Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus)
Abstract
1. Introduction
2. Results
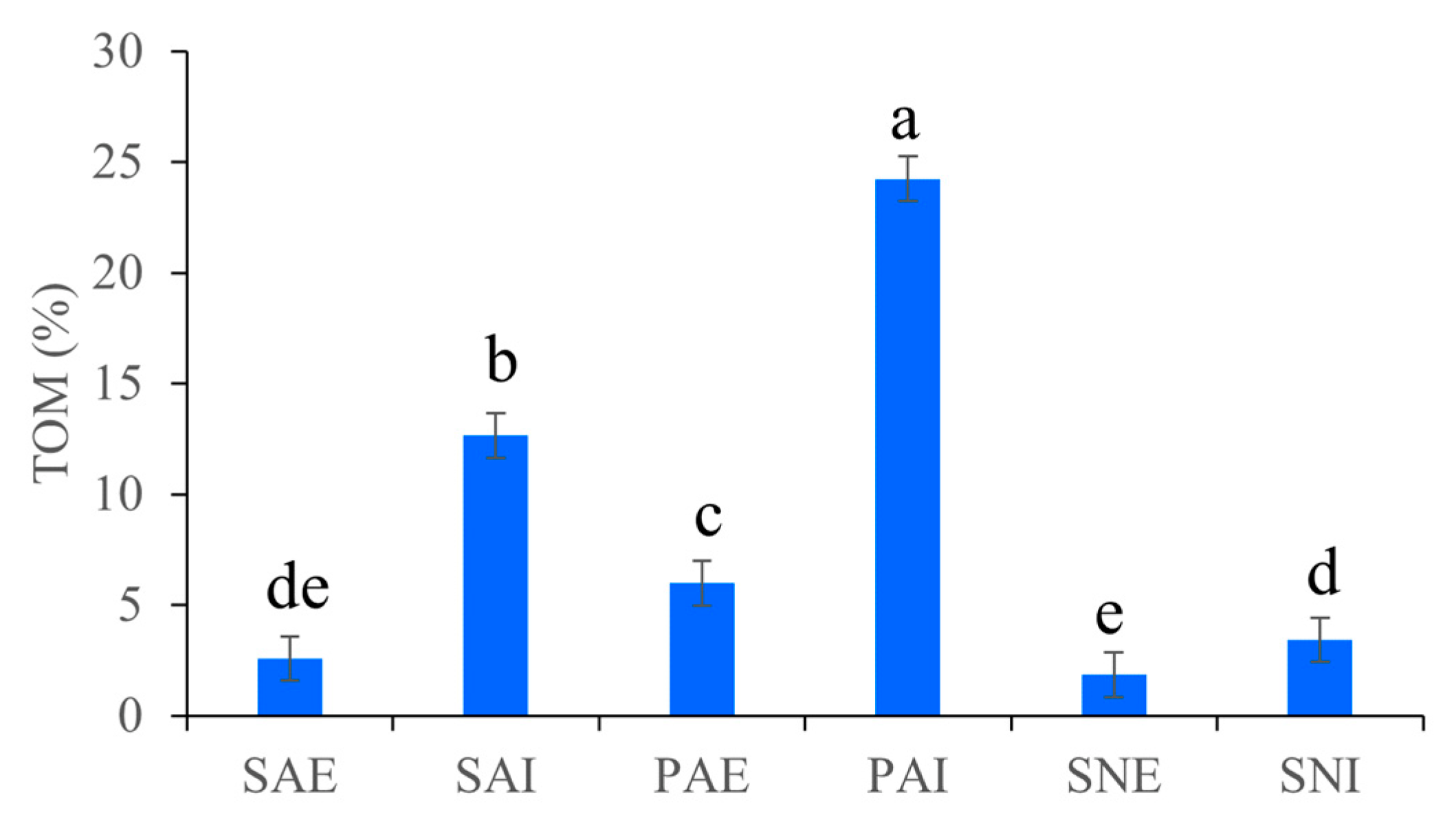
2.1. Grain Size Fraction and TOM of Gut Contents and Surrounding Sediments
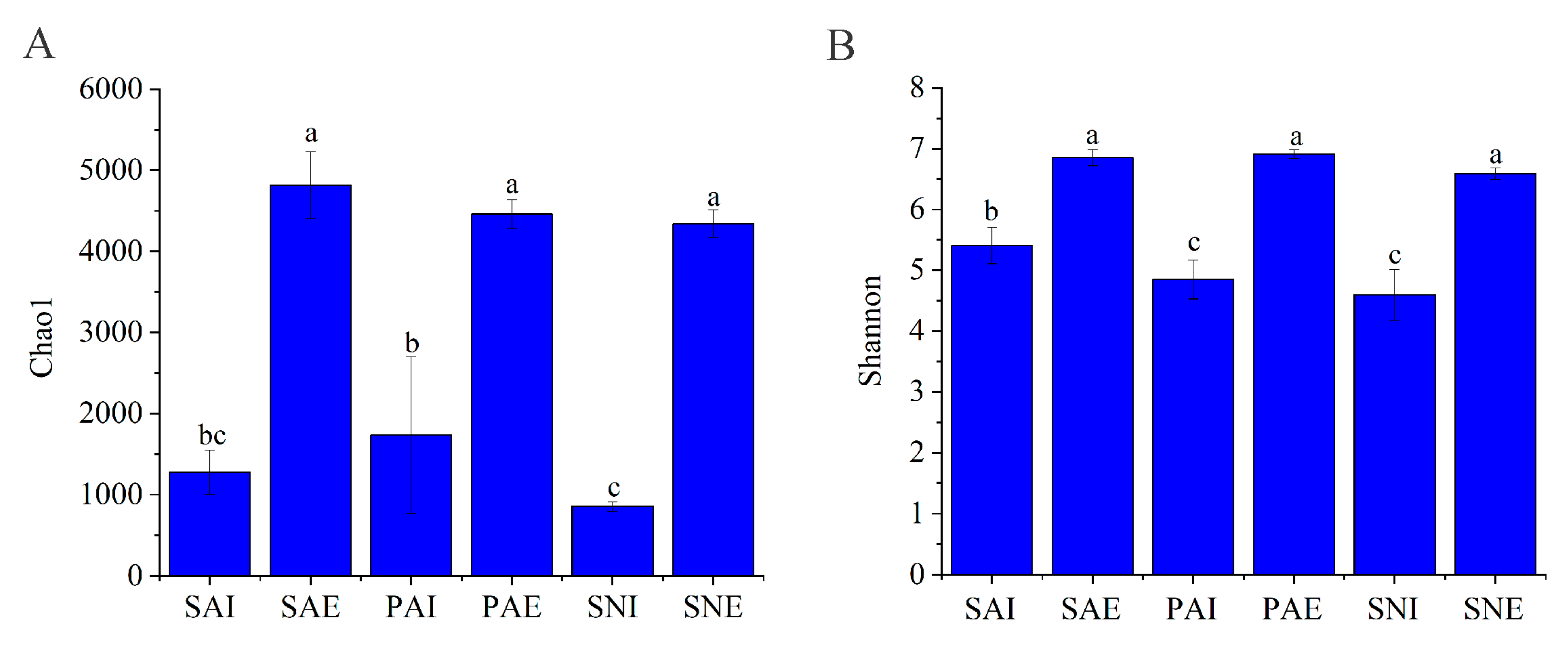
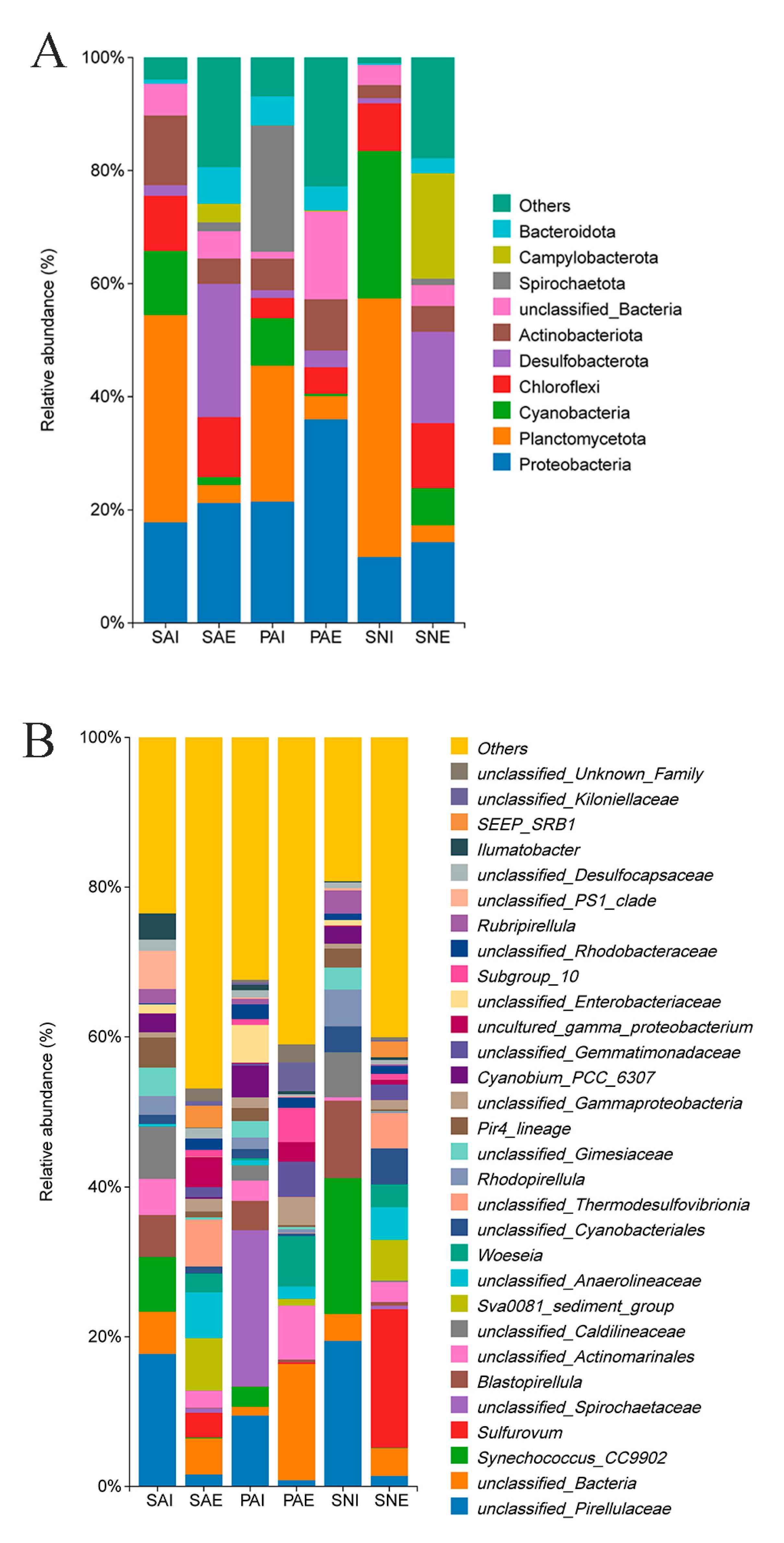
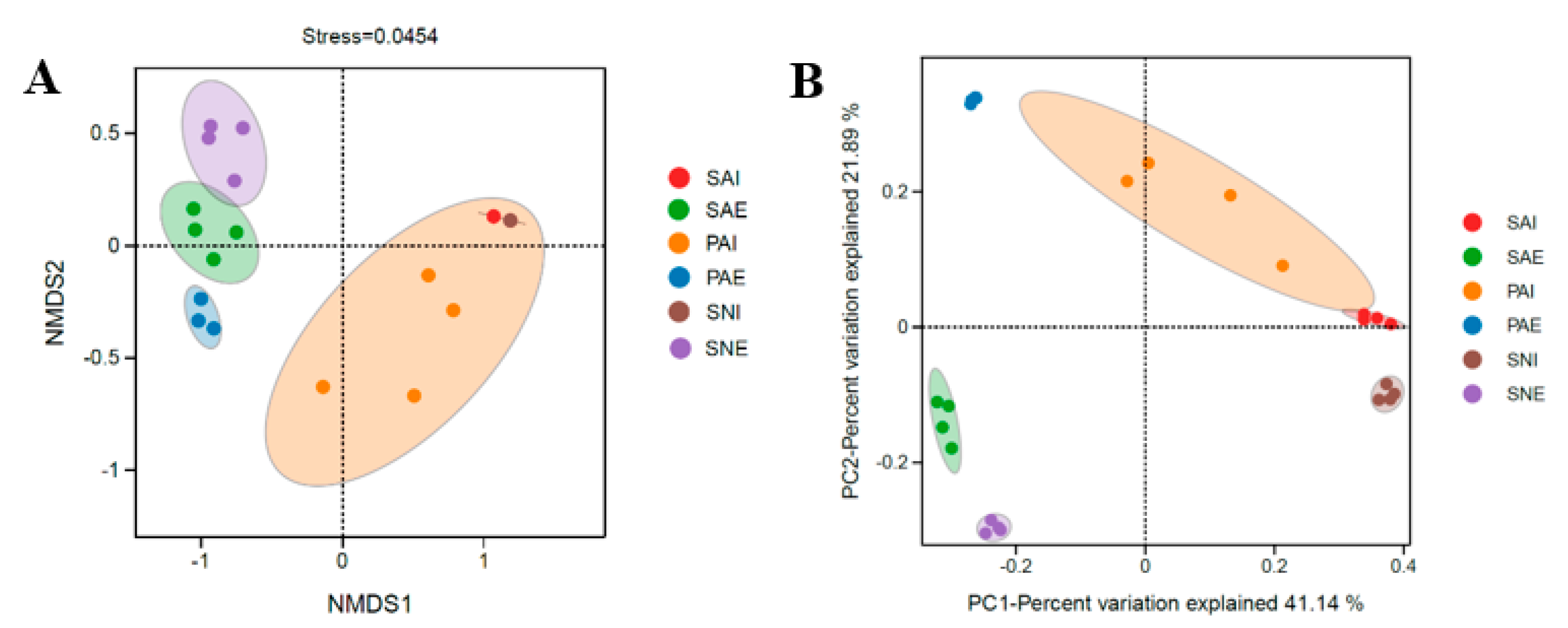
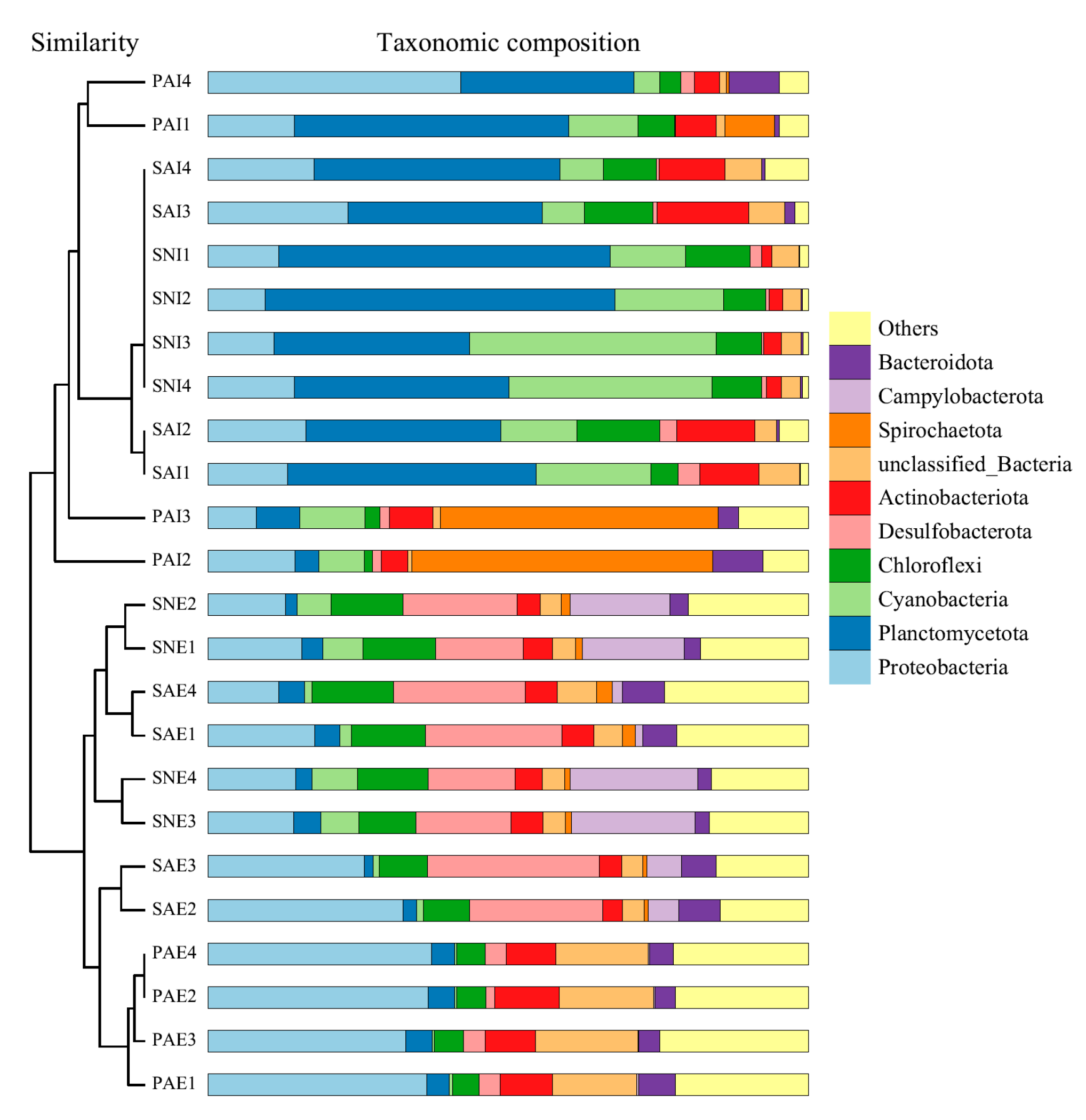
2.2. Bacterial Community of Gut Contents and Surrounding Sediments
3. Discussion
3.1. Habitat Differences of Three Sipunculan Species
3.2. Comparison of Gut Microbe Composition among Three Sipunculan Species
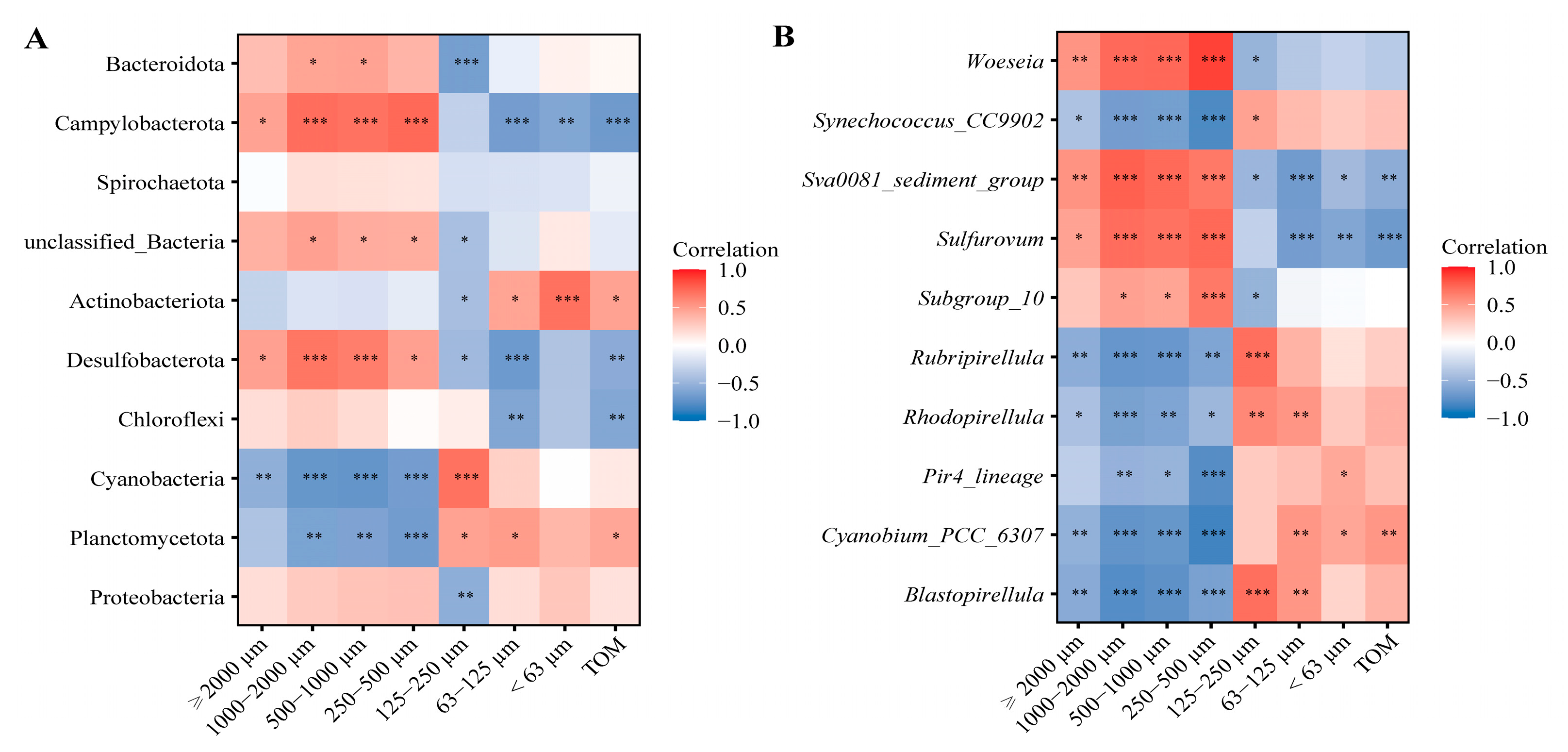
3.3. Selective Feeding of Three Sipunculan Species
4. Materials and Methods
4.1. Sample Collection
4.2. DNA Extraction and PCR Amplification
4.3. Library Preparation and Sequencing
4.4. Bioinformatic and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Li, F.; Zhou, H.; Wang, W. A checklist of Sipuncula from the China coasts. J. Ocean. Univ. Qingdao 1992, 22, 72–87, (In Chinese with English abstract). [Google Scholar]
- Li, J.; Xie, X.; Zhu, C.; Guo, Y.; Chen, S. Edible peanut worm (Sipunculus nudus) in the Beibu Gulf: Resource, aquaculture, ecological impact and counterplan. J. Ocean. Univ. China 2017, 16, 823–830. [Google Scholar] [CrossRef]
- Ying, X.; Dahms, H.; Liu, X.; Wu, H.; Zhang, Y.; Chen, C.; Zhou, Z.; Zeng, G.; Zhou, K.; Yang, W. Development of germ cells and reproductive biology in the sipunculid Phascolosoma esculenta. Aquac. Res. 2009, 40, 305–314. [Google Scholar] [CrossRef]
