Extracellular Vesicles Produced by the Cardiac Microenvironment Carry Functional Enzymes to Produce Lipid Mediators In Situ
Abstract
1. Introduction
2. Results
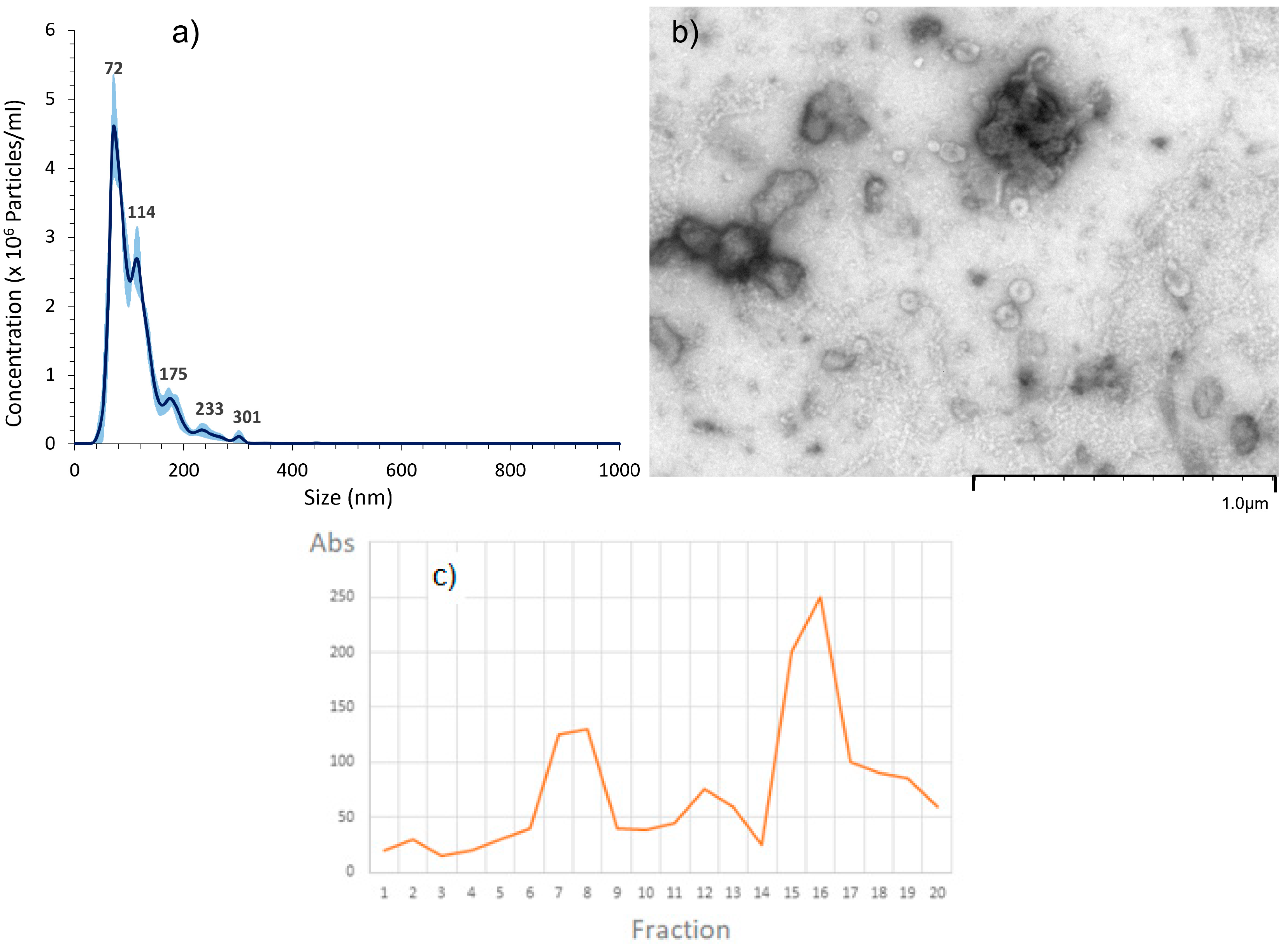
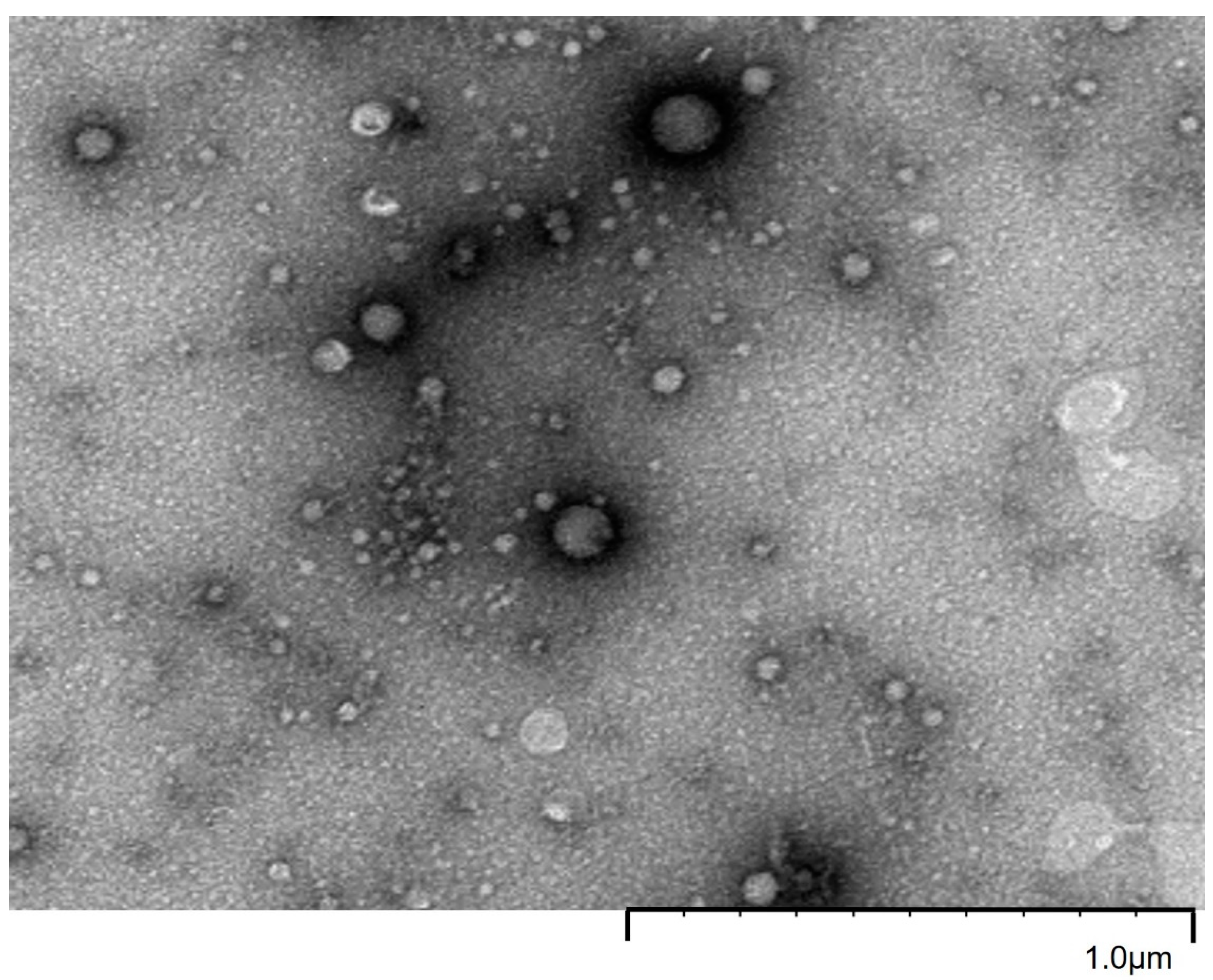
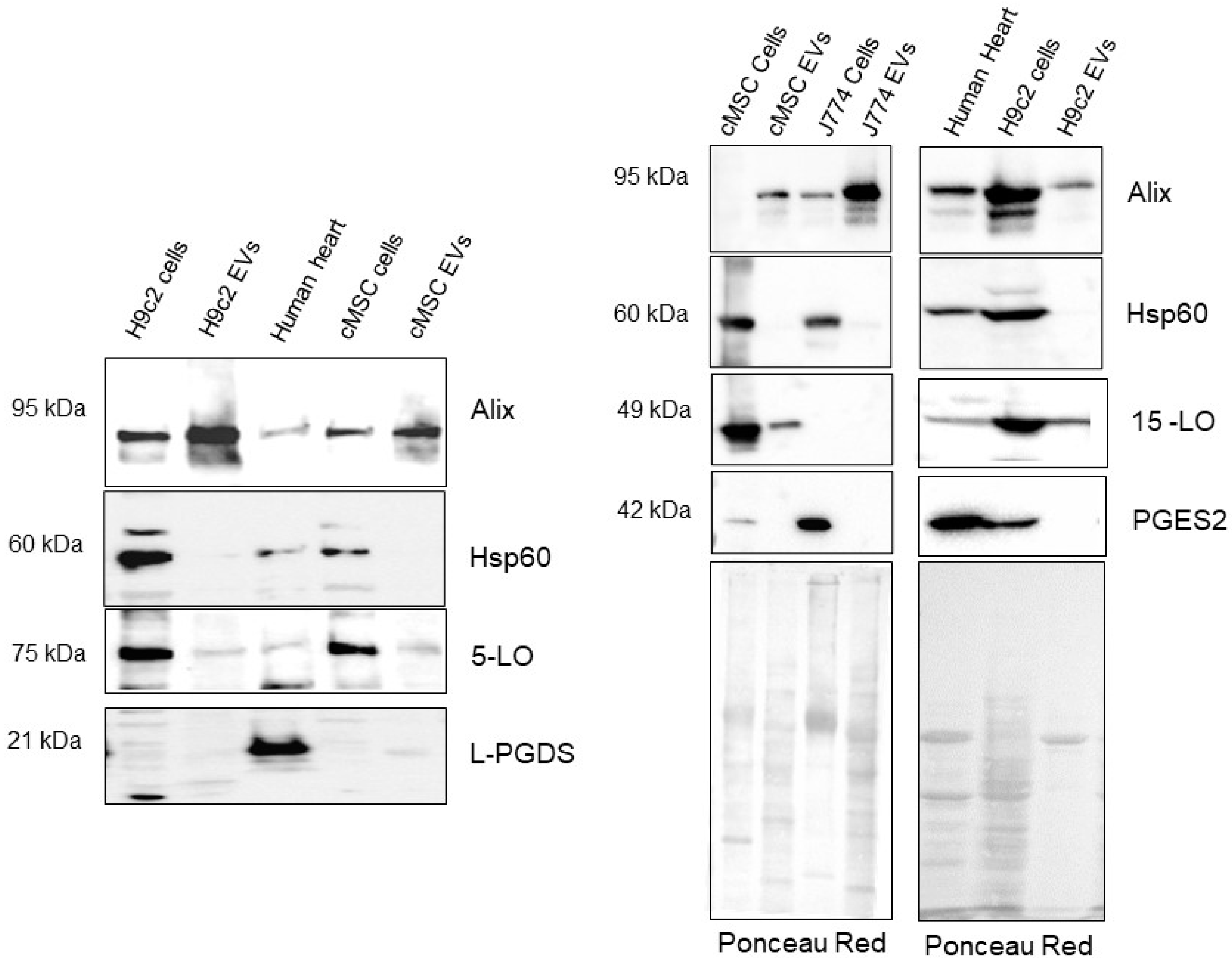
2.1. EVs Characterization
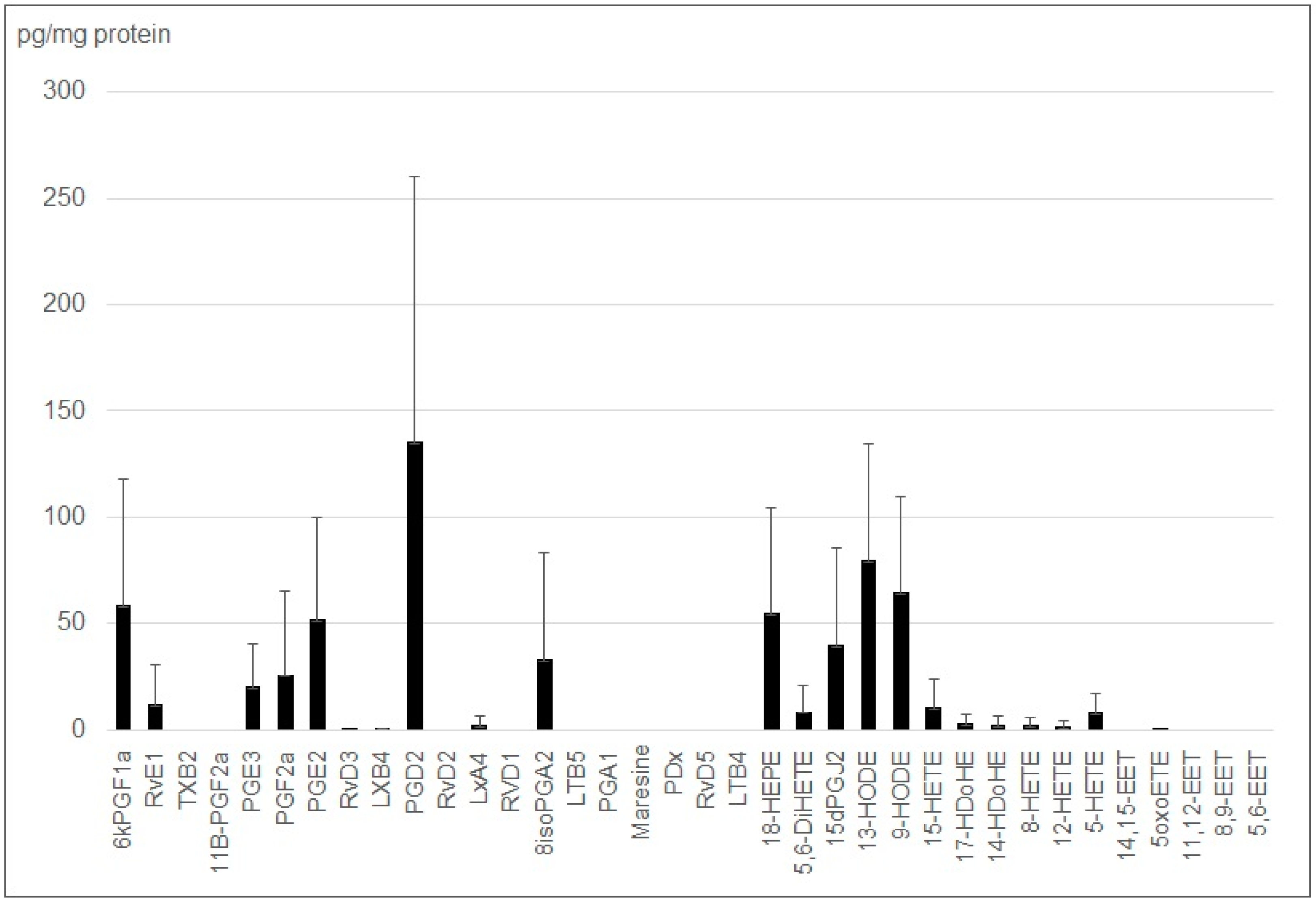
2.2. Small EVs from J774 Macrophages Carry Lipid Mediator
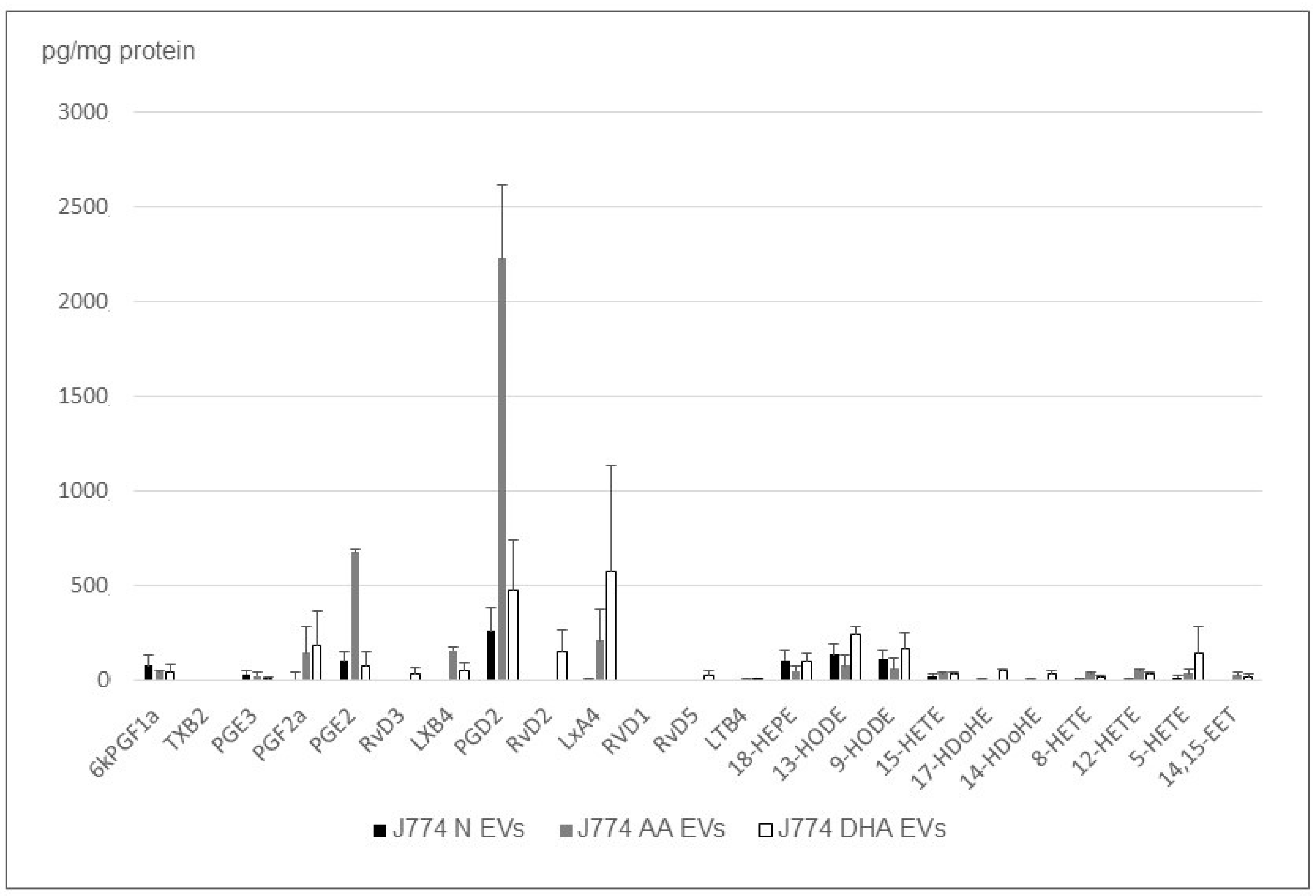
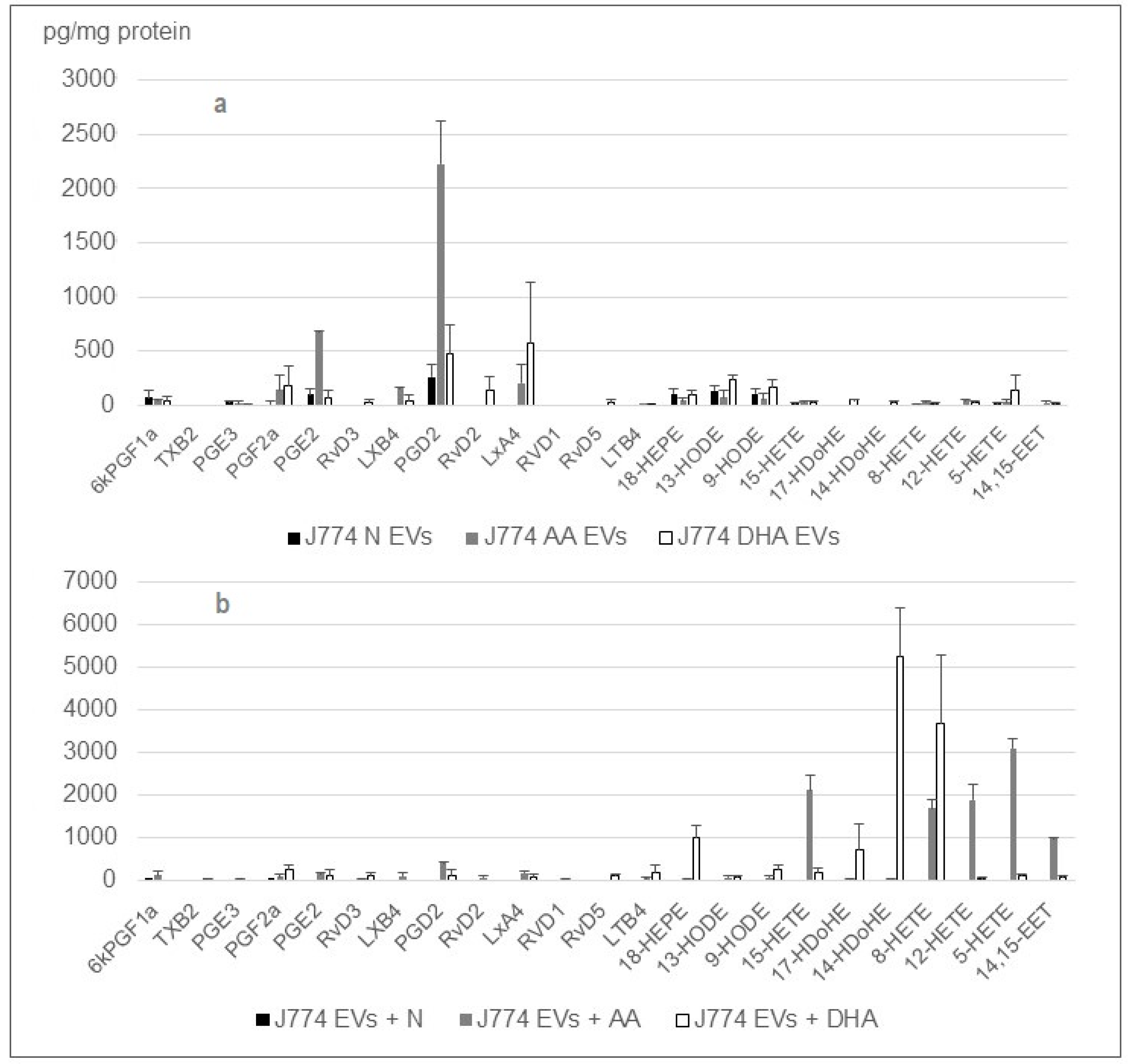
2.3. The Cargo of Small EV’s from J774 Macrophages Is Sensitive to PUFA
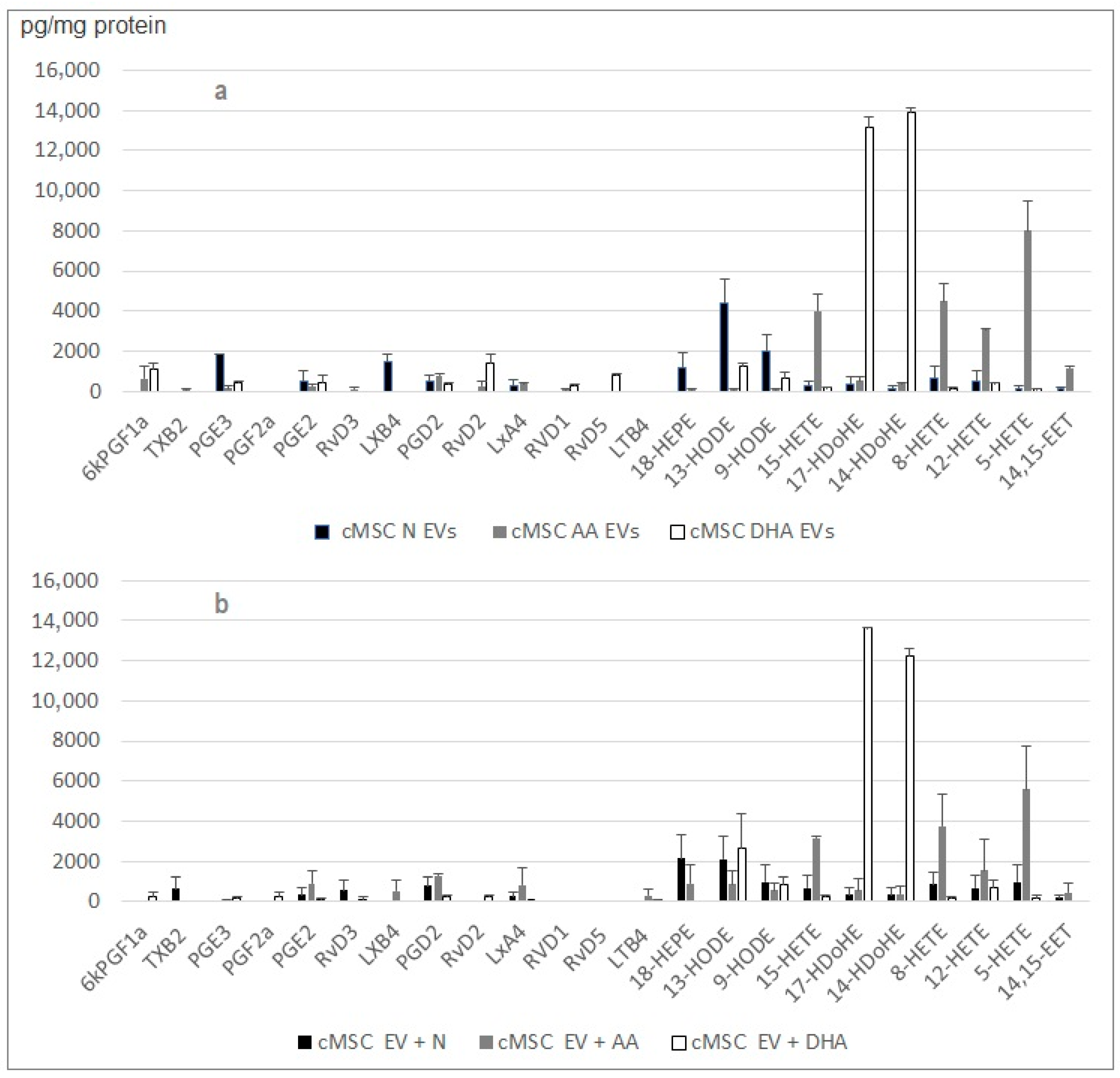
2.4. Small EV Cargo of Rat Cardiac MSC and Mouse Monocytes Changes when Cells Are Treated with PUFAs
2.5. Small EVs from the Cardiac Stroma Carry Functional Enzymes to Synthesize Lipid Mediators
3. Discussion
4. Materials and Methods
4.1. Biological Material and Cell Cultures
4.2. Protein Assay and Western Blotting
4.3. Sample Preparation
4.4. Small EV Characterization
4.4.1. Nanoparticle Tracking Analysis
4.4.2. Transmission Electron Microscopy
4.4.3. qEV Size Exclusion Chromatography (Izon)
4.5. Free-Lipid Extraction
4.6. HPLC/MS-MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adamo, L.; Rocha-Resende, C.; Prabhu, S.D.; Mann, D.L. Reappraising the role of inflammation in heart failure. Nat. Rev. Cardiol. 2020, 17, 269–285. [Google Scholar] [CrossRef]
- Sager, H.B.; Hulsmans, M.; Lavine, K.J.; Moreira, M.B.; Heidt, T.; Courties, G.; Sun, Y.; Iwamoto, Y.; Tricot, B.; Khan, O.; et al. Proliferation and recruitment contribute to myocardial macrophage expansion in chronic heart failure. Circ. Res. 2016, 119, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Nahrendorf, M. Myeloid