Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry?
Abstract
1. Introduction
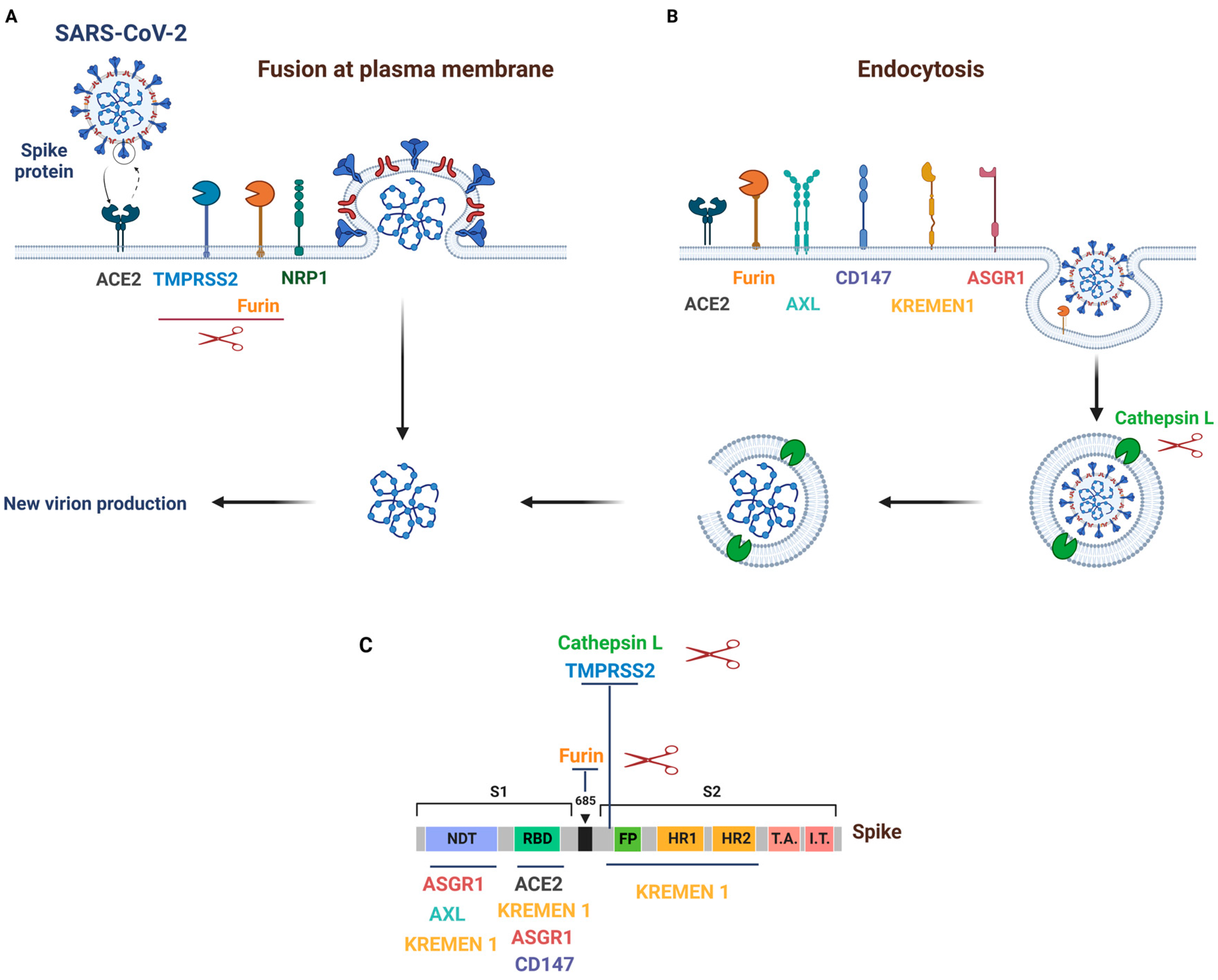
2. Mechanisms and Pathways of SARS-CoV-2 Entry into Host Cells
3. SARS-CoV-2 Primary Receptor and Cofactors
3.1. ACE2
3.1.1. Background
3.1.2. ACE2 Tissue Expression and Its Importance in SARS-CoV-2 Infection
3.2. TMPRSS2
3.2.1. Background
3.2.2. TMPRSS2 Tissue Expression and Its Importance in SARS-CoV-2 Infection
3.3. Furin
3.3.1. Background
3.3.2. Furin Tissue Expression and Its Importance in SARS-CoV-2 Infection
3.4. Cathepsin L
3.4.1. Background
3.4.2. Cathepsin L Tissue Expression and Its Importance in SARS-CoV-2 Infection
3.5. NRP1
3.5.1. Background
3.5.2. NRP1 Tissue Expression and Its Importance in SARS-CoV-2 Infection
4. SARS-CoV-2 Alternative Receptors
4.1. CD147
4.1.1. Background
4.1.2. CD147 Tissue Expression and Its Importance in SARS-CoV-2 Infection
4.2. AXL
4.2.1. Background
4.2.2. AXL Tissue Expression and Its Importance in SARS-CoV-2 Infection
4.3. KREMEN1 and ASGR1
4.3.1. Background
4.3.2. KREMEN1 and ASGR1 Tissue Expression and Its Importance in SARS-CoV-2 Infection
4.4. Additional Receptors Facilitating Virus Attachment
4.5. LRRC15, a Novel Receptor Binding SARS-CoV-2 Spike and Suppressing Viral Entry
4.5.1. Background
4.5.2. LRRC15 Tissue Expression and Its Importance in SARS-CoV-2 Infection
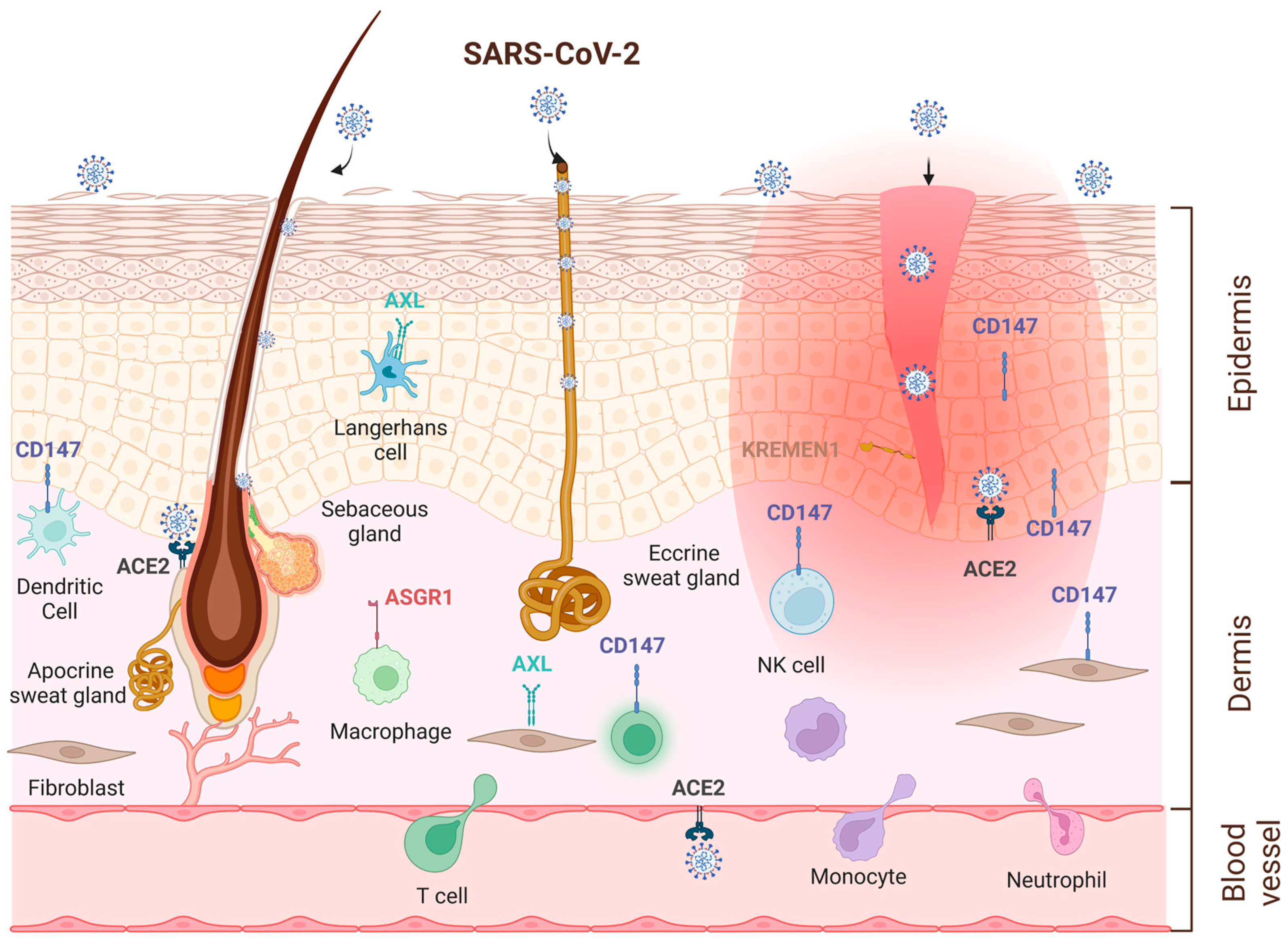
5. Skin as an Alternative Entry Route for SARS-CoV-2 Virus
| Receptors and Cofactors | Expression in Skin | References |
|---|---|---|
| ACE2 | mRNA: low levels (whole skin) | https://gtexportal.org, [139,144,145,146,147,149] |
| Protein: keratinocytes in the basal cell layer of epidermis, smooth muscle cells surrounding sebaceous glands, eccrine gland cells | [29,138,139,147] | |
| Upregulation by IFNs, IL-33, in AD, in psoriasis | [31,40,41,42,141,150,151] | |
| TMPRSS2 | mRNA: low levels (whole skin) | https://gtexportal.org, [47,139,144,145], |
| Protein: sweat glands | [138,141] | |
| Furin | mRNA: high levels (whole skin) | https://gtexportal.org, |
| Protein: keratinocytes | [161] | |
| Cathepsin L | mRNA: low levels (whole skin), higher in fibroblasts, immune cells | https://gtexportal.org, [159] |
| Protein: low levels in healthy skin. Upregulation in AD, psoriasis, lupus erythematosus, cSCC | [152,153] | |
| NRP1 | mRNA: intermediate levels (whole skin), high in cultured fibroblasts | https://gtexportal.org, |
| Protein: keratinocytes in the suprabasal cell layer of epidermis, immune cells | [77,78] | |
| CD147 | mRNA: high levels (whole skin), keratinocytes, fibroblasts, immune cells | https://gtexportal.org, [145] |
| Protein: keratinocytes, fibroblasts, immune cells Upregulation by IL-22, IL-1α, TGFβ1, EGF | [157] [83,157,158] | |
| AXL | mRNA: intermediate levels (whole skin), high in cultured fibroblasts | https://gtexportal.org, |
| Protein: Langerhans cells, keratinocytes | [155] | |
| KREMEN1 | mRNA: intermediate/high levels (whole skin), expression on keratinocytes | https://gtexportal.org, [159,160] |
| Protein: epidermis | https://www.proteinatlas.org | |
| ASGR1 | mRNA: low levels (whole skin) Protein: macrophages | https://gtexportal.org, [110,111] |
| LRRC15 | mRNA: high expression in skin Protein: fibroblasts | https://gtexportal.org, [124,162] |
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johansen, M.D.; Irving, A.; Montagutelli, X.; Tate, M.D.; Rudloff, I.; Nold, M.F.; Hansbro, N.G.; Kim, R.Y.; Donovan, C.; Liu, G.; et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020, 13, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Perez, P.; Kato, T.; Mikami, Y.; Okuda, K.; Gilmore, R.C.; Conde, C.D.; Gasmi, B.; Stein, S.; Beach, M.; et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021, 27, 892–903. [Google Scholar] [CrossRef]
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 317. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Zhang, A.L.; Wang, Y.; Molina, M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 14857–14863. [Google Scholar] [CrossRef]
- Lan, J.; Song, Z.; Miao, X.; Li, H.; Li, Y.; Dong, L.; Yang, J.; An, X.; Zhang, Y.; Yang, L.; et al. Skin damage among health care workers managing coronavirus disease-2019. J. Am. Acad. Dermatol. 2020, 82, 1215–1216. [Google Scholar] [CrossRef]