- Tan, Q.; Ke, C.; Wang, W. Rapid assessments of metal bioavailability in marine sediments using coelomic fluid of sipunculan worms. Environ. Sci. Technol. 2013, 47, 7499–7505. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, T.; Huang, G.; Gong, J. Molecular detection of eukaryotic diets and gut mycobiomes in two marine sediment-dwelling worms, Sipunculus nudus and Urechis unicinctus. Microbes Environ. 2018, 33, 290–300. [Google Scholar] [CrossRef]
- Shields, M.A.; Kedra, M. A deep burrowing sipunculan of ecological and geochemical importance. Deep Sea Res. Part I 2009, 56, 2057–2064. [Google Scholar] [CrossRef]
- Li, J.; Hu, R.; Guo, Y.; Chen, S.; Xie, X.; Qin, J.G.; Ma, Z.; Zhu, C.; Pei, S. Bioturbation of peanut worms Sipunculus nudus on the composition of prokaryotic communities in a tidal flat as revealed by 16S rRNA gene sequences. MicrobiologyOpen 2019, 8, e00802. [Google Scholar] [CrossRef] [PubMed]
- Cutler, E.B. The Sipuncula: Their Systematics, Biology, and Evolution; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Maiorova, A.; Adrianov, A. Distribution of peanut worms (Sipuncula) in the west Pacific. In Proceedings of the China-Russian bilateral Symposium on Comparison on Marine Biodiversity in the Northwest Pacific Ocean, Qingdao, China, 10–11 October 2010; pp. 13–17. [Google Scholar]
- Sokratis, P.; Trine, G.; Raymond, P.C.; Maria, T.; Erik, K. Sediment properties and bacterial community in burrows of the ghost shrimp Pestarella tyrrhena (Decapoda: Thalassinidea). Aquat. Microb. Ecol. 2005, 38, 181–190. [Google Scholar]
