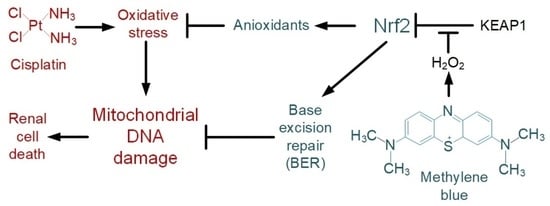
Methylene Blue Induces Antioxidant Defense and Reparation of Mitochondrial DNA in a Nrf2-Dependent Manner during Cisplatin-Induced Renal Toxicity
Abstract
1. Introduction
2. Results
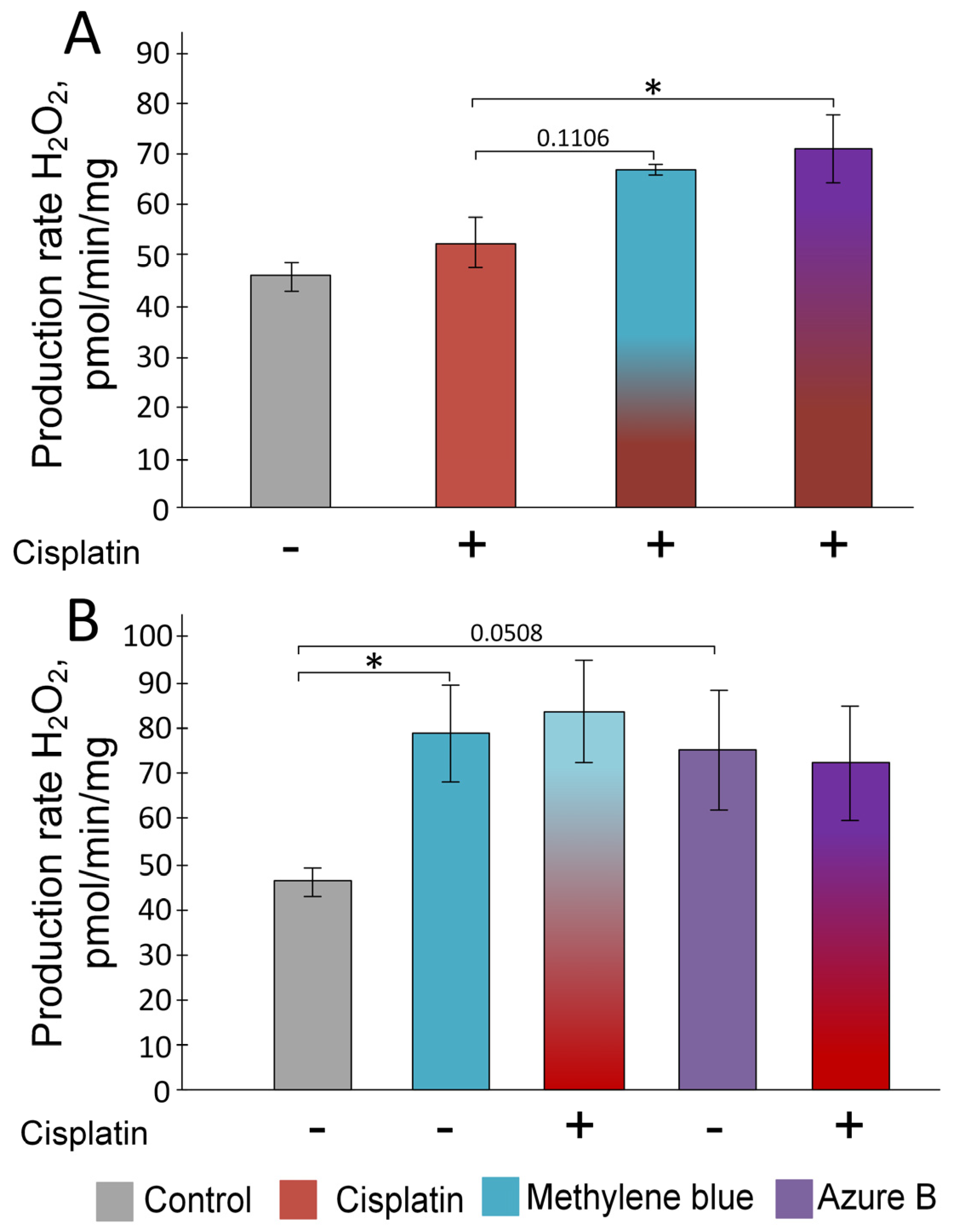
2.1. Effect of Cisplatin and Thiazine Dyes on the Rate of H2O2 Production in Kidney Mitochondria
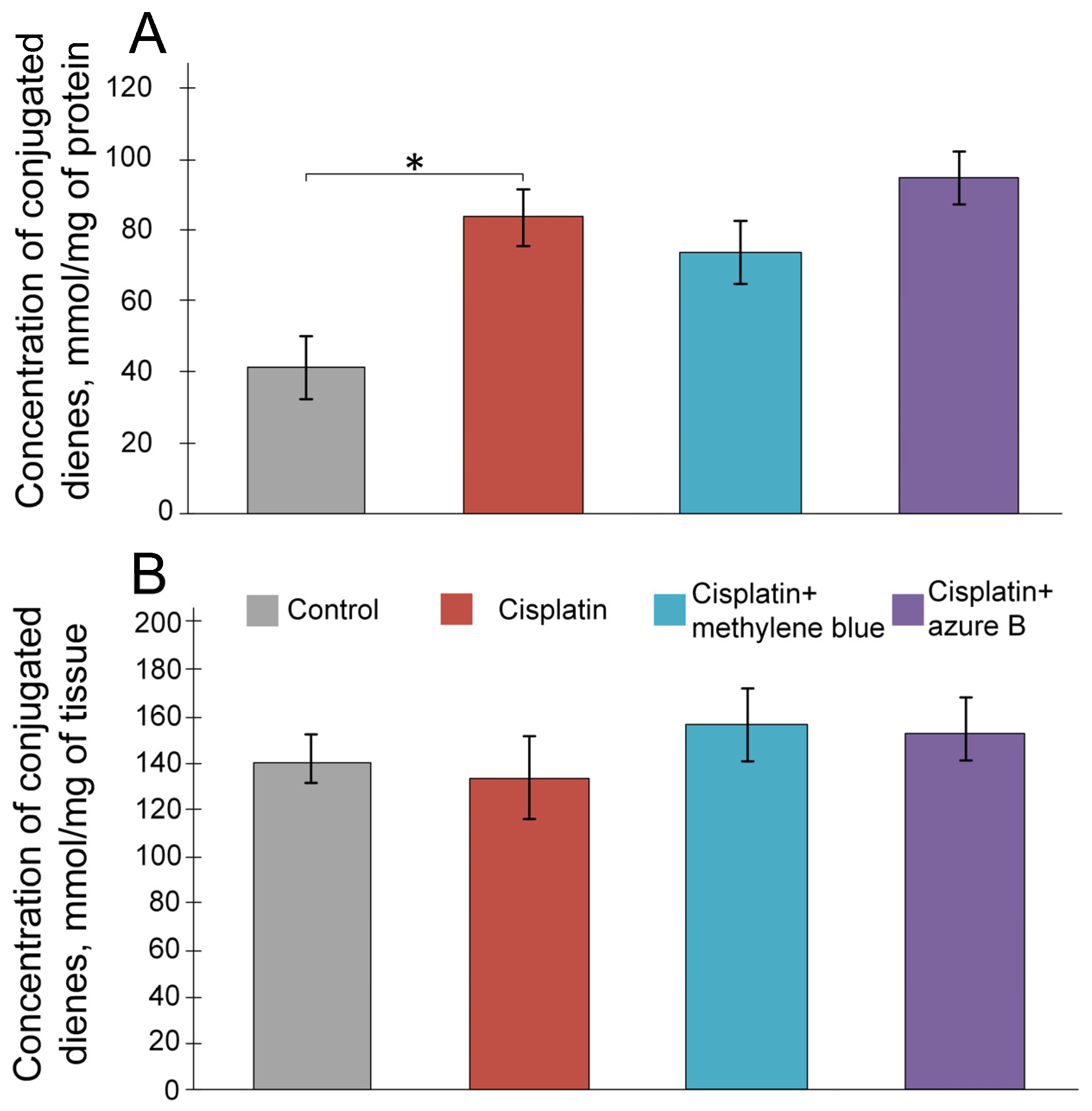
2.2. The Effect of Cisplatin and Thiazine Dyes on the Levels of Lipid Peroxidation Products in the Kidneys
2.3. Effect of In Vitro Cisplatin and Thiazine Dye Addition to Mitochondria on mtDNA Damage Levels
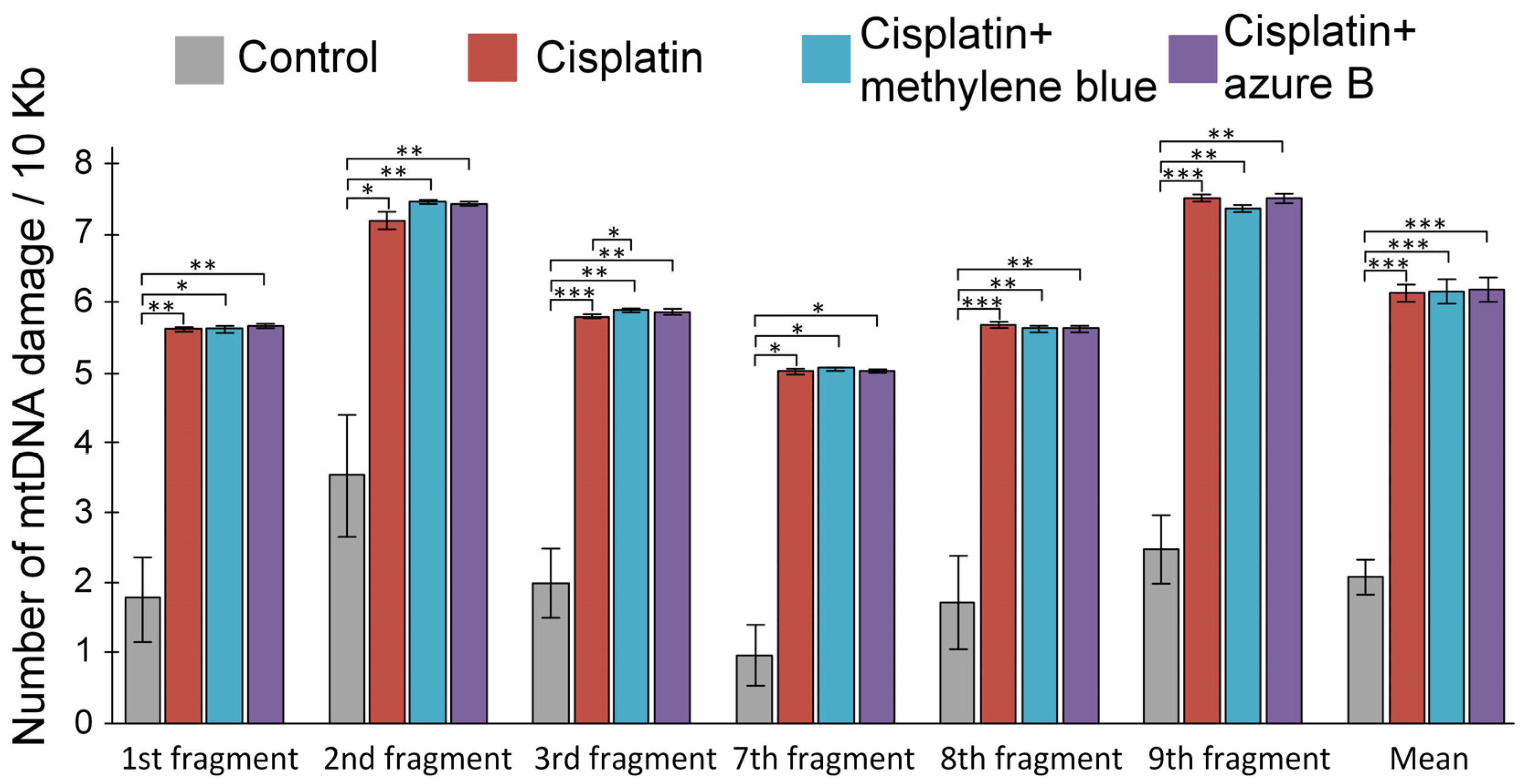
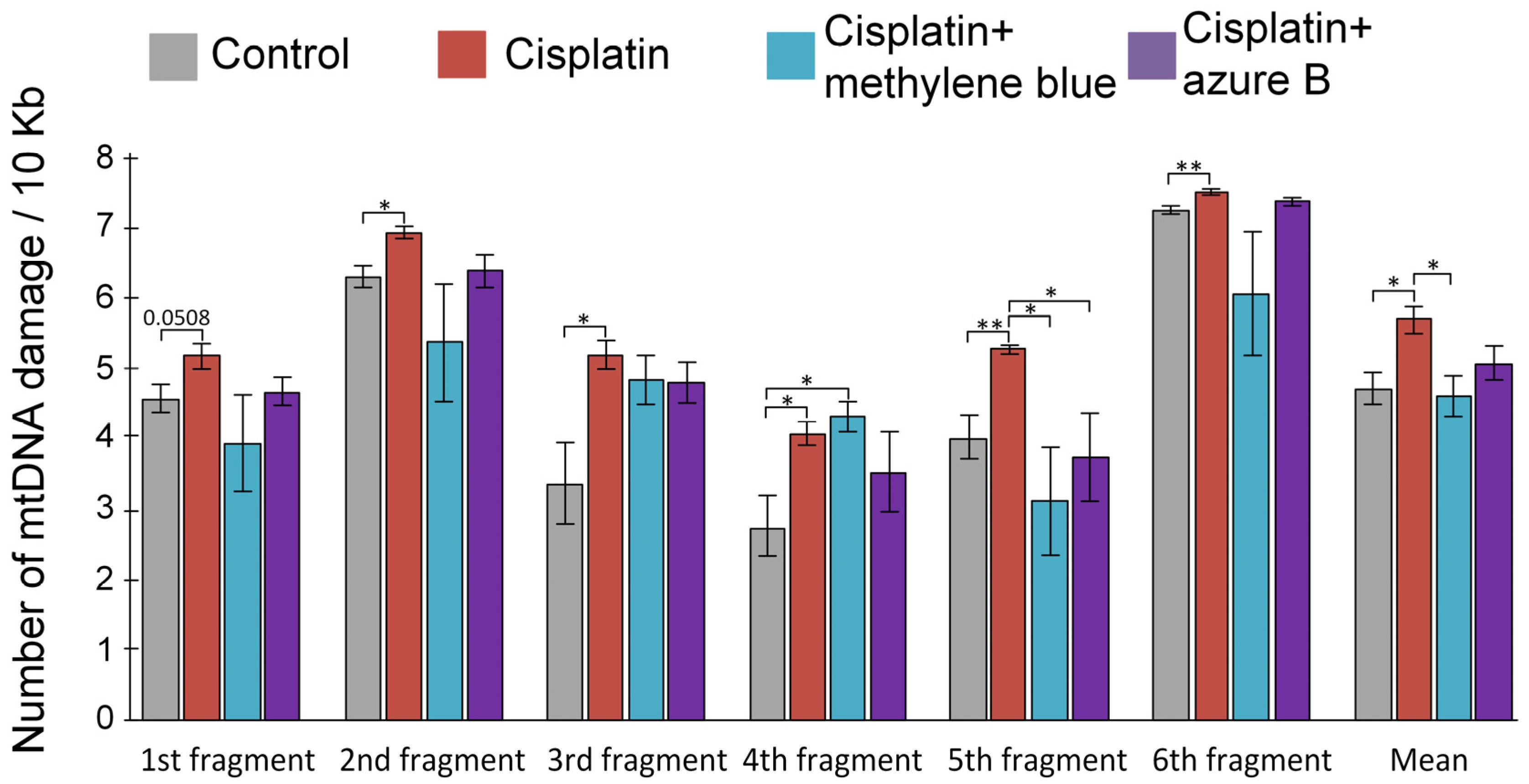
2.4. Effect of Cisplatin Injections and In Vivo Administration of Thiazine Dyes on Levels of mtDNA Damage
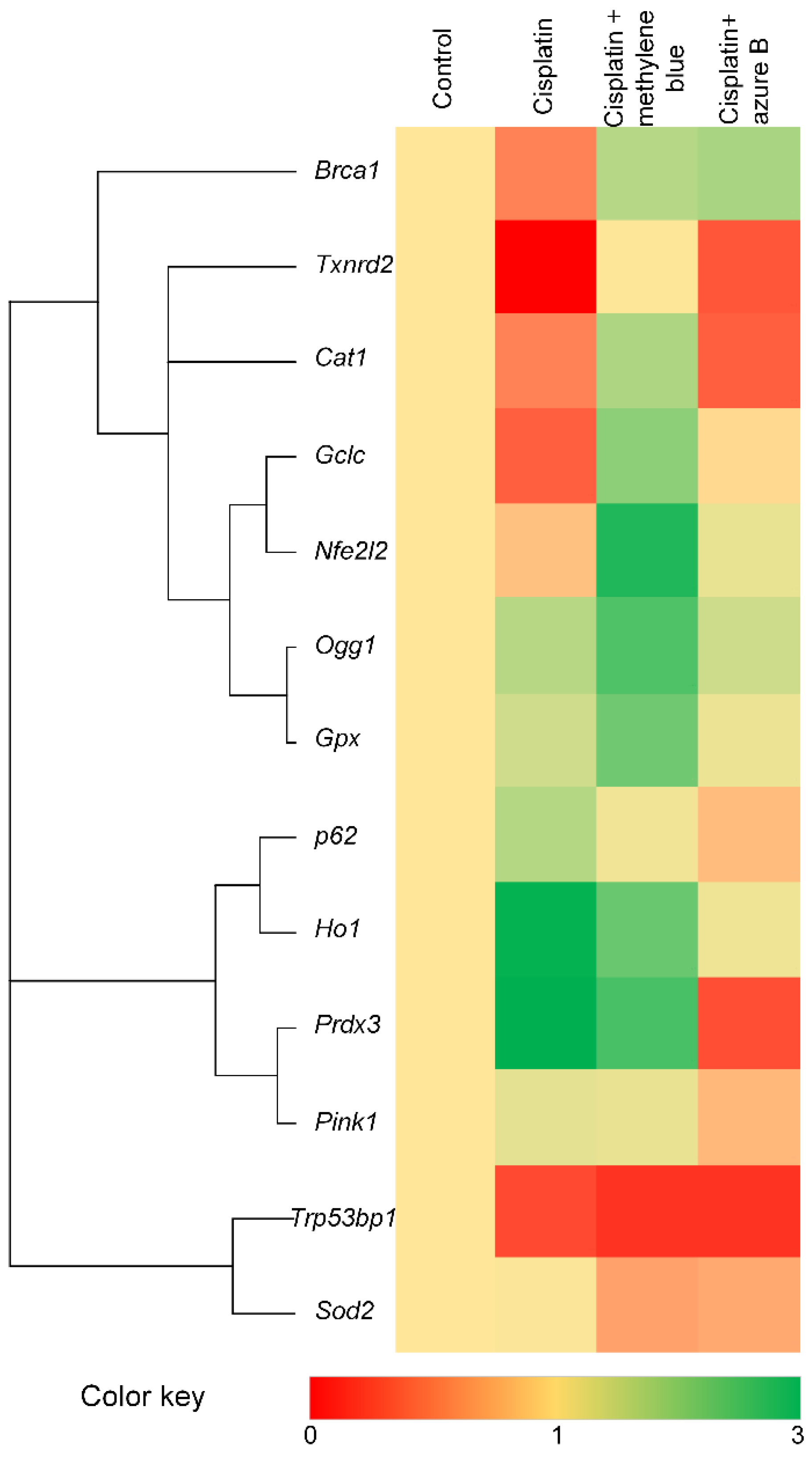
2.5. Effect of Cisplatin and Thiazine Dyes on Gene Expression Levels
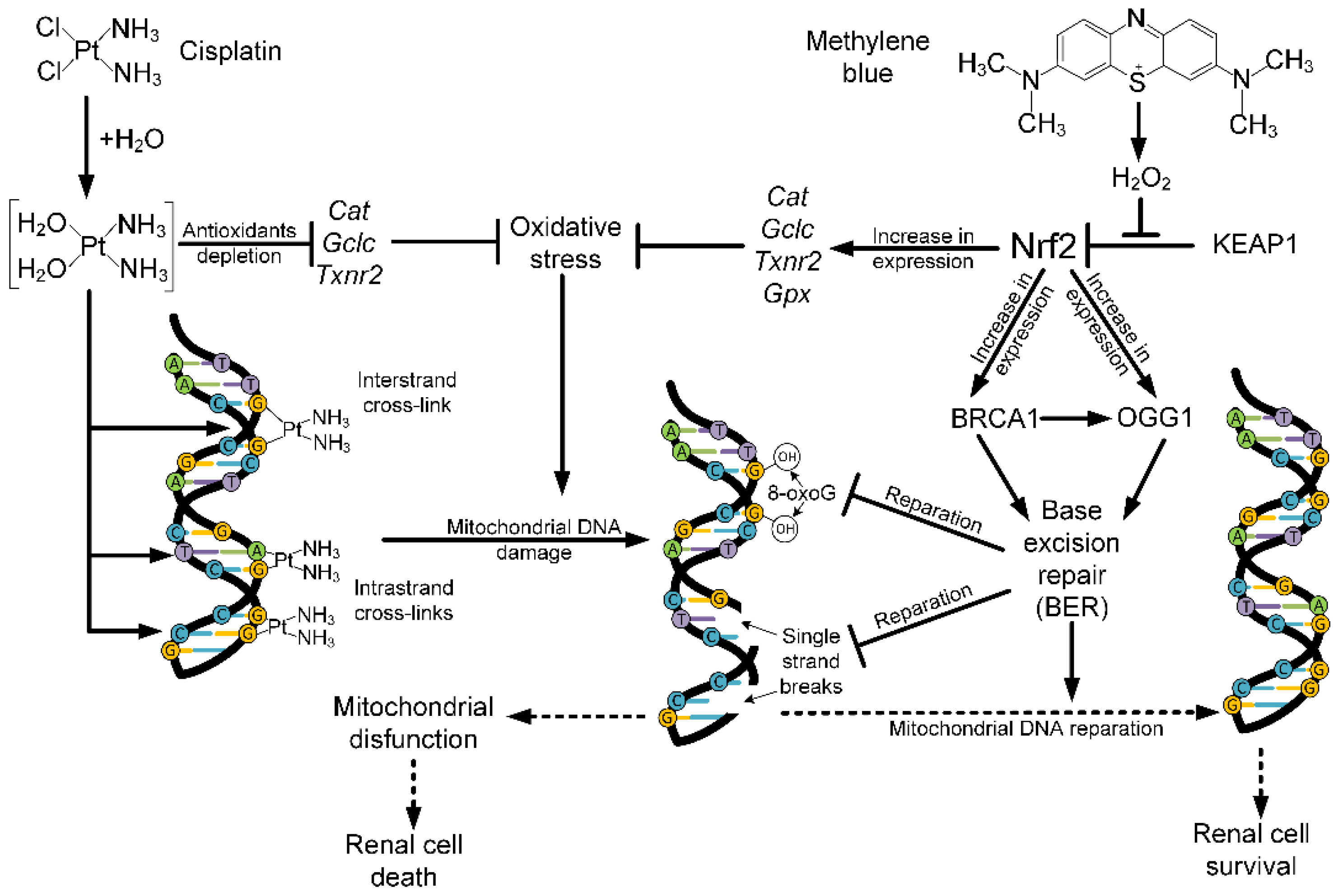
3. Discussion
4. Materials and Methods
4.1. Laboratory Animals
4.2. Designs of Experiment
4.3. Isolation of Mitochondria from the Kidneys
4.4. Assessment of the Rate of H2O2 Production in Mitochondria
4.5. Measurement of Lipid Peroxidation Products
4.6. Measuring the mtDNA Damage Level
4.7. Estimation of Gene Expression Level
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trudu, F.; Amato, F.; Vaňhara, P.; Pivetta, T.; Peña-Méndez, E.M.; Havel, J. Coordination compounds in cancer: Past, present and perspectives. J. Appl. Biomed. 2015, 13, 79–103. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Ekinci Akdemir, F.; Albayrak, M.; Çalik, M.; Bayir, Y.; Gülçin, İ. The protective effects of p-coumaric acid on acute liver and kidney damages induced by cisplatin. Biomedicines 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, B.P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef]
- Ali, R.; Aouida, M.; Sulaiman, A.A.; Madhusudan, S.; Ramotar, D. Can Cisplatin Therapy Be Improved? Pathways That Can Be Targeted. Int. J. Mol. Sci. 2022, 23, 7241. [Google Scholar] [CrossRef]
- Rancoule, C.; Guy, J.B.; Vallard, A.; Ben Mrad, M.; Rehailia, A.; Magné, N. Les 50 ans du cisplatine. Bull. Cancer 2017, 104, 167–176. [Google Scholar] [CrossRef]
- De Biasi, A.R.; Villena-Vargas, J.; Adusumilli, P.S. Cisplatin-Induced Antitumor Immunomodulation: A Review of Preclinical and Clinical Evidence. Clin. Cancer Res. 2014, 20, 5384–5391. [Google Scholar] [CrossRef]
- Chen, X.; Wei, W.; Li, Y.; Huang, J.; Ci, X. Hesperetin relieves cisplatin-induced acute kidney injury by mitigating oxidative stress, inflammation and apoptosis. Chem. Biol. Interact. 2019, 308, 269–278. [Google Scholar] [CrossRef]