cell contributions to cardiovascular health and disease. Nat. Med. 2018, 24, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Moskalik, A.; Niderla-Bielińska, J.; Ratajska, A. Multiple roles of cardiac macrophages in heart homeostasis and failure. Heart Fail Rev. 2022, 27, 1413–1430. [Google Scholar] [CrossRef] [PubMed]
- Bonaventura, A.; Montecucco, F.; Dallegri, F. Cellular recruitment in myocardial ischaemia/reperfusion injury. Eur. J. Clin. Investig. 2016, 46, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Nurakhayev, S.; Nurkesh, A.; Zharkinbekov, Z.; Saparov, A. Macrophage Polarization in Cardiac Tissue Repair Following Myocardial Infarction. Int. J. Mol. Sci. 2021, 22, 2715. [Google Scholar] [CrossRef] [PubMed]
- Dick, S.A.; Macklin, J.A.; Nejat, S.; Momen, A.; Clemente-Casares, X.; Althagafi, M.G.; Chen, J.; Kantores, C.; Hosseinzadeh, S.; Aronoff, L.; et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 2019, 20, 29–39. [Google Scholar] [CrossRef]
- Ben-Mordechai, T.; Holbova, R.; Landa-Rouben, N.; Harel-Adar, T.; Feinberg, M.S.; Abd-Elrahman, I.; Blum, G.; Epstein, F.H.; Silman, Z.; Cohen, S.; et al. Macrophage subpopulations are essential for infarct repair with and without stem cell therapy. J. Am. Coll. Cardiol. 2013, 62, 1890–1901. [Google Scholar] [CrossRef]
- De Couto, G.; Liu, W.; Tseliou, E.; Sun, B.; Makkar, N.; Kanazawa, H.; Arditi, M.; Marbán, E. Macrophages mediate cardioprotective cellular postconditioning in acute myocardial infarction. J. Clin. Investig. 2015, 125, 3147–3162. [Google Scholar] [CrossRef]