- Aller, R.C. Benthic fauna and biogeochemical processes in marine sediments: The role of burrow structures. In Nitrogen Cycling in Coastal Marine Environments; Blackburn, T.H., Soerensen, J., Eds.; John Wiley and Sons: Chichester, UK, 1988; pp. 301–339. [Google Scholar]
- Harris, J.M. The presence, nature, and role of gut microflora in aquatic invertebrates: A synthesis. Microb. Ecol. 1993, 25, 195–231. [Google Scholar] [CrossRef]
- Bogatyrenko, E.A.; Buzoleva, L.S. Characterization of the gut bacterial community of the Japanese sea cucumber Apostichopus japonicus. Microbiology 2016, 85, 116–123. [Google Scholar] [CrossRef]
- Li, F.; Gao, F.; Tan, J.; Fan, C.; Sun, H.; Yan, J.; Chen, S.; Wang, X. Characterization and identification of enzyme-producing microflora isolated from the gut of sea cucumber Apostichopus japonicus. Chin. J. Oceanol. Limn. 2016, 34, 153–162. [Google Scholar] [CrossRef]
- Amaro, T.; Harry, W.; Herndl, G.J.; Cunha, M.R.; Billett, D.S.M. Deep-sea bacterial communities in sediments and guts of deposit-feeding holothurians in Portuguese canyons (NE Atlantic). Deep Sea Res. Part I 2009, 56, 1834–1843. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, Y.; Wu, P.; Chen, M.; He, L.; Xu, Q.; Wang, A. Bacterial community composition in gut content and ambient sediment of two tropical wild sea cucumbers (Holothuria atra and H. leucospilota). J. Oceanol. Limnol. 2022, 40, 360–372. [Google Scholar] [CrossRef]