- Safirstein, R.; Winston, J.; Goldstein, M.; Moel, D.; Dikman, S.; Guttenplan, J. Cisplatin Nephrotoxicity. Am. J. Kidney Dis. 1986, 8, 356–367. [Google Scholar] [CrossRef]
- Safirstein, R.; Miller, P.; Guttenplan, J.B. Uptake and metabolism of cisplatin by rat kidney. Kidney Int. 1984, 25, 753–758. [Google Scholar] [CrossRef]
- Sweeney, K. Cisplatin induced Acute Kidney Injury. FEBS Lett. 2021, 2, 16–25. [Google Scholar]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef]
- Ozkok, A.; Edelstein, C.L. Pathophysiology of Cisplatin-Induced Acute Kidney Injury. Biomed. Res. Int. 2014, 2014, 967826. [Google Scholar] [CrossRef] [PubMed]
- Podratz, J.L.; Knight, A.M.; Ta, L.E.; Staff, N.P.; Gass, J.M.; Genelin, K.; Schlattau, A.; Lathroum, L.; Windebank, A.J. Cisplatin induced Mitochondrial DNA damage in dorsal root ganglion neurons. Neurobiol. Dis. 2011, 41, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Tu, P.; Xu, P.; Sun, Y.; Yu, F.; Tu, N.; Guo, L.; Yang, Y. The Mitochondrial Response to DNA Damage. Front. Cell Dev. Biol. 2021, 9, 669379. [Google Scholar] [CrossRef]
- Shokolenko, T.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef]
- Govers, L.P.; Toka, H.R.; Hariri, A.; Walsh, S.B.; Bockenhauer, D. Mitochondrial DNA mutations in renal disease: An overview. Pediatr. Nephrol. 2021, 36, 9–17. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Xing, Z.; Liu, D.; Sun, J.; Li, X.; Zhang, Y. Status of bi- and multi-nuclear platinum anticancer drug development. Anticancer. Agents Med. Chem. 2010, 10, 272–282. [Google Scholar] [CrossRef]
- Usefzay, O.; Yari, S.; Amiri, P.; Hasanein, P. Evaluation of protective effects of methylene blue on cisplatin-induced nephrotoxicity. Biomed. Pharmacother. 2022, 150, 113023. [Google Scholar] [CrossRef]
- Gholami Jourabi, F.; Yari, S.; Amiri, P.; Heidarianpour, A.; Hashemi, H. The ameliorative effects of methylene blue on testicular damage induced by cisplatin in rats. Andrologia 2020, 53, e13850. [Google Scholar] [CrossRef] [PubMed]
- Krutskikh, E.P.; Potanina, D.V.; Samoylova, N.A.; Gryaznova, M.V.; Sadovnikova, I.S.; Gureev, A.P.; Popov, V.N. Brain Protection by Methylene Blue and Its Derivative, Azur B, via Activation of the Nrf2/ARE Pathway in Cisplatin-Induced Cognitive Impairment. Pharmaceuticals 2022, 15, 815. [Google Scholar] [CrossRef]
- Turan, M.; Cayir, A.; Cetin, N.; Suleyman, H.; Turan, I.S.; Tan, H. An investigation of the effect of thiamine pyrophosphate on cisplatin-induced oxidative stress and DNA damage in rat brain tissue compared with thiamine: Thiamine and thiamine pyrophosphate effects on cisplatin neurotoxicity. Hum. Exp. Toxicol. 2013, 33, 14–21. [Google Scholar] [CrossRef]
