Abstract
Long non-coding RNAs (lncRNAs), a class of poorly conserved transcripts without protein-encoding ability, are widely involved in plant organogenesis and stress responses by mediating the transmission and expression of genetic information at the transcriptional, posttranscriptional, and epigenetic levels. Here, we cloned and characterized a novel lncRNA molecule through sequence alignment, Sanger sequencing, transient expression in protoplasts, and genetic transformation in poplar. lncWOX11a is a 215 bp transcript located on poplar chromosome 13, ~50 kbp upstream of PeWOX11a on the reverse strand, and the lncRNA may fold into a series of complex stem–loop structures. Despite the small open reading frame (sORF) of 51 bp within lncWOX11a, bioinformatics analysis and protoplast transfection revealed that lncWOX11a has no protein-coding ability. The overexpression of lncWOX11a led to a decrease in the quantity of adventitious roots on the cuttings of transgenic poplars. Further, cis-regulatory module prediction and CRISPR/Cas9 knockout experiments with poplar protoplasts demonstrated that lncWOX11a acts as a negative regulator of adventitious rooting by downregulating the WUSCHEL-related homeobox gene WOX11, which is supposed to activate adventitious root development in plants. Collectively, our findings imply that lncWOX11a is essential for modulating the formation and development of adventitious roots.
1. Introduction
The formation and development of adventitious roots (ARs) is the key to the large-scale vegetative propagation of elite genotypes in many economically important woody species [1]. As ecologically and economically important trees in the Northern Hemisphere [2,3], the species of the genus Populus in Salicaceae mainly rely on cutting propagation to produce a large number of genotypically uniform plant materials [2] for not only timber and pulp production, but also the construction of bioenergy forests and shelter forests [4]. The adventitious rooting of tree cuttings, as a key criterion for selecting superior genotypes in the forestry industry, is a complex quantitative trait regulated by polygenes [5,6,7,8]. Despite enormous efforts over the last few decades to understand the anatomical, physiological, and biochemical mechanisms of adventitious rooting in forest trees, the genetic and molecular mechanisms of AR formation remain elusive.
Our current understanding of the molecular regulation mechanism of AR formation mainly comes from the progress of Arabidopsis thaliana [9], Oryza sativa [10], and Zea mays [11]. The basis of AR organogenesis is the totipotency and plasticity of cells, and the essence is the fate transitions of cells induced by trauma [12], environment [13], and physiological signals [14]. Plant-specific WUSCHEL-related homeobox (WOX) transcription factors [15], especially WOX11 [16], play critical regulatory roles in this process. Arabidopsis WUSCHEL (AtWUS) is the prototypical member of the WOX gene family, while AtWOX5 [17], as a close homologue of AtWUS, is a root stem cell organizer. Both genes were reported to be functionally exchangeable in regulating stem cell maintenance in both shoot and root contexts. AtWOX11 and AtWOX12 control the first-step cell fate transition during AR organogenesis by activating LATERAL ORGAN BOUNDARIES DOMAIN 16 (LBD 16) and WOX5/7 [18]. AtWOX11 and auxin are involved in the transition of some vascular formative layers from xylem/woody stem cell production to the establishment of new lateral root primordia [19]. OsWOX11, the rice homologue of AtWOX11, has been reported to command the organogenesis of rice crown roots by participating in multiple regulatory pathways [20,21]. There are many conservative molecular regulatory pathways in the AR formation of trees and herbs; WOX11, WOX5, LBD, and other functional genes also play a central role in de novo AR primordia formation in trees. The overexpression of PeWOX11a and PeWOX11b not only increases the number of Ars, but also induces ectopic roots in transgenic poplar [22]. The WOX11/12a-SMALL AUXIN-UP RNA 36 (SAUR36) module can mediate AR formation via auxin or salt stress pathways in poplar [23]. In Populus, woody taproots were treated with bending to induce the formation of new roots. During this process, PnWOX11 was shown to be expressed during new lateral root formation at different developmental stages [24]. Overall, the rooting pathway mediated by WOX11 may help many plant species create plastic root systems.