- Gnecchi, M.; He, H.; Noiseux, N.; Liang, O.D.; Zhang, L.; Morello, F.; Mu, H.; Melo, L.G.; Pratt, R.E.; Ingwall, J.S.; et al. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006, 20, 661–669. [Google Scholar] [CrossRef]
- Uemura, R.; Xu, M.; Ahmad, N.; Ashraf, M. Bone marrow stem cells prevent left ventricular remodeling of ischemic heart through paracrine signaling. Circ. Res. 2006, 98, 1414–1421. [Google Scholar] [CrossRef]
- Loyer, X.; Zlatanova, I.; Devue, C.; Yin, M.; Howangyin, K.-Y.; Klaihmon, P.; Guerin, C.L.; Kheloufi, M.; Vilar, J.; Zannis, K.; et al. Intra-cardiac release of extracellular vesicles shapes inflammation following myocardial infarction. Circ. Res. 2018, 123, 100–106. [Google Scholar] [CrossRef]
- Vagnozzi, R.J.; Maillet, M.; Sargent, M.A.; Khalil, H.; Johansen, A.K.Z.; Schwanekamp, J.A.; York, A.J.; Huang, V.; Nahrendorf, M.; Sadayappan, S.; et al. An acute immune response underlies the benefit of cardiac stem cell therapy. Nat. Cell Biol. 2020, 577, 405–409. [Google Scholar] [CrossRef]
- Barile, L.; Lionetti, V.; Cervio, E.; Matteucci, M.; Gherghiceanu, M.; Popescu, L.M.; Torre, T.; Siclari, F.; Moccetti, T.; Vassalli, G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 2014, 103, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Nickoloff, E.; Abramova, T.; Johnson, J.; Verma, S.K.; Krishnamurthy, P.; Mackie, A.R.; Vaughan, E.; Garikipati, V.N.S.; Benedict, C.; et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ. Res. 2015, 117, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Zhu, Y.; Meng, X.; Zhang, C.; Yang, Y.; Bei, Y. Immunomodulation by Exosomes in myocardial Infarction. J. Cardiovasc. Transl. Res. 2018, 12, 28–36. [Google Scholar] [CrossRef]