- Starkov, A.A. The Role of Mitochondria in Reactive Oxygen Species Metabolism and Signaling. Ann. N. Y. Acad. Sci. 2008, 1147, 37–52. [Google Scholar] [CrossRef]
- Yilmaz, H.R.; Iraz, M.; Sogut, S.; Ozyurt, H.; Yildirim, Z.; Akyol, O.; Gergerlioglu, S. The effects of erdosteine on the activities of some metabolic enzymes during cisplatin-induced nephrotoxicity in rats. Pharmacol. Res. 2004, 50, 287–290. [Google Scholar] [CrossRef]
- Ognjanović, B.I.; Djordjević, N.Z.; Matić, M.M.; Obradović, J.M.; Mladenović, J.M.; Štajn, A.Š.; Saičić, Z.S. Lipid Peroxidative Damage on Cisplatin Exposure and Alterations in Antioxidant Defense System in Rat Kidneys: A Possible Protective Effect of Selenium. Int. J. Mol. Sci. 2012, 13, 1790–1803. [Google Scholar] [CrossRef] [PubMed]
- Eftekhari, A.; Hasanzadeh, A.; Khalilov, R.; Hosainzadegan, H.; Ahmadian, E.; Eghbal, M.A. Hepatoprotective role of berberine against paraquat-induced liver toxicity in rat. Environ. Sci. Pollut. Res. Int. 2019, 27, 4969–4975. [Google Scholar] [CrossRef] [PubMed]
- Fard, J.; Hamzeiy, H.; Sattari, M.; Eftekhari, A.; Ahmadian, E.; Eghbal, M. Triazole rizatriptan induces liver toxicity through lysosomal/mitochondrial dysfunction. Drug Res. 2016, 66, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zhu, D.; Chen, M.; Wang, T.; Gao, Y.; Liu, W. Mubritinib enhanced the inhibiting function of cisplatin in lung cancer by interfering with mitochondrial function. Thorac. Cancer 2022, 13, 1513–1524. [Google Scholar] [CrossRef]
- Glorieux, C.; Zamocky, M.; Sandoval, J.M.; Verrax, J.; Calderon, P.B. Regulation of catalase expression in healthy and cancerous cells. Free Radic. Biol. Med. 2015, 87, 84–97. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Kiermayer, C.; Northrup, E.; Schrewe, A.; Walch, A.; Hrabe de Angelis, M.; Schoensiege, F.; Zischka, H.; Prehn, C.; Adamski, J.; Bekeredjian, R.; et al. Heart-Specific Knockout of the Mitochondrial Thioredoxin Reductase (Txnrd2) Induces Metabolic and Contractile Dysfunction in the Aging Myocardium. J. Am. Heart Assoc. 2015, 4, e002153. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Roy, S.S.; Sk, U.H.; Bhattacharya, S. Amelioration of cisplatin-induced nephrotoxicity in mice by oral administration of diphenylmethyl selenocyanate. Free Radic. Res. 2010, 45, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Hanigan, M.H.; Devarajan, P. Cisplatin nephrotoxicity: Molecular mechanisms. Cancer Ther. 2003, 1, 47–61. [Google Scholar]