- Mushtaq, S.; Terzi, E.; Recalcati, S.; Salas-Alanis, J.C.; Amin, S.; Faizi, N. Cutaneous adverse effects due to personal protective measures during COVID-19 pandemic: A study of 101 patients. Int. J. Dermatol. 2021, 60, 327–331. [Google Scholar] [CrossRef]
- Kabashima, K.; Honda, T.; Ginhoux, F.; Egawa, G. The immunological anatomy of the skin. Nat. Rev. Immunol. 2019, 19, 19–30. [Google Scholar] [CrossRef]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. New Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Hirose, R.; Ikegaya, H.; Naito, Y.; Watanabe, N.; Yoshida, T.; Bandou, R.; Daidoji, T.; Itoh, Y.; Nakaya, T. Survival of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Influenza Virus on Human Skin: Importance of Hand Hygiene in Coronavirus Disease 2019 (COVID-19). Clin. Infect. Dis. 2021, 73, e4329–e4335. [Google Scholar] [CrossRef] [PubMed]
- Hirose, R.; Itoh, Y.; Ikegaya, H.; Miyazaki, H.; Watanabe, N.; Yoshida, T.; Bandou, R.; Daidoji, T.; Nakaya, T. Differences in environmental stability among SARS-CoV-2 variants of concern: Both omicron BA.1 and BA.2 have higher stability. Clin. Microbiol. Infect. 2022, 28, 1486–1491. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Rosen, O.; Wang, L.; Turner, H.L.; Stevens, L.J.; Corbett, K.S.; Bowman, C.A.; Pallesen, J.; Shi, W.; Zhang, Y.; et al. Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD. Cell Rep. 2019, 28, 3395–3405.e3396. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.-W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van, T.V.L.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef]
- Jocher, G.; Grass, V.; Tschirner, S.K.; Riepler, L.; Breimann, S.; Kaya, T.; Oelsner, M.; Hamad, M.S.; Hofmann, L.I.; Blobel, C.P.; et al. ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion. EMBO Rep. 2022, 23, e54305. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.-E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, J.A.; Tremblay, B.J.; Mansfield, M.J.; Woody, O.; Lobb, B.; Banerjee, A.; Chandiramohan, A.; Tiessen, N.; Cao, Q.; Dvorkin-Gheva, A.; et al. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur. Respir. J. 2020, 56, 2001123. [Google Scholar] [CrossRef]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A Novel Angiotensin-Converting Enzyme–Related Carboxypeptidase (ACE2) Converts Angiotensin I to Angiotensin 1–9. Circ. Res. 2000, 87, e1–e9. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A Human Homolog of Angiotensin-converting Enzyme: CLONING AND FUNCTIONAL EXPRESSION AS A CAPTOPRIL-INSENSITIVE CARBOXYPEPTIDASE *. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Harmer, D.; Gilbert, M.; Borman, R.; Clark, K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002, 532, 107–110. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.; Lely, A.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Cardot-Leccia, N.; Hubiche, T.; Dellamonica, J.; Burel-Vandenbos, F.; Passeron, T. Pericyte alteration sheds light on micro-vasculopathy in COVID-19 infection. Intensive Care Med. 2020, 46, 1777–1778. [Google Scholar] [CrossRef]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e1019. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef]
- Li, M.Y.; Li, L.; Zhang, Y.; Wang, X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 2020, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Triana, S.; Metz-Zumaran, C.; Ramirez, C.; Kee, C.; Doldan, P.; Shahraz, M.; Schraivogel, D.; Gschwind, A.R.; Sharma, A.K.; Steinmetz, L.M.; et al. Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut. Mol. Syst. Biol. 2021, 17, e10232. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.W.; Cheng, Y.; Zhang, J.; Jiang, X.M.; Wang, L.; Deng, J.; Wang, P.H. Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection. J. Med. Virol. 2020, 92, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thurmann, L.; Kurth, F.; Volker, M.T.; et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Heuberger, J.; Trimpert, J.; Vladimirova, D.; Goosmann, C.; Lin, M.; Schmuck, R.; Mollenkopf, H.J.; Brinkmann, V.; Tacke, F.; Osterrieder, N.; et al. Epithelial response to IFN-gamma promotes SARS-CoV-2 infection. EMBO Mol. Med. 2021, 13, e13191. [Google Scholar] [CrossRef]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef]