- Rubin-Blum, M.; Shemesh, E.; Goodman-Tchernov, B.; Coleman, D.F.; Ben-Avraham, Z.; Tchernov, D. Cold seep biogenic carbonate crust in the Levantine basin is inhabited by burrowing Phascolosoma aff. turnerae, a sipunculan worm hosting a distinctive microbiota. Deep Sea Res. Part I 2014, 90, 17–26. [Google Scholar] [CrossRef]
- Pagola-Carte, S.; Saiz-Salinas, J.I. Sipuncula from Hainan Island (China). J. Nat. Hist. 2000, 34, 2187–2207. [Google Scholar] [CrossRef]
- Ouyang, Y.C.; Chen, F.L.; Sun, C.B.; Zhou, J.Z. Study on the diversity of bacteria from peanut worm (Sipunculus nudus). Guangdong Agric. Sci. 2012, 39, 178–181, (In Chinese with English absrtact). [Google Scholar]
- Zhong, R.; Huang, J.; Liao, Y.; Yang, C.; Wang, Q.; Deng, Y. Insights into the bacterial community compositions of peanut worm (Sipunculus nudus) and their association with the surrounding environment. Front. Mar. Sci. 2022, 9, 1076804. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.; Wu, P.; Zhu, C.; Hu, R.; Li, T.; Guo, Y. Insights into the relationship between intestinal microbiota of the aquaculture worm Sipunculus nudus and surrounding sediments. Fishes 2023, 8, 32. [Google Scholar] [CrossRef]
- Açik, S. Distribution of sipunculans along the Aegean and Levantine coasts of Turkey. Cah. Biol. Mar. 2017, 58, 317–330. [Google Scholar]
- Ip, Y.K.; Tan, G.Q.; Kuah, S.S.L.; Chew, S.F. Detoxification of environmental sulfide to sulfane sulfur in the intertidal sipunculid Phascolosoma arcuatum. J. Comp. Physiol. B 1997, 167, 213–220. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.; He, Y.; Wu, J.; Jiang, Y.; Tam, N.F.; Zhou, H. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of Illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wang, L.; Liu, L. Bacterial community structure and its regulating factors in the intertidal sediment along the Liaodong Bay of Bohai Sea, China. Microbiol. Res. 2014, 169, 585–592. [Google Scholar] [CrossRef]
- Guo, X.; Lu, D.; Niu, Z.; Feng, J.; Chen, Y.; Tou, F.; Liu, M.; Yang, Y. Bacterial community structure in response to environmental impacts in the intertidal sediments along the Yangtze Estuary, China. Mar. Pollut. Bull. 2018, 126, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Menge, B.A.; Branch, G.M. Rocky intertidal communities. In Marine Community Ecology; Bertness, M.D., Gaines, S.D., Hay, M.E., Eds.; Sinauer Associates: Sunderland, UK, 2001; pp. 221–251. [Google Scholar]
- Lasher, C.; Dyszynski, G.; Everett, K.; Edmonds, J.; Ye, W.; Sheldon, W.; Wang, S.; Joye, S.B.; Moran, M.A.; Whitman, W.B. The diverse bacterial community in intertidal, anaerobic sediments at Sapelo Island, Georgia. Environ. Microbiol. 2009, 58, 244–261. [Google Scholar] [CrossRef]
- Lee, H.; Heo, Y.; Kwon, S.L.; Yoo, Y.; Kim, D.; Lee, J.; Kwon, B.O.; Khim, J.S.; Kim, J.J. Environmental drivers affecting the bacterial community of intertidal sediments in the Yellow Sea. Sci. Total Environ. 2021, 755, 142726. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Wiegand, S.; Kohn, T.; Boedeker, C.; Jeske, O.; Rast, P.; Müller, R.