- Kalinec, G.M.; Gao, L.; Cohn, W.; Whitelegge, J.P.; Faull, K.F.; Kalinec, F. Extracellular Vesicles from Auditory Cells as Nanocarriers for Anti-inflammatory Drugs and Pro-resolving Mediators. Front. Cell. Neurosci. 2019, 13, 530. [Google Scholar] [CrossRef]
- Bahmani, L.; Mujib, U. Different Sourced Extracellular Vesicles and Their Potential Applications in Clinical Treatments. Cells 2022, 11, 1989. [Google Scholar] [CrossRef] [PubMed]
- Slomka, A.; Kornek, M.; Cho, W.C. Small Extracellular Vesicles and Their Involvement in Cancer Resistance: An Up-to-Date Review. Cells 2022, 11, 2913. [Google Scholar] [CrossRef]
- Deng, S.; Zhou, X.; Ge, Z.; Song, Y.; Wang, H.; Liu, X.; Zhang, D. Exosomes from adipose-derived mesenchymal stem cells ameliorate cardiac damage after myocardial infarction by activating S1P/SK1/S1PR1 signaling and promoting macrophage M2 polarization. Int. J. Biochem. Cell Biol. 2019, 114, 105564. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Hu, J.; Chen, F.; Qiao, S.; Sun, X.; Gao, L.; Xie, J.; Xu, B. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc. Res. 2019, 115, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Mentkowski, K.; Mursleen, A.; Snitzer, J.D.; Euscher, L.M.; Lang, J.K. CDC-derived extracellular vesicles reprogram inflammatory macrophages to an arginase 1-dependent proangiogenic phenotype. Am. J. Physiol. Circ. Physiol. 2020, 318, H1447–H1460. [Google Scholar] [CrossRef] [PubMed]
- De Couto, G.; Jaghatspanyan, E.; Deberge, M.; Liu, W.; Luther, K.; Wang, Y.; Tang, J.; Thorp, E.B.; Marbán, E. Mechanism of Enhanced MerTK-Dependent Macrophage Efferocytosis by Extracellular Vesicles. Arter. Thromb. Vasc. Biol. 2019, 39, 2082–2096. [Google Scholar] [CrossRef]