- Blume, C.; Jackson, C.L.; Spalluto, C.M.; Legebeke, J.; Nazlamova, L.; Conforti, F.; Perotin, J.M.; Frank, M.; Butler, J.; Crispin, M.; et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021, 53, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.W.; Attig, J.; Bolland, W.; Young, G.R.; Major, J.; Wrobel, A.G.; Gamblin, S.; Wack, A.; Kassiotis, G. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat. Genet. 2020, 52, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 2020, 27, 876–889.e812. [Google Scholar] [CrossRef] [PubMed]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef]
- Mantzourani, C.; Vasilakaki, S.; Gerogianni, V.E.; Kokotos, G. The discovery and development of transmembrane serine protease 2 (TMPRSS2) inhibitors as candidate drugs for the treatment of COVID-19. Expert Opin. Drug Discov. 2022, 17, 231–246. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Mucci, L.A.; Antonarakis, E.S.; Nelson, P.S.; Kantoff, P.W. TMPRSS2 and COVID-19: Serendipity or opportunity for intervention? Cancer Discov. 2020, 10, 779–782. [Google Scholar] [CrossRef]
- Mikkonen, L.; Pihlajamaa, P.; Sahu, B.; Zhang, F.-P.; Jänne, O.A. Androgen receptor and androgen-dependent gene expression in lung. Mol. Cell. Endocrinol. 2010, 317, 14–24. [Google Scholar] [CrossRef]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Zhou, L.; Niu, Z.; Jiang, X.; Zhang, Z.; Zheng, Y.; Wang, Z.; Zhu, Y.; Gao, L.; Huang, H.; Wang, X.; et al. SARS-CoV-2 Targets by the pscRNA Profiling of ACE2, TMPRSS2 and Furin Proteases. iScience 2020, 23, 101744. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93, e01815–e01818. [Google Scholar] [CrossRef] [PubMed]
- Hatesuer, B.; Bertram, S.; Mehnert, N.; Bahgat, M.M.; Nelson, P.S.; Pöhlmann, S.; Pöhlman, S.; Schughart, K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013, 9, e1003774. [Google Scholar] [CrossRef]
- Tarnow, C.; Engels, G.; Arendt, A.; Schwalm, F.; Sediri, H.; Preuss, A.; Nelson, P.S.; Garten, W.; Klenk, H.-D.; Gabriel, G.; et al. TMPRSS2 Is a Host Factor That Is Essential for Pneumotropism and Pathogenicity of H7N9 Influenza A Virus in Mice. J. Virol. 2014, 88, 4744–4751. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Ami, Y.; Tahara, M.; Kubota, T.; Anraku, M.; Abe, M.; Nakajima, N.; Sekizuka, T.; Shirato, K.; Suzaki, Y.; et al. The Host Protease TMPRSS2 Plays a Major Role in In Vivo Replication of Emerging H7N9 and Seasonal Influenza Viruses. J. Virology 2014, 88, 5608–5616. [Google Scholar] [CrossRef] [PubMed]
- Ruffin, M.; Bigot, J.; Calmel, C.; Mercier, J.; Givelet, M.; Oliva, J.; Pizzorno, A.; Rosa-Calatrava, M.; Corvol, H.; Balloy, V.; et al. Flagellin From Pseudomonas aeruginosa Modulates SARS-CoV-2 Infectivity in Cystic Fibrosis Airway Epithelial Cells by Increasing TMPRSS2 Expression. Front. Immunol. 2021, 12, 714027. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Do Anti-androgens Have Potential as Therapeutics for COVID-19? Endocrinology 2021, 162, bqab114. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Kruger, N.; Arora, P.; Sorensen, L.K.; Sogaard, O.S.; Hasselstrom, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef]
- Osman, E.E.A.; Rehemtulla, A.; Neamati, N. Why All the Fury over Furin? J. Med. Chem. 2022, 65, 2747–2784. [Google Scholar] [CrossRef]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e775. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, H.; Eerola, R.; Hopsu-Havu, V.K.; Bromme, D.; Vuorio, E. Antisense RNA inhibition of cathepsin L expression reduces tumorigenicity of malignant cells. Eur. J. Cancer 2000, 36, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The Ins and Outs of Cathepsins: Physiological Function and Role in Disease Management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef]
- Nakagawa, T.; Roth, W.; Wong, P.; Nelson, A.; Farr, A.; Deussing, J.; Villadangos, J.A.; Ploegh, H.; Peters, C.; Rudensky, A.Y. Cathepsin L: Critical role in Ii degradation and CD4 T cell selection in the thymus. Science 1998, 280, 450–453. [Google Scholar] [CrossRef]
- Gomes, C.P.; Fernandes, D.E.; Casimiro, F.; da Mata, G.F.; Passos, M.T.; Varela, P.; Mastroianni-Kirsztajn, G.; Pesquero, J.B. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front. Cell. Infect. Microbiol. 2020, 10, 589505. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef]
- Huang, I.-C.; Bosch, B.J.; Li, F.; Li, W.; Lee, K.H.; Ghiran, S.; Vasilieva, N.; Dermody, T.S.; Harrison, S.C.; Dormitzer, P.R.; et al. SARS Coronavirus, but Not Human Coronavirus NL63, Utilizes Cathepsin L to Infect ACE2-expressing Cells. J. Biol. Chem. 2006, 281, 3198–3203. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e116. [Google Scholar] [CrossRef]