- Kim, Y.K.; Jung, J.S.; Lee, S.H.; Kim, Y.W. Effects of Antioxidants and Ca2+ in Cisplatin-Induced Cell Injury in Rabbit Renal Cortical Slices. Toxicol. Appl. Pharmacol. 1997, 146, 261–269. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Horváth, B.; Zsengellér, Z.; Zielonka, J.; Tanchian, G.; Holovac, E.; Kechrid, M.; Patel, V.; Stillman, I.E.; Parikh, S.M.; et al. Mitochondrial-targeted antioxidants represent a promising approach for prevention of cisplatin-induced nephropathy. Free Radic. Biol. Med. 2012, 52, 497–506. [Google Scholar] [CrossRef]
- Situnayake, R.D.; Crump, B.J.; Zezulka, A.V.; Davis, M.; McConkey, B.; Thurnham, D.I. Measurement of conjugated diene lipids by derivative spectroscopy in heptane extracts of plasma. Ann. Clin. Biochem. 1990, 27, 258–266. [Google Scholar] [CrossRef]
- Sergent, O.; Morel, I.; Cogrel, P.; Chevanne, M.; Pasdeloup, N.; Brissot, P.; Lescoat, G.; Cillard, P.; Cillard, J. Simultaneous measurements of conjugated dienes and free malondialdehyde, used as a micromethod for the evaluation of lipid peroxidation in rat hepatocyte cultures. Chem. Phys. Lipids 1993, 65, 133–139. [Google Scholar] [CrossRef]
- Fuertes, M.; Castilla, J.; Alonso, C.; Pérez, J. Cisplatin Biochemical Mechanism of Action: From Cytotoxicity to Induction of Cell Death Through Interconnections Between Apoptotic and Necrotic Pathways. Curr. Med. Chem. 2003, 10, 257–266. [Google Scholar] [CrossRef]
- McDonald, E.C.; Randon, K.R.; Knight, A.; Windebank, A.J. Cisplatin preferentially binds to DNA in dorsal root ganglion neurons in vitro and in vivo: A potential mechanism for neurotoxicity. Neurobiol. Dis. 2005, 18, 305–313. [Google Scholar] [CrossRef]
- Volarevic, V.; Djokovic, B.; Jankovic, M.G.; Harrell, C.R.; Fellabaum, C.; Djonov, V.; Arsenijevic, N. Molecular mechanisms of cisplatin-induced nephrotoxicity: A balance on the knife edge between renoprotection and tumor toxicity. J. Biomed. Sci. 2019, 26, 25. [Google Scholar] [CrossRef] [PubMed]
- Hayati, F.; Hossainzadeh, M.; Shayanpour, S.; Abedi-Gheshlaghi, Z.; Beladi Mousavi, S.S. Prevention of cisplatin nephrotoxicity. J. Nephropharmacol. 2015, 5, 57–60. [Google Scholar] [PubMed]
- Ali, B.H.; Al Moundhri, M.S. Agents ameliorating or augmenting the nephrotoxicity of cisplatin and other platinum compounds: A review of some recent research. Food Chem. Toxicol. 2006, 44, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Nematbakhsh, M.; Pezeshki, Z.; Jazi, F.E.; Mazaheri, B.; Moeini, M.; Safari, T.; Azarkish, F.; Moslemi, F.; Maleki, M.; Rezaei, A.; et al. Cisplatin-Induced Nephrotoxicity; Protective Supplements and Gender Differences. Asian Pac. J. Cancer Prev. 2017, 18, 295–314. [Google Scholar] [CrossRef] [PubMed]