In recent years, increasing studies have focused on the involvement of long non-coding RNAs (lncRNAs) in plant organogenesis by mediating the transmission and expression of genetic information at the transcriptional, posttranscriptional, and epigenetic levels [25]. Unlike well-known microRNAs (miRNAs), lncRNAs are a class of poorly conserved transcripts with a length of more than 200 nucleotides and no protein-coding ability [26,27]. Most plant lncRNAs have specific secondary structures and spatiotemporal expression patterns, and their regulatory mechanisms are complex and diverse [28]. With the continuous updating of new sequencing technologies and biological prediction algorithms, the action trace of lncRNA has been found in many links between plant growth [29,30,31,32,33] and stress response [25,34]. The authenticity identification and annotation of these lncRNAs have been enormously challenging, and there is still a long way to go for further experimental verification and validation. To date, despite a large number of lncRNAs predicted in trees, knowledge about the biological functions and mechanisms of tree lncRNAs is still very limited.
According to the current study, lncRNAs play an important role mainly in the response of poplar to various abiotic stresses and in the development of woody tissue. An lncRNA–gene interaction network was constructed in poplar, deciphering that lncRNAs can respond to heat stress by regulating heat shock protein (HSP) family genes [35]. Another study showed that lncRNAs can regulate their target genes through RNA interference to increase photosynthetic recovery and protection while reducing DNA damage in plants from heat stress [36]. Exploring the physiological mechanisms of wood properties in poplars as they adapt to low-nitrogen environments provides a strategy for growing poplars in nitrogen-poor soils. Several studies have demonstrated the involvement of lncRNAs in this biological process, either directly or through the miRNAs–lncRNAs–mRNA pathway [37,38]. A number of lncRNAs have been identified in poplar that can participate in biological processes such as carbon metabolism, cellulose and lignin synthesis, and signal transduction by regulating their target genes [39,40]. Among them, lncRNA PMAT can control the tolerance and uptake of Pb2+ in root organs through the PMAT–PtoMYB46–PtoMATE–PtoARF2 pathway [41]. The mechanism of the role of lncRNAs in poplar development still needs to be studied in depth.
In our previous study [22], two genes encoding WOX11 transcription factors were identified and characterized through the rapid amplification of cDNA ends and genetic transformation in Populus. Overexpressing PeWOX11a or PeWOX11b in poplar would encourage more AR formation and stimulate ectopic root formation in aerial parts. Further research based on strand-specific RNA sequencing and preliminary validation experiments indicated that a large number of lncRNAs were involved in regulating the organogenesis of poplar ARs, and several potentially important lncRNAs may affect adventitious rooting by regulating the expression of PeWOX11s [42]. In this study, lncWOX11a, a 215 bp transcript located on poplar chromosome 13, ~50 kbp upstream of PeWOX11a on the reverse strand, was cloned and characterized, and detailed information about the stem–loop structures, expression patterns, coding ability, and biological functions of lncWOX11a was revealed. The overexpression of lncWOX11a led to a decrease in the quantity of ARs on the cuttings of transgenic poplars. Further cis-regulatory module prediction and CRISPR/Cas9 knockout experiments with poplar protoplasts demonstrated that lncWOX11a acts as a negative regulator of adventitious rooting by downregulating PeWOX11a.