- Zhao, M.-M.; Yang, W.-L.; Yang, F.-Y.; Zhang, L.; Huang, W.-J.; Hou, W.; Fan, C.-F.; Jin, R.-H.; Feng, Y.-M.; Wang, Y.-C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Smieszek, S.P.; Przychodzen, B.P.; Polymeropoulos, M.H. Amantadine disrupts lysosomal gene expression: A hypothesis for COVID19 treatment. Int. J. Antimicrob. Agents 2020, 55, 106004. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Mosbauer, K.; Hofmann-Winkler, H.; Kaul, A.; Kleine-Weber, H.; Kruger, N.; Gassen, N.C.; Muller, M.A.; Drosten, C.; Pohlmann, S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 2020, 585, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, M.; Gagnon, M.L.; Klagsbrun, M. Genomic Organization of Human Neuropilin-1 and Neuropilin-2 Genes: Identification and Distribution of Splice Variants and Soluble Isoforms. Genomics 2000, 70, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Wild, J.R.; Staton, C.A.; Chapple, K.; Corfe, B.M. Neuropilins: Expression and roles in the epithelium. Int. J. Exp. Pathol. 2012, 93, 81–103. [Google Scholar] [CrossRef]
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted Role of Neuropilins in the Immune System: Potential Targets for Immunotherapy. Front. Immunol. 2017, 8, 1228. [Google Scholar] [CrossRef] [PubMed]
- Chuckran, C.A.; Liu, C.; Bruno, T.C.; Workman, C.J.; Vignali, D.A. Neuropilin-1: A checkpoint target with unique implications for cancer immunology and immunotherapy. J. Immunother. Cancer 2020, 8, e000967. [Google Scholar] [CrossRef]
- Shahrabi-Farahani, S.; Wang, L.; Zwaans, B.M.; Santana, J.M.; Shimizu, A.; Takashima, S.; Kreuter, M.; Coultas, L.; D’Amore, P.A.; Arbeit, J.M.; et al. Neuropilin 1 expression correlates with differentiation status of epidermal cells and cutaneous squamous cell carcinomas. Lab. Invest. 2014, 94, 752–765. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef]
- Balistreri, G.; Yamauchi, Y.; Teesalu, T. A widespread viral entry mechanism: The C-end Rule motif-neuropilin receptor interaction. Proc. Natl. Acad. Sci. USA 2021, 118, e2112457118. [Google Scholar] [CrossRef]
- Iacono, K.T.; Brown, A.L.; Greene, M.I.; Saouaf, S.J. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp. Mol. Pathol. 2007, 83, 283–295. [Google Scholar] [CrossRef]
- Guindolet, D.; Gabison, E.E. Role of CD147 (EMMPRIN/Basigin) in Tissue Remodeling. Anat. Rec. 2020, 303, 1584–1589. [Google Scholar] [CrossRef] [PubMed]
- Barillari, G.; Melaiu, O.; Gargari, M.; Pomella, S.; Bei, R.; Campanella, V. The Multiple Roles of CD147 in the Development and Progression of Oral Squamous Cell Carcinoma: An Overview. Int. J. Mol. Sci. 2022, 23, 8336. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 2016, 159, 481–490. [Google Scholar] [CrossRef]
- Grass, G.D.; Toole, B.P. How, with whom and when: An overview of CD147-mediated regulatory networks influencing matrix metalloproteinase activity. Biosci. Rep. 2015, 36, e00283. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Kaur, I.; Aleya, L.; Sehgal, A.; Singh, S.; Sharma, N.; Bhatia, S.; Al-Harrasi, A.; Bungau, S. CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target. Sci. Total Environ. 2022, 808, 152072. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.-Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 2021, 10, 1434. [Google Scholar] [CrossRef]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in Invasion of Host Cells by Severe Acute Respiratory Syndrome Coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef]
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021, 11, 413. [Google Scholar] [CrossRef]
- Ragotte, R.J.; Pulido, D.; Donnellan, F.R.; Hill, M.L.; Gorini, G.; Davies, H.; Brun, J.; McHugh, K.; King, L.D.W.; Skinner, K.; et al. Human Basigin (CD147) Does Not Directly Interact with SARS-CoV-2 Spike Glycoprotein. mSphere 2021, 6, e0064721. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, A.; Geng, K.; Honnen, W.; Wang, X.; Bruiners, N.; Singh, S.; Ferrara, F.; D’Angelo, S.; Bradbury, A.R.M.; et al. Human Immunodeficiency Viruses Pseudotyped with SARS-CoV-2 Spike Proteins Infect a Broad Spectrum of Human Cell Lines through Multiple Entry Mechanisms. Viruses 2021, 13, 953. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Zheng, Z.H.; Wei, D.; Wen, A.; Zhang, Z.; Lian, J.Q.; Kang, W.Z.; Hao, C.Q.; Wang, J.; Xie, R.H.; et al. Safety and efficacy of meplazumab in healthy volunteers and COVID-19 patients: A randomized phase 1 and an exploratory phase 2 trial. Signal Transduct. Target. Ther. 2021, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Chen, L.; Yuan, Y.; Wang, K.; Wang, Y.; Qin, C.; Wu, G.; Chen, R.; Zhang, Z.; Wei, D.; et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021, 6, 347. [Google Scholar] [CrossRef]