- Cambier, L.; De Couto, G.; Ibrahim, A.; Echavez, A.K.; Valle, J.; Liu, W.; Kreke, M.; Smith, R.R.; Marbán, L.; Marbán, E. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol. Med. 2017, 9, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular Vesicles—Connecting Kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yip, K.W.; Spence, T.; Liu, F.F. MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J. Hum. Genet. 2017, 62, 67–74. [Google Scholar] [CrossRef]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Vera, M.P.; Blackwell, T.S.; Baron, R.M.; et al. MicroRNA-181b regulates NF-_B–mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar]
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: Role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 2018, 114, 992–1005. [Google Scholar] [CrossRef]
- Boilard, E. Thematic Review Series: Exosomes and Microvesicles: Lipids as Key Components of their Biogenesis and Functions Extracellular vesicles and their content in bioactive lipid mediators: More than a sack of microRNA. J. Lipid Res. 2018, 59, 2037–2046. [Google Scholar] [CrossRef]
- Muraoka, N.; Nara, K.; Tamura, F.; Kojima, H.; Yamakawa, H.; Sadahiro, T.; Miyamoto, K.; Isomi, M.; Haginiwa, S.; Tani, H.; et al. Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac Reprogramming. Nat. Commun. 2019, 10, 674. [Google Scholar] [CrossRef]
- Bosviel, R.; Joumard-Cubizolles, L.; Chinetti-Gbaguidi, G.; Bayle, D.; Copin, C.; Hennuyer, N.; Duplan, I.; Staels, B.; Zanoni, G.; Porta, A.; et al. DHA-derived oxilipins, neuroprostanes and protectins, differentially and dose-dependently modulate the inflammatory response in human macrophages: Putative mechanisms through PPAR activation. Free Radic. Biol. Med. 2017, 103, 146–154. [Google Scholar] [CrossRef]