-W.; Brümmer, F.; Heuer, A.; Jetten, M.S.M.; et al. Cultivation-independent analysis of the bacterial community associated with the calcareous sponge Clathrina clathrus and isolation of Poriferisphaera corsica Gen. Nov., Sp. Nov., belonging to the barely studied class Phycisphaerae in the phylum Planctomycetota. Front. Microbiol. 2020, 11, 602250. [Google Scholar] [PubMed]
- Kohn, T.; Wiegand, S.; Boedeker, C.; Rast, P.; Heuer, A.; Jetten, M.; Schüler, M.; Becker, S.; Rohde, C.; Müller, R.-W. Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Sys. Appl. Microbiol. 2020, 43, 126022. [Google Scholar] [CrossRef]
- Sheikh, M.; Rathore, D.; Gohel, S.; Singh, S. Marine Actinobacteriota associated with the invertebrate hosts: A rich source of bioactive compounds: A review. J. Cell Tissue Res. 2018, 18, 6361–6374. [Google Scholar]
- Taghon, G.L.; Jumars, P.A. Variable ingestion rate and its role in optimal foraging behavior of marine deposit feeders. Ecology 1984, 65, 549–558. [Google Scholar] [CrossRef]
- Zamora, L.N.; Jeffs, A.G. Feeding, selection, digestion and absorption of the organic matter from mussel waste by juveniles of the deposit-feeding sea cucumber, Australostichopus mollis. Aquaculture 2011, 317, 223–228. [Google Scholar] [CrossRef]
- Jumars, P.A.; Dorgan, K.M.; Lindsay, S.M. Diet of worms emended: An update of polychaete feeding guilds. Ann. Rev. Mar. Sci. 2015, 7, 497. [Google Scholar] [CrossRef]
- Hansen, M.D. Nahrung und Freßverhalten bei Sedimentfressern dargestellt am Beispiel von Sipunculiden und Holothurien. Helgoländer Wiss. Meeresunters. 1978, 31, 191–221. [Google Scholar] [CrossRef]