- Geng, J.; Yang, X.; Wang, K.; Wang, K.; Chen, R.; Chen, Z.N.; Qin, C.; Wu, G.; Wang, Y.; Xu, K.; et al. Immunological and metabolic characteristics of the Omicron variants infection. Signal Transduct. Target. Ther. 2023, 8, 42. [Google Scholar] [CrossRef]
- Zhang, L.; Richard, A.S.; Jackson, C.B.; Ojha, A.; Choe, H. Phosphatidylethanolamine and Phosphatidylserine Synergize to Enhance GAS6/AXL-Mediated Virus Infection and Efferocytosis. J. Virol. 2020, 95, e02079-20. [Google Scholar] [CrossRef] [PubMed]
- Morizono, K.; Chen, I.S. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 2014, 88, 4275–4290. [Google Scholar] [CrossRef]
- Linger, R.M.A.; Keating, A.K.; Earp, H.S.; Graham, D.K. Taking aim at Mer and Axl receptor tyrosine kinases as novel therapeutic targets in solid tumors. Expert Opin. Ther. Targets 2010, 14, 1073–1090. [Google Scholar] [CrossRef]
- Berk, B.C. Vascular smooth muscle growth: Autocrine growth mechanisms. Physiol. Rev. 2001, 81, 999–1030. [Google Scholar] [CrossRef]
- Neubauer, A.; Fiebeler, A.; Graham, D.K.; O’Bryan, J.P.; Schmidt, C.A.; Barckow, P.; Serke, S.; Siegert, W.; Snodgrass, H.R.; Huhn, D.; et al. Expression of axl, a transforming receptor tyrosine kinase, in normal and malignant hematopoiesis. Blood 1994, 84, 1931–1941. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Bohan, D.; Maury, W. Enveloped RNA virus utilization of phosphatidylserine receptors: Advantages of exploiting a conserved, widely available mechanism of entry. PLoS Pathog. 2021, 17, e1009899. [Google Scholar] [CrossRef] [PubMed]
- Bohan, D.; Van Ert, H.; Ruggio, N.; Rogers, K.J.; Badreddine, M.; Aguilar Briseno, J.A.; Elliff, J.M.; Rojas Chavez, R.A.; Gao, B.; Stokowy, T.; et al. Phosphatidylserine receptors enhance SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009743. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Aoki, S.; Kitajima, K.; Takahashi, T.; Matsumoto, K.; Nakamura, T. Molecular cloning and characterization of Kremen, a novel kringle-containing transmembrane protein. Biochim. Et Biophys. Acta 2001, 1518, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Zebisch, M.; Jackson, V.A.; Zhao, Y.; Jones, E.Y. Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight into Ternary Complex Formation with LRP6 and Dickkopf. Structure 2016, 24, 1599–1605. [Google Scholar] [CrossRef]
- Mao, B.; Wu, W.; Davidson, G.; Marhold, J.; Li, M.; Mechler, B.M.; Delius, H.; Hoppe, D.; Stannek, P.; Walter, C.; et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 2002, 417, 664–667. [Google Scholar] [CrossRef]
- Staring, J.; van den Hengel, L.G.; Raaben, M.; Blomen, V.A.; Carette, J.E.; Brummelkamp, T.R. KREMEN1 Is a Host Entry Receptor for a Major Group of Enteroviruses. Cell Host Microbe 2018, 23, 636–643.e635. [Google Scholar] [CrossRef]
- Xu, Y.; Tao, J.; Yu, X.; Wu, Y.; Chen, Y.; You, K.; Zhang, J.; Getachew, A.; Pan, T.; Zhuang, Y.; et al. Hypomorphic ASGR1 modulates lipid homeostasis via INSIG1-mediated SREBP signaling suppression. JCI Insight 2022, 6, e147038. [Google Scholar] [CrossRef]
- Hoober, J.K. ASGR1 and Its Enigmatic Relative, CLEC10A. Int. J. Mol. Sci. 2020, 21, 4818. [Google Scholar] [CrossRef]
- Kanemaru, K.; Noguchi, E.; Tahara-Hanaoka, S.; Mizuno, S.; Tateno, H.; Fujisawa, Y.; Nakamura, Y.; Denda-Nagai, K.; Irimura, T.; Matsuda, H.; et al. Clec10a regulates mite-induced dermatitis. Sci. Immunol. 2019, 4, eaax6908. [Google Scholar] [CrossRef]
- Harris, R.L.; van den Berg, C.W.; Bowen, D.J. ASGR1 and ASGR2, the Genes that Encode the Asialoglycoprotein Receptor (Ashwell Receptor), Are Expressed in Peripheral Blood Monocytes and Show Interindividual Differences in Transcript Profile. Mol. Biol. Int. 2012, 2012, 283974. [Google Scholar] [CrossRef]
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; He, J.; Jiang, X.; Zhang, J.; Shen, G.; et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2022, 32, 24–37. [Google Scholar] [CrossRef]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, W.D.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef]