- Bhatt, D.L.; Steg, P.G.; Miller, M.; Brinton, E.A.; Jacobson, T.A.; Ketchum, S.B.; Doyle, R.T., Jr.; Juliano, R.A.; Jiao, L.; Granowitz, C.; et al. Cardiovascular Risk Reduction with Icosapent Ethyl for Hypertriglyceridemia. N. Engl. J. Med. 2019, 380, 11. [Google Scholar] [CrossRef]
- Mourmoura, E.; Chate, V.; Couturier, K.; Laillet, B.; Vial, G.; Rigaudiere, J.P.; Morio, B.; Malpuech-Brugere, C.; Azarnoush, K.; Demaison, L. Body adiposity dictates different mechanisms of increased coronary reactivity related to improve in vivo cardiac function. Cardiovasc. Diabetol. 2014, 13, 54. [Google Scholar] [CrossRef]
- Reinicke, M.; Shamkeeva, S.; Hell, M.; Isermann, B.; Ceglarek, U.; Heinemann, M.L. Targeted Lipidomics for Characterization of PUFAs and Eicosanoids in Extracellular Vesicles. Nutrients 2022, 14, 1319. [Google Scholar] [CrossRef]
- Holopainen, M.; Colas, A.R.; Valkonen, S.; Tigistu-Sahle, F.; Hyvarinen, K.; Mazzacuva, F.; Lehenkari, P.; Kakela, R.; Dalli, J.; Kerkela, E.; et al. Polyunsaturated fatty acids modify the extracellular vesicle membranes and increase the production of proresolving lipid mediators of human mesenchymal stromal cells. BBA Mol. Cell Biol. Lipids 2019, 1864, 1350–1362. [Google Scholar] [CrossRef] [PubMed]
- Pizzinat, N.; Ong-Meang, V.; Bourgailh-Tortosa, F.; Blanzat, M.; Perquis, L.; Cussac, D.; Parini, A.; Poinsot, V. Extracellular vesicles of MSCs and cardiomyoblasts are vehicles for lipid mediators. Biochimie 2020, 178, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D. Minimal information for studies of extracellular