- Edmonds, S.J. Some notes on the abundance, environment, and nutrition of Sipunculus nudus L. (Sipunculoidea) at Morgat, Brittany. Cah. Biol. Mar. 1962, 3, 183–190. [Google Scholar]
- Amon, R.M.W.; Herndl, G.J. Deposit feeding and sediment I. interrelationship between Holofhuria fubulosa (Holofhurioida, Echinodermata) and the sediment microbial community. Mar. Ecol. 1991, 12, 163–174. [Google Scholar] [CrossRef]
- Andrei, A.; Baricz, A.; Robeson, M.S.; Păuşan, M.R.; Tămaş, T.; Chiriac, C.; Szekeres, E.; Barbu-Tudoran, L.; Levei, E.A.; Coman, C.; et al. Hypersaline sapropels act as hotspots for microbial dark matter. Sci. Rep. 2017, 7, 6150. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Jiang, G.; Wan, X.; Li, H.; Qiao, Y.; Thrush, S.; He, P. Response of the microbial community to bioturbation by benthic macrofauna on intertidal flats. J. Exp. Mar. Biol. Ecol. 2017, 488, 44–51. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetota and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef]
- Fuerst, J.A.; Sagulenko, E. Beyond the bacterium: Planctomycetota challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
| Grain Size | ≥2000 μm | 1000–2000 μm | 500–1000 μm | 250–500 μm | 125–250 μm | 63–125 μm | <63 μm |
|---|---|---|---|---|---|---|---|
| SAE | 6.50 ± 1.40 a | 11.57 ± 2.45 a | 28.92 ± 2.69 a | 29.59 ± 0.36 b | 16.41 ± 1.10 e | 5.93 ± 1.26 c | 1.09 ± 0.49 d |
| SAI | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 1.24 ± 0.33 e | 17.11 ± 1.23 d | 41.33 ± 2.15 c | 27.33 ± 1.09 a | 12.97 ± 0.34 a |