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Thepaut, M.; Luczkowiak, J.; Vives, C.; Labiod, N.; Bally, I.; Lasala, F.; Grimoire, Y.; Fenel, D.; Sattin, S.; Thielens, N.; et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021, 17, e1009576. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, Y.; Zhou, Z.; Zhang, Z.; Xiao, X.; Liu, Z.; Chen, A.; Dong, X.; Tian, F.; Chen, S.; et al. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry. Sci. China Life Sci. 2022, 65, 701–717. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Kim, A.S.; Kafai, N.M.; Earnest, J.T.; Shah, A.P.; Case, J.B.; Basore, K.; Gilliland, T.C.; Sun, C.; Nelson, C.A.; et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 2020, 588, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.G.; Mukhtar, T.; Eze, U.C.; Simoneau, C.R.; Ross, J.; Parikshak, N.; Wang, S.; Zhou, L.; Koontz, M.; Velmeshev, D.; et al. Tropism of SARS-CoV-2 for human cortical astrocytes. Proc. Natl. Acad. Sci. USA 2022, 119, e2122236119. [Google Scholar] [CrossRef]
- Solis, O.; Beccari, A.R.; Iaconis, D.; Talarico, C.; Ruiz-Bedoya, C.A.; Nwachukwu, J.C.; Cimini, A.; Castelli, V.; Bertini, R.; Montopoli, M.; et al. The SARS-CoV-2 spike protein binds and modulates estrogen receptors. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ben-Ami, E.; Perret, R.; Huang, Y.; Courgeon, F.; Gokhale, P.C.; Laroche-Clary, A.; Eschle, B.K.; Velasco, V.; Le Loarer, F.; Algeo, M.P.; et al. LRRC15 Targeting in Soft-Tissue Sarcomas: Biological and Clinical Implications. Cancers 2020, 12, 757. [Google Scholar] [CrossRef]
- Satoh, K.; Hata, M.; Shimizu, T.; Yokota, H.; Akatsu, H.; Yamamoto, T.; Kosaka, K.; Yamada, T. Lib, transcriptionally induced in senile plaque-associated astrocytes, promotes glial migration through extracellular matrix. Biochem. Biophys. Res. Commun. 2005, 335, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurty, A.T.; Shyer, J.A.; Thai, M.; Gandham, V.; Buechler, M.B.; Yang, Y.A.; Pradhan, R.N.; Wang, A.W.; Sanchez, P.L.; Qu, Y.; et al. LRRC15(+) myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature 2022, 611, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Buechler, M.B.; Pradhan, R.N.; Krishnamurty, A.T.; Cox, C.; Calviello, A.K.; Wang, A.W.; Yang, Y.A.; Tam, L.; Caothien, R.; Roose-Girma, M.; et al. Cross-tissue organization of the fibroblast lineage. Nature 2021, 593, 575–579. [Google Scholar] [CrossRef]
- Loo, L.; Waller, M.A.; Moreno, C.L.; Cole, A.J.; Stella, A.O.; Pop, O.T.; Jochum, A.K.; Ali, O.H.; Denes, C.E.; Hamoudi, Z.; et al. Fibroblast-expressed LRRC15 is a receptor for SARS-CoV-2 spike and controls antiviral and antifibrotic transcriptional programs. PLoS Biol. 2023, 21, e3001967. [Google Scholar] [CrossRef] [PubMed]
- Shilts, J.; Crozier, T.W.M.; Teixeira-Silva, A.; Gabaev, I.; Gerber, P.P.; Greenwood, E.J.D.; Watson, S.J.; Ortmann, B.M.; Gawden-Bone, C.M.; Pauzaite, T.; et al. LRRC15 mediates an accessory interaction with the SARS-CoV-2 spike protein. PLoS Biol. 2023, 21, e3001959. [Google Scholar] [CrossRef]
- Song, J.; Chow, R.D.; Pena-Hernandez, M.A.; Zhang, L.; Loeb, S.A.; So, E.Y.; Liang, O.D.; Ren, P.; Chen, S.; Wilen, C.B.; et al. LRRC15 inhibits SARS-CoV-2 cellular entry in trans. PLoS Biol. 2022, 20, e3001805. [Google Scholar] [CrossRef]
- Behzadinasab, S.; Chin, A.W.H.; Hosseini, M.; Poon, L.L.M.; Ducker, W.A. SARS-CoV-2 virus transfers to skin through contact with contaminated solids. Sci Rep. 2021, 11, 22868. [Google Scholar] [CrossRef]
- Smythe, P.; Wilkinson, H.N. The Skin Microbiome: Current Landscape and Future Opportunities. Int. J. Mol. Sci. 2023, 24, 3950. [Google Scholar] [CrossRef]
- Recalcati, S. Cutaneous manifestations in COVID-19: A first perspective. J. Eur. Acad. Dermatol. Venereol. 2020, 34, e212–e213. [Google Scholar] [CrossRef]
- Galvan Casas, C.; Catala, A.; Carretero Hernandez, G.; Rodriguez-Jimenez, P.; Fernandez-Nieto, D.; Rodriguez-Villa Lario, A.; Navarro Fernandez, I.; Ruiz-Villaverde, R.; Falkenhain-Lopez, D.; Llamas Velasco, M.; et al. Classification of the cutaneous manifestations of COVID-19: A rapid prospective nationwide consensus study in Spain with 375 cases. Br. J. Dermatol. 2020, 183, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Polly, S.; Fernandez, A.P. Common skin signs of COVID-19 in adults: An update. Cleve Clin. J. Med. 2022, 89, 161–167. [Google Scholar] [CrossRef]