vesicles, (MISEV 2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV 2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Sahlén, G.E.; Egevad, L.; Ahlander, A.; Norlén, B.J.; Ronquist, G.; Nilsson, B.O. Ultrastructure of the secretion of prostasomes from benign and malignant epithelial cells in the prostate. Prostate 2002, 53, 192. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.F.; van Adrichem, N.P.H.; van Wijnen, M.; Wauben, M.H.M.; Stout, T.A.E.; Stoorvogel, W. Identification of Distinct Populations of Prostasomes That Differentially Express Prostate Stem Cell Antigen, Annexin A1, and GLIPR2 in Humans. Biol. Reprod. 2012, 86, 1. [Google Scholar] [CrossRef]
- Edelmann, M.J.; Kima, P.E. Current understanding of extracellular vesicle homing/tropism. Zoonoses 2022, 2, 14. [Google Scholar] [CrossRef]
- Zhang, M.; Xin, W.; Ma, C.; Zhang, H.; Mao, M.; Liu, Y.; Zheng, X.; Zhang, L.; Yu, X.; Li, H.; et al. Exosomal 15-LO2 mediates hypoxia-induced pulmonary artery hypertension in vivo and in vitro. Cell Death Dis. 2018, 9, 1022. [Google Scholar] [CrossRef]
- Lukic, A.; Wahlund, C.J.E.; Gómez, C.; Brodin, D.; Samuelsson, B.; Wheelock, C.E.; Gabrielsson, S.; Rådmark, O. Exosomes and cells from lung cancer pleural exudates transform LTC4 to LTD4, promoting cell migration and survival39 via CysLT1. Cancer Lett. 2019, 444, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Laroumanie, F.; Douin-Echinard, V.; Pozzo, J.; Lairez, O.; Tortosa, F.; Vinel, C.; Delage, C.; Calise, D.; Dutaur, M.; Parini, A.; et al. CD4+ T cells promote the transition from hypertrophy to heart failure during chronic pressure overload. Circulation 2014, 129, 2111–2124. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Leung, T.C.N.; Ngai, S.-M.; Tsai, S.-N.; Thakur, A.; Li, W.-K.; Lee, Y.; Leung, L.; Ng, T.-H.; Yam, J.; et al. Proteomic Analysis of Circulating Extracellular Vesicles Identifies Potential Biomarkers for Lymph Node Metastasis in Oral Tongue Squamous. Cells 2021, 10, 2179. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong-Meang, V.; Blanzat, M.; Savchenko, L.; Perquis, L.; Guardia, M.; Pizzinat, N.; Poinsot, V. Extracellular Vesicles Produced by the Cardiac Microenvironment Carry Functional Enzymes to Produce Lipid Mediators In Situ. Int. J. Mol. Sci. 2023, 24, 5866. https://doi.org/10.3390/ijms24065866
Ong-Meang V, Blanzat M, Savchenko L, Perquis L, Guardia M, Pizzinat N, Poinsot V. Extracellular Vesicles Produced by the Cardiac Microenvironment Carry Functional Enzymes to Produce Lipid Mediators In Situ. International Journal of Molecular Sciences. 2023; 24(6):5866. https://doi.org/10.3390/ijms24065866
Chicago/Turabian StyleOng-Meang, Varravaddheay, Muriel Blanzat, Lesia Savchenko, Lucie Perquis, Mégane Guardia, Nathalie Pizzinat, and Verena Poinsot. 2023. "Extracellular Vesicles Produced by the Cardiac Microenvironment Carry Functional Enzymes to Produce Lipid Mediators In Situ" International Journal of Molecular Sciences 24, no. 6: 5866. https://doi.org/10.3390/ijms24065866
APA StyleOng-Meang, V., Blanzat, M., Savchenko, L., Perquis, L., Guardia, M., Pizzinat, N., & Poinsot, V. (2023). Extracellular Vesicles Produced by the Cardiac Microenvironment Carry Functional Enzymes to Produce Lipid Mediators In Situ. International Journal of Molecular Sciences, 24(6), 5866. https://doi.org/10.3390/ijms24065866





