2019, 74, S38–S44. [Google Scholar] [CrossRef]
- Ciaglia, E.; Montella, F.; Lopardo, V.; Scala, P.; Ferrario, A.; Cattaneo, M.; Carrizzo, A.; Malovini, A.; Madeddu, P.; Vecchione, C.; et al. Circulating BPIFB4 Levels Associate with and Influence the Abundance of Reparative Monocytes and Macrophages in Long Living Individuals. Front. Immunol. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Ligthart, S.; Marzi, C.; Aslibekyan, S.; Mendelson, M.M.; Conneely, K.N.; Tanaka, T.; Colicino, E.; Waite, L.L.; Joehanes, R.; Guan, W.; et al. DNA Methylation Signatures of Chronic Low-Grade Inflammation Are Associated with Complex Diseases. Genome Biol. 2016, 17, 255. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Jaramillo, V.; Portilla-Fernandez, E.; Glisic, M.; Voortman, T.; Ghanbari, M.; Bramer, W.; Chowdhury, R.; Nijsten, T.; Dehghan, A.; Franco, O.H.; et al. Epigenetics and Inflammatory Markers: A Systematic Review of the Current Evidence. Int. J. Inflam. 2019, 2019, 6273680. [Google Scholar] [CrossRef] [PubMed]
- Di Pardo, A.; Ciaglia, E.; Cattaneo, M.; Maciag, A.; Montella, F.; Lopardo, V.; Ferrario, A.; Villa, F.; Madonna, M.; Amico, E.; et al. The Longevity-Associated Variant of BPIFB4 Improves a CXCR4-Mediated Striatum–Microglia Crosstalk Preventing Disease Progression in a Mouse Model of Huntington’s Disease. Cell Death Dis. 2020, 11, 546. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Hu, Q.; Wang, L.; Liu, J.; Zheng, Z.; Zhang, W.; Ren, J.; Zhu, F.; Liu, G.-H. Epigenetic Regulation of Aging: Implications for Interventions of Aging and Diseases. Signal. Transduct. Target. Ther. 2022, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Hou, Y.; Fang, C.; Ma, K.; Zhang, S.; Guo, Z.; Kang, N.; Niu, K.; Shen, X.; Li, Y.; et al. Bazi Bushen Mitigates Epigenetic Aging and Extends Healthspan in Naturally Aging Mice. Biomed. Pharmacother. 2023, 160, 114384. [Google Scholar] [CrossRef] [PubMed]
- Clegg, A.; Young, J.; Iliffe, S.; Rikkert, M.O.; Rockwood, K. Frailty in Elderly People. Lancet 2013, 381, 752–762. [Google Scholar] [CrossRef]
- Kuo, P.L.; Schrack, J.A.; Levine, M.E.; Shardell, M.D.; Simonsick, E.M.; Chia, C.W.; Moore, A.Z.; Tanaka, T.; An, Y.; Karikkineth, A.; et al. Longitudinal Phenotypic Aging Metrics in the Baltimore Longitudinal Study of Aging. Nat. Aging 2022, 2, 635–643. [Google Scholar] [CrossRef]
- Soysal, P.; Stubbs, B.; Lucato, P.; Luchini, C.; Solmi, M.; Peluso, R.; Sergi, G.; Isik, A.T.; Manzato, E.; Maggi, S.; et al. Inflammation and Frailty in the Elderly: A Systematic Review and Meta-Analysis. Ageing Res. Rev. 2016, 31, 1–8. [Google Scholar] [CrossRef]
- Whitehead, J.C.; Hildebrand, B.A.; Sun, M.; Rockwood, M.R.; Rose, R.A.; Rockwood, K.; Howlett, S.E. A Clinical Frailty Index in Aging Mice: Comparisons with Frailty Index Data in Humans. J. Gerontol.-Ser. A Biol. Sci. Med. Sci. 2014, 69, 621–632. [Google Scholar] [CrossRef]
| Females | Males | |||
|---|---|---|---|---|
| F | p | F | p | |
| ΔCFI (t1 − t0) | 1.143 | 0.145 | 4.124 | 0.048 |
| ΔCFI (t2 − t0) | 6.019 | 0.020 | 1.879 | 0.177 |
| ΔCFI (t3 − t0) | 0.295 | 0.592 | 1.372 | 0.248 |
| ΔPFS (t1 − t0) | 0.834 | 0.368 | 2.966 | 0.091 |
| ΔPFS (t2 − t0) | 13.235 | 0.001 | 6.689 | 0.013 |
| ΔPFS (t3 − t0) | 0.001 | 0.991 | 10.210 | 0.003 |
| ΔVS (t1 − t0) | 1.378 | 0.250 | 5.221 | 0.027 |
| ΔVS (t2 − t0) | 13.146 | 0.001 | 6.098 | 0.017 |
| ΔVS (t3 − t0) | 0.036 | 0.852 | 5.117 | 0.029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giuliani, M.E.; Barbi, V.; Bigossi, G.; Marcozzi, S.; Giacconi, R.; Cardelli, M.; Piacenza, F.; Orlando, F.; Ciaglia, E.; Cattaneo, M.; et al. Effects of Human LAV-BPIFB4 Gene Therapy on the Epigenetic Clock and Health of Aged Mice. Int. J. Mol. Sci. 2023, 24, 6464. https://doi.org/10.3390/ijms24076464
Giuliani ME, Barbi V, Bigossi G, Marcozzi S, Giacconi R, Cardelli M, Piacenza F, Orlando F, Ciaglia E, Cattaneo M, et al. Effects of Human LAV-BPIFB4 Gene Therapy on the Epigenetic Clock and Health of Aged Mice. International Journal of Molecular Sciences. 2023; 24(7):6464. https://doi.org/10.3390/ijms24076464
Chicago/Turabian StyleGiuliani, Maria Elisa, Veronica Barbi, Giorgia Bigossi, Serena Marcozzi, Robertina Giacconi, Maurizio Cardelli, Francesco Piacenza, Fiorenza Orlando, Elena Ciaglia, Monica Cattaneo, and et al. 2023. "Effects of Human LAV-BPIFB4 Gene Therapy on the Epigenetic Clock and Health of Aged Mice" International Journal of Molecular Sciences 24, no. 7: 6464. https://doi.org/10.3390/ijms24076464
APA StyleGiuliani, M. E., Barbi, V., Bigossi, G., Marcozzi, S., Giacconi, R., Cardelli, M., Piacenza, F., Orlando, F., Ciaglia, E., Cattaneo, M., Mongelli, A., Gaetano, C., Provinciali, M., Puca, A. A., & Malavolta, M. (2023). Effects of Human LAV-BPIFB4 Gene Therapy on the Epigenetic Clock and Health of Aged Mice. International Journal of Molecular Sciences, 24(7), 6464. https://doi.org/10.3390/ijms24076464