- Humanes, B.; Camaño, S.; Lara, J.M.; Sabbisetti, V.; González-Nicolás, M.A.; Bonventre, J.V.; Tejedor, A.; Lazaro, A. Cisplatin-induced renal inflammation is ameliorated by cilastatin nephroprotection. Nephrol. Dial. Transplant. 2017, 32, 1645–1655. [Google Scholar] [CrossRef]
- Zaballos, M.; Power, M.; Canal-Alonso, M.I.; González-Nicolás, M.A.; Vasquez-Jimenez, W.; Lozano-Lominchar, P.; Cabrerizo-Torrente, P.; Palencia-García, N.; Gago-Quiroga, S.; Ginel-Feito, M.D.; et al. Effect of Cilastatin on Cisplatin-Induced Nephrotoxicity in Patients Undergoing Hyperthermic Intraperitoneal Chemotherapy. Int. J. Mol. Sci. 2021, 22, 1239. [Google Scholar] [CrossRef]
- El-Sayed, R.M.; Abo, E.l.; Gheit, R.E.; Badawi, G.A. Vincamine protects against cisplatin induced nephrotoxicity via activation of Nrf2/HO-1 and hindering TLR4/ IFN-γ/CD44 cells inflammatory cascade. Life Sci. 2021, 272, 119224. [Google Scholar] [CrossRef]
- Abd El-Rhman, R.H.; El-Naga, R.N.; Gad, A.M.; Tadros, M.G.; Hassaneen, S.K. Dibenzazepine Attenuates Against Cisplatin-Induced Nephrotoxicity in Rats: Involvement of NOTCH Pathway. Front. Pharmacol. 2020, 11, 567852. [Google Scholar] [CrossRef]
- Akca, G.; Eren, H.; Tumkaya, L.; Mercantepe, T.; Horsanali, M.O.; Deveci, E.; Dila, E.; Yilmaze, A. The protective effect of astaxanthin against cisplatin-induced nephrotoxicity in rats. Biomed. Pharmacother. 2018, 100, 575–582. [Google Scholar] [CrossRef]
- Beladi Mousavi, S.S.; Zadeh, M.H.; Shahbazian, H.; Khanzadeh, A.; Hayati, F.; Ghorbani, A.; Golzari, K.; Valavi, E.; Motemednia, F.; Beladi Mousavi, M. The protective effect of theophyline in cisplatin nephrotoxicity. Saudi J. Kidney Dis. Transpl. 2014, 25, 333–337. [Google Scholar] [CrossRef]
- Gureev, A.P.; Sadovnikova, I.S.; Popov, V.N. Molecular Mechanisms of the Neuroprotective Effect of Methylene Blue. Biochemistry 2022, 87, 940–956. [Google Scholar] [CrossRef]
- Gureev, A.P.; Samoylova, N.A.; Potanina, D.V.; Popov, V.N. Effect of methylene blue and its metabolite—Azure I—On bioenergetic parameters of intact mice brain mitochondria. Biomed. Khim. 2021, 67, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Gureev, A.P.; Syromyatnikov, M.Y.; Gorbacheva, T.M.; Starkov, A.A.; Popov, V.N. Methylene blue improves sensorimotor phenotype and decreases anxiety in parallel with activating brain mitochondria biogenesis in mid-age mice. Neurosci. Res. 2016, 113, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Gureev, A.P.; Shaforostova, E.A.; Popov, V.N.; Starkov, A.A. Methylene blue does not bypass Complex III antimycin block in mouse brain mitochondria. FEBS Lett. 2019, 593, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Tretter, L.; Horvath, G.; Hölgyesi, A.; Essek, F.; Adam-Vizi, V. Enhanced hydrogen peroxide generation accompanies the beneficial bioenergetic effects of methylene blue in isolated brain mitochondria. Free Radic. Biol. Med. 2014, 77, 317–330. [Google Scholar] [CrossRef]