2. Results
2.1. Isolation and Sequence Analysis of lncWOX11a
Based on our previous strand-specific RNA sequencing data [42], DNA and cDNA sequences of the target lncRNA were cloned from P. deltoides × P. euramericana cv. ‘Nanlin 895’ roots, which were the same, with a length of 215 bp. lncWOX11a was located 54293 bp upstream of PeWOX11a (Figure 1a) on chromosome 13. The subcellular localization was predicted using lncLocator, and the results showed that lncWOX11a was localized in the nucleus. To investigate the conservation of the sequence, homology searches were performed on the NCBI, TAIR, and NONCODE databases. No similar sequences were retrieved, indicating that lncWOX11a was poorly conserved. The online RNAfold software was used to analyze the secondary structure of lncWOX11a, which had a complex secondary structure composed of multiple stem–loop structures. The minimum free energy (MFE) of lncWOX11a was predicted to be −11.40 kcal/mol, which indicates that it is relatively stable (Figure 1b). The prediction of miRNA binding sites for lncWOX11a using a plant miRNA target gene analysis tool revealed that lncWOX11a cannot bind to all the miRNAs in poplar, indicating that lncWOX11a cannot function by binding to miRNAs.
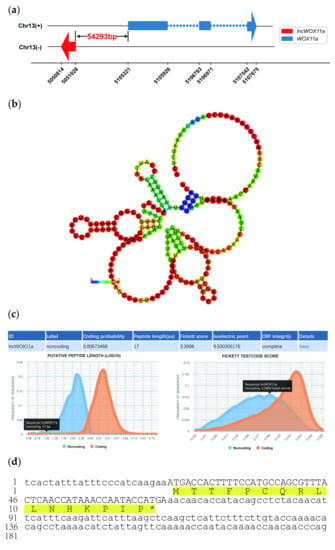
Figure 1.
Sequence analysis of lncWOX11a. (a) Schematic representation of the lncWOX11a–PeWOX11a genomic region. (b) Prediction of the secondary structure of lncWOX11a. The different colors represent the level of minimum free energy. (c) Prediction of the coding ability of lncWOX11a by CPC2 software. (d) Nucleotide sequence of lncWOX11a.
The coding ability is a crucial indicator for identifying lncRNAs. The results obtained with CPC2 software showed that lncWOX11a has no coding ability (Figure 1c). The ribosome is where mRNAs are translated into proteins, and the presence or absence of ribosome binding sites (RBSs) in an RNA sequence determines whether it can be translated successfully. The RBS was predicted using IRESite software, and no RBS was detected on lncWOX11a (Figure S1). As shown in Figure 1d, the small open reading frame (sORF) of lncWOX11a was only 51 bp in length and may encode 16 amino acids. A search of the BLASTP database for the 16 amino acids revealed no matching homologous sequences; in addition, a search of the Pfam database for the amino acids did not reveal any functional structural domains, suggesting, from a bioinformatics perspective, that the small peptide of lncWOX11a may not have the capacity to encode the protein.
2.2. Verification of the Coding Ability of lncWOX11a
Recent studies have reported that some lncRNAs predicted by bioinformatics software have false positives, and these were later experimentally verified to have the ability to encode peptides and then act as peptides. Therefore, we further verified the coding ability of lncWOX11a by transient expression in protoplasts. We constructed a 35S::lncWOX11a 5’UTR+sORF-GFP fusion vector using Gateway technology and transferred it into poplar protoplasts. The results showed fluorescence of the control 35S::GFP construct in the nucleus, cell membrane, and cytoplasm of poplar protoplasts, whereas after 35S::lncWOX11a 5’UTR+sORF-GFP transfection, only chloroplast autofluorescence and no green fluorescent protein (GFP) fluorescence were detected (Figure 2). All results were replicated in three independent experiments, proving that lncWOX11a is a long non-coding RNA.
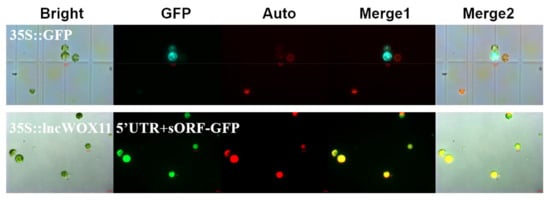
Figure 2.
Verification of the coding ability of lncWOX11a. Protoplasts transiently expressing lncWOX11a were observed under a fluorescence microscope. Bright-field, green fluorescent protein (GFP), chlorophyll autofluorescence (Auto), merged 1, and merged 2 images are shown. Scale bar = 10 µm.