- Visconti, A.; Bataille, V.; Rossi, N.; Kluk, J.; Murphy, R.; Puig, S.; Nambi, R.; Bowyer, R.C.E.; Murray, B.; Bournot, A.; et al. Diagnostic value of cutaneous manifestation of SARS-CoV-2 infection. Br. J. Dermatol. 2021, 184, 880–887. [Google Scholar] [CrossRef]
- Magro, C.M.; Mulvey, J.J.; Laurence, J.; Seshan, S.; Crowson, A.N.; Dannenberg, A.J.; Salvatore, S.; Harp, J.; Nuovo, G.J. Docked severe acute respiratory syndrome coronavirus 2 proteins within the cutaneous and subcutaneous microvasculature and their role in the pathogenesis of severe coronavirus disease 2019. Hum. Pathol. 2020, 106, 106–116. [Google Scholar] [CrossRef]
- Liu, F.; Han, K.; Blair, R.; Kenst, K.; Qin, Z.; Upcin, B.; Worsdorfer, P.; Midkiff, C.C.; Mudd, J.; Belyaeva, E.; et al. SARS-CoV-2 Infects Endothelial Cells In Vivo and In Vitro. Front. Cell Infect. Microbiol. 2021, 11, 701278. [Google Scholar] [CrossRef] [PubMed]
- Magro, C.M.; Mulvey, J.; Kubiak, J.; Mikhail, S.; Suster, D.; Crowson, A.N.; Laurence, J.; Nuovo, G. Severe COVID-19: A multifaceted viral vasculopathy syndrome. Ann. Diagn. Pathol. 2021, 50, 151645. [Google Scholar] [CrossRef]
- Ma, J.; Liu, J.; Gao, D.; Li, X.; Zhang, Q.; Lv, L.; Wang, Y.; Li, J.; Zhu, Y.; Wu, Z.; et al. Establishment of Human Pluripotent Stem Cell-Derived Skin Organoids Enabled Pathophysiological Model of SARS-CoV-2 Infection. Adv. Sci. 2022, 9, e2104192. [Google Scholar] [CrossRef] [PubMed]
- Colmenero, I.; Santonja, C.; Alonso-Riano, M.; Noguera-Morel, L.; Hernandez-Martin, A.; Andina, D.; Wiesner, T.; Rodriguez-Peralto, J.L.; Requena, L.; Torrelo, A. SARS-CoV-2 endothelial infection causes COVID-19 chilblains: Histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br. J. Dermatol. 2020, 183, 729–737. [Google Scholar] [CrossRef]
- Cazzato, G.; Cascardi, E.; Colagrande, A.; Foti, C.; Stellacci, A.; Marrone, M.; Ingravallo, G.; Arezzo, F.; Loizzi, V.; Solimando, A.G.; et al. SARS-CoV-2 and Skin: New Insights and Perspectives. Biomolecules 2022, 12, 1212. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, R.; Zhang, H.; Rong, L.; Zhou, W.; Liang, Y.; Li, Q. Skin is a potential host of SARS-CoV-2: A clinical, single-cell transcriptome-profiling and histologic study. J. Am. Acad. Dermatol. 2020, 83, 1755–1757. [Google Scholar] [CrossRef]
- Jamiolkowski, D.; Muhleisen, B.; Muller, S.; Navarini, A.A.; Tzankov, A.; Roider, E. SARS-CoV-2 PCR testing of skin for COVID-19 diagnostics: A case report. Lancet 2020, 396, 598–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Liu, L.; Hu, X.; Wang, X.; Hu, H.; Hu, Z.; Zhou, Y.; Wang, M. Infection of human sweat glands by SARS-CoV-2. Cell Discov. 2020, 6, 84. [Google Scholar] [CrossRef]
- Chudakova, D.; Klopot, A.; Shi, B.; Bhalla, P.; Tsoi, L.C.; Perez White, B.E.; Budunova, I. Potential role of skin in SARS-CoV-2 infection. J. Investig. Dermatol. 2022, 142, S147. [Google Scholar] [CrossRef]
- Zupin, L.; Moltrasio, C.; Tricarico, P.M.; Del Vecchio, C.; Fontana, F.; Marzano, A.V.; Crovella, S. Paraviral cutaneous manifestations associated to SARS-CoV-2 Omicron variant. Infect. Dis. 2023, 55, 181–188. [Google Scholar] [CrossRef]
- Baughn, L.B.; Sharma, N.; Elhaik, E.; Sekulic, A.; Bryce, A.H.; Fonseca, R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin. Proc. 2020, 95, 1989–1999. [Google Scholar] [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhang, L.; Chen, L.; Zhao, X.; Wang, X.; Hu, M.; Le, Y.; Xue, F.; Li, X.; Zheng, J. SARS-CoV-2 might transmit through the skin while the skin barrier function could be the mediator. Med. Hypotheses 2022, 159, 110752. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Mi, Z.; Wang, Z.; Pang, Z.; Liu, H.; Zhang, F. High Expression of ACE2 on Keratinocytes Reveals Skin as a Potential Target for SARS-CoV-2. J. Investig. Dermatol. 2021, 141, 206–209.e201. [Google Scholar] [CrossRef] [PubMed]
- Al-Benna, S. Association of high level gene expression of ACE2 in adipose tissue with mortality of COVID-19 infection in obese patients. Obes. Med. 2020, 19, 100283. [Google Scholar] [CrossRef]
- Ganier, C.; Harun, N.; Peplow, I.; Du-Harpur, X.; Arthurs, C.; Watt, F.M.; Lynch, M.D. Angiotensin-Converting Enzyme 2 Expression Is Detectable in Keratinocytes, Cutaneous Appendages, and Blood Vessels by Multiplex RNA In Situ Hybridization. Adv. Skin Wound Care 2022, 35, 219–223. [Google Scholar] [CrossRef]