- Sváb, G.; Kokas, M.; Sipos, I.; Ambrus, A.; Tretter, L. Methylene Blue Bridges the Inhibition and Produces Unusual Respiratory Changes in Complex III-Inhibited Mitochondria. Studies on Rats, Mice and Guinea Pigs. Antioxidants 2021, 10, 305. [Google Scholar] [CrossRef]
- Purdom-Dickinson, S.E.; Sheveleva, E.V.; Sun, H.; Chen, Q.M. Translational Control of Nrf2 Protein in Activation of Antioxidant Response by Oxidants. Mol. Pharmacol. 2007, 72, 1074–1081. [Google Scholar] [CrossRef]
- Kobayashi, M.; Li, L.; Iwamoto, N.; Nakajima-Takagi, Y.; Kaneko, H.; Nakayama, Y.; Eguchi, M.; Wada, Y.; Kumagai, Y.; Yamamoto, M. The Antioxidant Defense System Keap1-Nrf2 Comprises a Multiple Sensing Mechanism for Responding to a Wide Range of Chemical Compounds. Mol. Cell. Biol. 2009, 29, 493–502. [Google Scholar] [CrossRef]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [CrossRef]
- Chodari, L.; Aytemir, M.D.; Vahedi, P.; Alipour, M.; Vahed, S.Z.; Hosseiniyan Khatibi, S.M.; Ahmadian, E.; Ardalan, M.; Eftekhari, A. Targeting mitochondrial biogenesis with polyphenol compounds. Oxid. Med. Cell. Longev. 2021, 12, 4946711. [Google Scholar] [CrossRef] [PubMed]
- Stack, C.; Jainuddin, S.; Elipenahli, C.; Gerges, M.; Starkova, N.; Starkov, A.A.; Jove, M.; Portero-Otin, M.; Launay, N.; Pujol, A.; et al. Methylene blue upregulates Nrf2/ARE genes and prevents tau-related neurotoxicity. Hum. Mol. Genet. 2014, 23, 3716–3732. [Google Scholar] [CrossRef]
- Sadovnikova, I.S.; Gureev, A.P.; Ignatyeva, D.A.; Gryaznova, M.V.; Chernyshova, E.V.; Krutskikh, E.P.; Novikova, A.G.; Popov, V.N. Nrf2/ARE Activators Improve Memory in Aged Mice via Maintaining of Mitochondrial Quality Control of Brain and the Modulation of Gut Microbiome. Pharmaceuticals 2021, 14, 607. [Google Scholar] [CrossRef]
- Aleksunes, L.M.; Goedken, M.J.; Rockwell, C.E.; Thomale, J.; Manautou, J.E.; Klaassen, C.D. Transcriptional Regulation of Renal Cytoprotective Genes by Nrf2 and Its Potential Use as a Therapeutic Target to Mitigate Cisplatin-Induced Nephrotoxicity. J. Pharmacol. Exp. Ther. 2010, 335, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, K.R.; Freeman, M.L. Nrf2 promotes survival following exposure to ionizing radiation. Free Radic. Biol. Med. 2015, 88, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjoras, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Rih, J.K.; Roy, R.; Ballal, R.; Rosen, E.M. Transcriptional Regulation of the Base Excision Repair Pathway by BRCA1. J. Biol. Chem. 2010, 285, 19092–19105. [Google Scholar] [CrossRef]