2.3. Organ-Specific Expression of lncWOX11a
The abundance of lncWOX11a transcripts in various poplar organs was assessed by quantitative real-time polymerase chain reaction (qRT–PCR). The results showed that lncWOX11a was mainly expressed in 1-week-old roots from cuttings and showed weak expression in 2-week-old roots and stems (Figure 3a). In addition, the expression patterns of lncWOX11a were ubiquitous in nine organs, and no significant organ-specific expression pattern was detected in the roots. Despite being more highly expressed in 1-week-old roots, lncWOX11a was very highly expressed in other organs. The lowest lncWOX11a expression was found in young stems, followed by young leaves. The expression of lncWOX11a was significantly higher in the other seven organs and was highest in the mature petiole (Figure 3b).
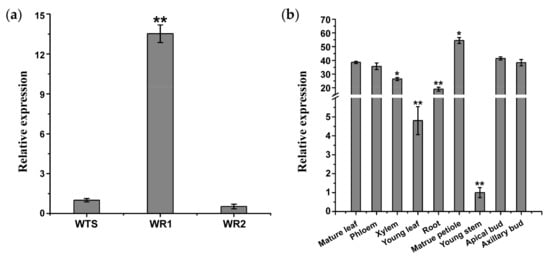
Figure 3.
Expression patterns of lncWOX11a in different organs of poplar. (a) lncWOX11a was specifically expressed in 1-week-old roots. WTS represents the stems of 30-day-old tissue-cultured seedlings. WR1 and WR2 represent 1-week-old roots and 2-week-old roots of tissue-cultured seedlings, respectively. (b) lncWOX11a was mainly expressed in mature organs. Mature leaves were picked in July of the study year, and all other organs were picked at the end of March of the test year, One-way ANOVA was used to identify statistically significant differences. The data are presented as the means ± SEs. The “*” above the histogram indicates significance, “*” represents p < 0.05, and “**” represents p < 0.01.
2.4. The Overexpression of lncWOX11a Inhibits AR Development
Previous studies have shown that WOX11a has a function in AR formation [22]. To further confirm the potential role of lncWOX11a in woody plants, we transformed the 35S::lncWOX11a construct into the hybrid poplar ‘Shanxin’. The overexpression vector 35S::lncWOX11a was constructed using the gateway method, as shown in Figure 4a. The lncWOX11a sequence was ligated to the entry vector by a BP reaction and was then transferred into the overexpression vector by an LR reaction. Afterwards, the correctly sequenced 35S::lncWOX11a vector was transferred into Agrobacterium EHA105. Nine lines of transgenic plants showing healthy growth were chosen for DNA detection. The results of agarose gel electrophoresis showed that the wild type (WT) and OE1# lines did not show any amplified bands after PCR amplification, whereas bands were detected with the other eight lines (Figure 4c). We randomly selected four OE transgenic lines for further detection. According to the qRT–PCR results, we found that the expression level of lncWOX11a in the four OE transgenic lines was 2–8 times higher than that in the WT, which tentatively implies that the 35S::lncWOX11a construct was integrated into the poplar genome (Figure 4d).
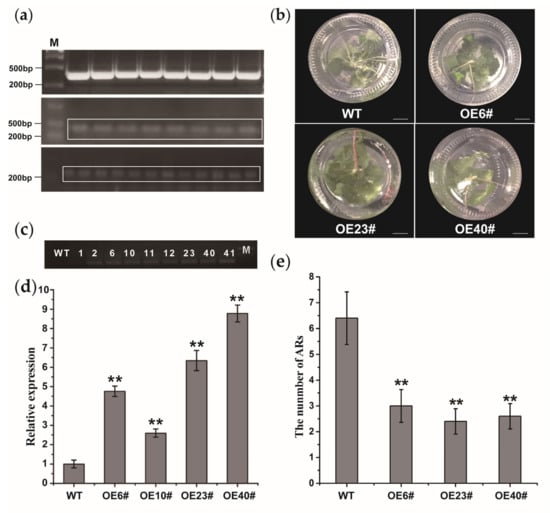
Figure 4.