- Lin, E.C.; Hong, C.H. IL-33 Enhances ACE2 Expression on Epidermal Keratinocytes in Atopic Dermatitis: A Plausible Issue for SARS-CoV-2 Transmission in Inflamed Atopic Skin. Biomedicines 2022, 10, 1183. [Google Scholar] [CrossRef]
- Tembhre, M.K.; Parihar, A.S.; Sharma, V.K.; Imran, S.; Bhari, N.; Lakshmy, R.; Bhalla, A. Enhanced expression of angiotensin-converting enzyme 2 in psoriatic skin and its upregulation in keratinocytes by interferon-gamma: Implication of inflammatory milieu in skin tropism of SARS-CoV-2. Br. J. Dermatol. 2021, 184, 577–579. [Google Scholar] [CrossRef]
- Bylaite, M.; Moussali, H.; Marciukaitiene, I.; Ruzicka, T.; Walz, M. Expression of cathepsin L and its inhibitor hurpin in inflammatory and neoplastic skin diseases. Exp. Dermatol. 2006, 15, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Z.A.; El Ashmawy, A.A.; Abd El-Naby, N.M.; Ghoraba, H.M. Immunohistochemical expression of cathepsin L in atopic dermatitis and lichen planus. Indian J. Dermatol. 2015, 60, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Blauer, M.; Vaalasti, A.; Pauli, S.L.; Ylikomi, T.; Joensuu, T.; Tuohimaa, P. Location of androgen receptor in human skin. J. Investig. Dermatol. 1991, 97, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.; Zagorska, A.; Jurkin, J.; Yasmin, N.; Koffel, R.; Richter, S.; Gesslbauer, B.; Lemke, G.; Strobl, H. Identification of Axl as a downstream effector of TGF-beta1 during Langerhans cell differentiation and epidermal homeostasis. J. Exp. Med. 2012, 209, 2033–2047. [Google Scholar] [CrossRef]
- Green, J.; Ikram, M.; Vyas, J.; Patel, N.; Proby, C.M.; Ghali, L.; Leigh, I.M.; O’Toole, E.A.; Storey, A. Overexpression of the Axl tyrosine kinase receptor in cutaneous SCC-derived cell lines and tumours. Br. J. Cancer 2006, 94, 1446–1451. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, S.; Lei, L.; Zhang, X.; Jia, X.; Luo, Z.; Huang, X.; Kuang, Y.; Zeng, W.; Su, J.; et al. Epidermal CD147 expression plays a key role in IL-22-induced psoriatic dermatitis. Sci. Rep. 2017, 7, 44172. [Google Scholar] [CrossRef]
- Wang, W.; Xiong, H.; Hu, Z.; Zhao, R.; Hu, Y.; Chen, W.; Han, Y.; Yang, L.; Hu, X.; Wang, C.; et al. Experimental study on TGF-beta1-mediated CD147 expression in oral submucous fibrosis. Oral Dis. 2018, 24, 993–1000. [Google Scholar] [CrossRef]
- He, H.; Suryawanshi, H.; Morozov, P.; Gay-Mimbrera, J.; Del Duca, E.; Kim, H.J.; Kameyama, N.; Estrada, Y.; Der, E.; Krueger, J.G.; et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J. Allergy Clin. Immunol. 2020, 145, 1615–1628. [Google Scholar] [CrossRef]
- Finnegan, A.; Cho, R.J.; Luu, A.; Harirchian, P.; Lee, J.; Cheng, J.B.; Song, J.S. Single-Cell Transcriptomics Reveals Spatial and Temporal Turnover of Keratinocyte Differentiation Regulators. Front. Genet. 2019, 10, 775. [Google Scholar] [CrossRef]
- Pearton, D.J.; Nirunsuksiri, W.; Rehemtulla, A.; Lewis, S.P.; Presland, R.B.; Dale, B.A. Proprotein convertase expression and localization in epidermis: Evidence for multiple roles and substrates. Exp. Dermatol. 2001, 10, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Sole-Boldo, L.; Raddatz, G.; Schutz, S.; Mallm, J.P.; Rippe, K.; Lonsdorf, A.S.; Rodriguez-Paredes, M.; Lyko, F. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol. 2020, 3, 188. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barthe, M.; Hertereau, L.; Lamghari, N.; Osman-Ponchet, H.; Braud, V.M. Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry? Int. J. Mol. Sci. 2023, 24, 6253. https://doi.org/10.3390/ijms24076253
Barthe M, Hertereau L, Lamghari N, Osman-Ponchet H, Braud VM. Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry? International Journal of Molecular Sciences. 2023; 24(7):6253. https://doi.org/10.3390/ijms24076253
Chicago/Turabian StyleBarthe, Manon, Leslie Hertereau, Noura Lamghari, Hanan Osman-Ponchet, and Véronique M. Braud. 2023. "Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry?" International Journal of Molecular Sciences 24, no. 7: 6253. https://doi.org/10.3390/ijms24076253
APA StyleBarthe, M., Hertereau, L., Lamghari, N., Osman-Ponchet, H., & Braud, V. M. (2023). Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry? International Journal of Molecular Sciences, 24(7), 6253. https://doi.org/10.3390/ijms24076253