| PAE | 0.97 ± 0.36 b | 2.85 ± 1.02 b | 14.72 ± 2.63 b | 35.53 ± 1.40 a | 28.03 ± 2.14 d | 14.65 ± 1.51 b | 3.27 ± 1.01 c |
| PAI | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 1.38 ± 0.43 e | 20.31 ± 1.12 c | 42.36 ± 2.22 c | 29.32 ± 2.10 a | 6.63 ± 0.68 b |
| SNE | 0.30 ± 0.16 c | 1.11 ± 0.09 b | 8.77 ± 1.01 c | 32.36 ± 1.74 ab | 51.11 ± 2.21 b | 6.25 ± 1.06 c | 0.10 ± 0.09 e |
| SNI | 0.50 ± 0.20 bc | 0.32 ± 0.11 c | 3.78 ± 0.077 d | 22.44 ± 1.43 c | 60.13 ± 1.17 a | 11.96 ± 1.34 b | 0.52 ± 0.21 de |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, C.; Gao, F.; Wang, A.; Wang, H.; Yang, Y.; He, L. Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus). Int. J. Mol. Sci. 2023, 24, 6001. https://doi.org/10.3390/ijms24066001
Liu C, Liu C, Gao F, Wang A, Wang H, Yang Y, He L. Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus). International Journal of Molecular Sciences. 2023; 24(6):6001. https://doi.org/10.3390/ijms24066001
Chicago/Turabian StyleLiu, Chunsheng, Chuang Liu, Fei Gao, Aimin Wang, Haiqing Wang, Yumei Yang, and Linwen He. 2023. "Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus)" International Journal of Molecular Sciences 24, no. 6: 6001. https://doi.org/10.3390/ijms24066001
APA StyleLiu, C., Liu, C., Gao, F., Wang, A., Wang, H., Yang, Y., & He, L. (2023). Composition of Particulate Matter and Bacterial Community in Gut Contents and Surrounding Sediments of Three Sipunculan Species (Siphonosoma australe, Phascolosoma arcuatum, and Sipunculus nudus). International Journal of Molecular Sciences, 24(6), 6001. https://doi.org/10.3390/ijms24066001