Molecular identification of transgenic poplar overexpressing lncWOX11a. (a) Construction of the lncWOX11a overexpression vector. The graph shows the BP reaction, LR reaction, and Agrobacterium tumefaciens detection. (b) The number of ARs in transgenic poplars at 30 days was lower than in WT plants at the same age. Scale bar = 10 mm. (c) DNA amplification was used to identify nine transgenic plants. (d) Detection of lncWOX11a expression in WT and OE transgenic lines. (e) Statistics on the number of ARs in the OE transgenic lines. One-way ANOVA was used to identify statistically significant differences. The data are presented as the means ± SEs. The “*” above the histogram indicates significance, and “**” represents p < 0.01.
To determine the effect of lncWOX11a overexpression on the growth of transgenic lines, we selected four independent lines (OE6#, OE23#, and OE40# lines) for subsequent phenotypic observation experiments. For each strain, we set up five biological replicas separately. One-month-old transgenic and WT lines were continuously cultured in MS medium. Then, we observed the growth of ARs in the transgenic lines and counted the number of roots produced. The root architecture of the OE lines was significantly different from that of the WT plants, and a 1-fold reduction in the average number of ARs (including taproots and lateral roots) was detected in the OE lines (Figure 4b,e). Specifically, 10 days after regeneration, the WT line began to form ARs near the stem, and no growth dominance was observed in the transgenic lines. After 30 days, the number of ARs continued to increase in the WT and OE transgenic lines. However, the OE transgenic lines had certain advantages in terms of plant height. These results indicated that lncWOX11a might negatively modulate AR formation.
2.5. The cis-Regulation Module for lncWOX11a
Based on the four target sites (Figure 5a), we constructed knockout vectors. The four target sequences were constructed into four sgRNA expression cassettes using the overlapping PCR method (Figure 5b); the four expression cassettes were then cloned into knockout vectors, and colony PCR was performed using the universal primers SP-L1 and SP-R to detect positive clones (Figure 5c,d). Genomic DNA from protoplasts was sequenced, and the sequencing data were subsequently analyzed using Synthego software to identify base insertions or deletions near the target sites. The knockout efficiency of lncWOX11a in protoplasts using the two vectors did not differ considerably. At targets 1 and 2, the knockout efficiency of the pYLCRISPR/Cas9pUbi-N vector was more noticeable, and at target 3, the knockout efficiency of the pYLCRISPR/Cas9p35S-N vector was higher. However, no editing events were discovered close to the sequence of target T4 (Figure 6).
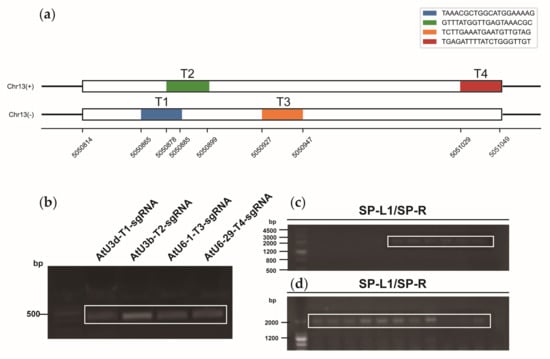
Figure 5.
Construction of the CRISPR/Cas9 vectors for lncWOX11a. (a) Target sequences of CRISPR/Cas9 in lncWOX11a. The white region represents the exon of lncWOX11a. Two separate targets were designed for the positive and negative sense strands, and each had a length of 20 bp. (b) Construction of the sgRNA intermediate vector. The second round of PCR products was detected by agarose gel electrophoresis. (c) Construction of the pYLCRISPR/Cas9pUbi-N vector. (d) Construction of the pYLCRISPR/Cas9p35S-N vector. Positive clones were detected by colony PCR using primers from the vector with the universal primers SP-R and SP-L1, and the white box indicates the fragment containing four tandem sgRNA expression cassettes.