- Singh, B.; Chatterjee, A.; Ronghe, A.M.; Bhat, N.K.; Bhat, H.K. Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer 2013, 13, 253. [Google Scholar] [CrossRef]
- Wang, Q.; Li, J.; Yang, X.; Sun, H.; Gao, S.; Zhu, H.; Wu, J.; Jin, W. Nrf2 is associated with the regulation of basal transcription activity of the BRCA1 gene. Acta Biochim. Biophys. Sin. 2013, 45, 179–187. [Google Scholar] [CrossRef]
- Williams, T.; Yang, L.; Estrada-Bernal, A.; Chatterjee, M.; Robb, R. A KRAS-NRF2-53BP1 Nonhomologous End-Joining Repair Pathway Mediates Oncogenic KRAS Radioresistance. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, S31. [Google Scholar] [CrossRef]
- Petzer, A.; Harvey, B.H.; Wegener, G.; Petzer, J.P. Azure B, a metabolite of methylene blue, is a high-potency, reversible inhibitor of monoamine oxidase. Toxicol. Appl. Pharmacol. 2012, 258, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Delport, A.; Harvey, B.N.; Petzer, A.; Petzer, J.P. Azure B and a synthetic structural analogue of methylene blue, ethylthioninium chloride, present with antidepressant-like properties. Life Sci. 2014, 117, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Haouzi, P.; McCann, M.; Tubbs, N. Azure B as a novel cyanide antidote: Preclinical in-vivo studie. Toxicol. Rep. 2020, 7, 1459–1464. [Google Scholar] [CrossRef] [PubMed]
- Čulo, F.; Sabolović, D.; Somogyi, L.; Marušić, M.; Berbiguier, N.; Galey, L. Anti-tumoral and anti-inflammatory effects of biological stains. Agents Actions 1991, 34, 424–428. [Google Scholar] [CrossRef]
- Basist, P.; Zahiruddin, S.; Khan, M.U.; Gautam, G.; Jan, B.; Khan, M.A.; Parveen, R.; Sayeed Ahmad, S. Metabolite profiling and nephroprotective potential of Glycyrrhiza glabra L. roots against cisplatin-induced nephrotoxicity in vitro and in vivo. Iran. J. Basic Med. Sci. 2022, 25, 1286–1298. [Google Scholar] [CrossRef]
- Starkov, A.A. Measurement of Mitochondrial ROS Production. Methods Mol. Biol. 2010, 648, 245–255. [Google Scholar] [CrossRef]
- Gureev, A.P.; Shaforostova, E.A.; Starkov, A.A.; Popov, V.N. Simplified qPCR method for detecting excessive mtDNA damage induced by exogenous factors. Toxicology 2017, 382, 67–74. [Google Scholar] [CrossRef]
| Fragment Name | Primers Sequence |
|---|---|
| 2 short | Forward: 5′-ACGAGGGTCCAACTGTCTCTTA-3′ Reserve: 5′-AGCTCCATAGGGTCTTCTCGT-3′ |
| 1 long | Forward: 5′-TAAATTTCGTGCCAGCCACC-3′ Reserve: 5′-ATGCTACCTTTGCACGGTCA-3′ |
| 2 long | Forward: 5′-ACGAGGGTCCAACTGTCTCTTA-3′ Reserve: 5′-CCGGCTGCGTATTCTACGTT-3′ |
| 3 long | Forward: 5′-CTAGCAGAAACAAACCGGGC-3′ Reserve: 5′-TTAGGGCTTTGAAGGCTCGC-3′ |
| 7 long | Forward: 5′-TCATTCTTCTACTATCCCCAATCC-3′ Reserve: 5′-TGGTTTGGGAGATTGGTTGATG-3′ |
| 8 long | Forward: 5′-CCCCAATCCCTCCTTCCAAC-3′ Reserve: 5′-GGTGGGGAGTAGCTCCTTCTT-3′ |
| 9 long | Forward: 5′-AAGAAGGAGCTACTCCCCACC-3′ Reserve: 5′-GTTGACACGTTTTACGCCGA-3′ |
| Gene Name | Primers Sequence |
|---|---|
| Gapdh | Forward: 5′-GGCTCCCTAGGCCCCTCCTG-3′; Reserve: 5′-TCCCAACTCGGCCCCCAACA-3′; |
| Nfe2l2 | Forward: 5′-CTCTCTGAACTCCTGGACGG-3′ Reserve: 5′-GGGTCTCCGTAAATGGAAG-3′ |
| Ho1 | Forward: 5′-CACGCATATACCCGCTACCT-3′ Reserve: 5′-CCAGAGTGTTCATTCGAGCA-3′ |
| p62 | Forward: 5′-GCCAGAGGAACAGATGGAGT-3′ Reserve: 5′-TCCGATTCTGGCATCTGTAG-3′ |
| Pink1 | Forward: 5′-GAGCAGACTCCCAGTTCTCG-3′ Reserve: 5′-GTCCCACTCCACAAGGATGT-3′ |
| Prdx3 | Forward: 5′-GTGGTTTGGGCCACATGAAC-3′ Reserve: 5′-TGGCTTGATCGTAGGGGACT-3′ |
| Gpx | Forward: 5′-AGTCCACCGTGTATGCCTTCT-3′ Reserve: 5′-GAGACGCGACATTCTCAATGA-3′ |
| Txnrd2 | Forward: 5′-GATCCGGTGGCCTAGCTTG-3′ Reserve: 5′-TCGGGGAGAAGGTTCCACAT-3′ |
| Gclc | Forward: 5′-GGGGTGACGAGGTGGAGTA-3′ Reserve: 5′-GTTGGGGTTTGTCCTCTCCC-3′ |
| Sod2 | Forward: 5′-CAGACCTGCCTTACGACTATGG-3′ Reserve: 5′-CTCGGTGGCGTTGAGATTGTT-3′ |
| Cat | Forward: 5′-AGCGACCAGATGAAGCAGTG-3′ Reserve: 5′-TCCGCTCTCTGTCAAAGTGTG-3′ |
| Ogg1 | Forward: 5′-GAGACGACAGCCAGGTGTGAG-3′ Reserve: 5′-CCGTTCCACCATGCCAGTA-3′ |
| Trp53bp1 | Forward: 5′-GAAGGAAAGCACAGATGAGGATT-3′ Reserve: 5′-CTAGAGGTTTCTGCACGCTG-3′ |
| Brca1 | Forward: 5′-AGGTGATTGCAGTGTGAGAGA-3′ Reserve: 5′-GTATCCGGATGCCTCCTCTTC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samoylova, N.A.; Gureev, A.P.; Popov, V.N. Methylene Blue Induces Antioxidant Defense and Reparation of Mitochondrial DNA in a Nrf2-Dependent Manner during Cisplatin-Induced Renal Toxicity. Int. J. Mol. Sci. 2023, 24, 6118. https://doi.org/10.3390/ijms24076118
Samoylova NA, Gureev AP, Popov VN. Methylene Blue Induces Antioxidant Defense and Reparation of Mitochondrial DNA in a Nrf2-Dependent Manner during Cisplatin-Induced Renal Toxicity. International Journal of Molecular Sciences. 2023; 24(7):6118. https://doi.org/10.3390/ijms24076118
Chicago/Turabian StyleSamoylova, Natalia A., Artem P. Gureev, and Vasily N. Popov. 2023. "Methylene Blue Induces Antioxidant Defense and Reparation of Mitochondrial DNA in a Nrf2-Dependent Manner during Cisplatin-Induced Renal Toxicity" International Journal of Molecular Sciences 24, no. 7: 6118. https://doi.org/10.3390/ijms24076118
APA StyleSamoylova, N. A., Gureev, A. P., & Popov, V. N. (2023). Methylene Blue Induces Antioxidant Defense and Reparation of Mitochondrial DNA in a Nrf2-Dependent Manner during Cisplatin-Induced Renal Toxicity. International Journal of Molecular Sciences, 24(7), 6118. https://doi.org/10.3390/ijms24076118