Figure 6.
Evaluation of the knockout efficiency of the pYLCRISPR/Cas9pubi-N and pYLCRISPR/Cas9p35S-N vectors for lncWOX11a. The first column of data represents missing or added cases, and the second column of data shows the knockout efficiency. The red font represents target sequences, and the grey font indicates deletions. Protospacer-adjacent motifs (PAMs; NGGs) are shown in red boxes.
Based on a genome annotation analysis within 100 kb upstream or downstream of lncWOX11a, we detected eight target genes cis-regulated by lncWOX11a. These putative targets included the PeWOX11a gene, which has been shown to be important for AR formation [22]. We used qRT–PCR to monitor the expression of these eight genes in the OE40# line to detect the regulation between lncWOX11a and the target genes. As shown in Figure 7a, the expression of these target genes varied significantly in the OE40# line. Among them, the expression of three target genes increased significantly after the overexpression of lncWOX11a; specifically, POPTR_0013s06270 showed the highest expression, and the expression of PeWOX11a and POPTR-0013s06300 was markedly lower than that in the WT. The results illustrated a suppressive effect of the overexpression of lncWOX11a on the expression of POPTR_0013s06300 and PeWOX11a.
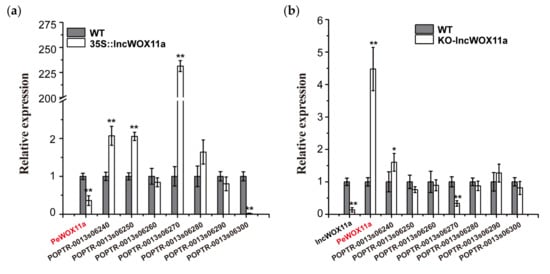
Figure 7.
Expression levels of cis-regulated target genes of lncWOX11a. (a) Expression levels of cis-regulated target genes of lncWOX11a in 35S::lncWOX11a poplar. (b) Expression level of the cis-regulated target gene of lncWOX11a in poplar protoplasts after the knockout of lncWOX11a. One-way ANOVA was used to identify statistically significant differences. The data are presented as the means ± SEs. The “*” above the histogram indicates significance, “*” indicates p < 0.05, and “**” indicates p < 0.01.
The expression levels of lncWOX11a and the target genes were examined in protoplasts that had been transduced into knockout vectors by qRT–PCR. First, the knockout experiment was successful, as evidenced by the decreased expression of lncWOX11a. Among the eight target genes, PeWOX11a showed a large increase in expression, whereas the target gene POPTR_0013s06240 showed a small increase (Figure 7b). POPTR_0013s06270 exhibited a decreasing trend relative to the expression of WT, whereas the others did not change significantly and may not be regulated by lncWOX11a. Based on these results, combined with the target gene expression quantity after overexpression, we found that lncWOX11a showed the same trend as POPTR_0013s06270, implying a positive regulatory relationship between them. PeWOX11a exhibited the opposite pattern, demonstrating that they were negatively regulated by one another. In summary, we preliminarily suppose that lncWOX11a may inhibit AR development in poplar by downregulating PeWOX11a.
3. Discussion
Exploring the molecular mechanisms of AR development provides ideas for large-scale asexual reproduction strategies for some woody species. A growing body of research suggests that lncRNAs are essential regulators of a wide range of biological processes. To understand how lncRNAs play a role in AR formation and development, we investigated the biological function of lncWOX11a in hybrid poplars and the regulatory relationship between lncWOX11a and WOX11a.
In this study, a 215 bp transcript located on poplar chromosome 13 was cloned from ‘Nanlin 895’, and a sequence analysis of lncWOX11a was performed using online bioinformatics software. The ability to code is a critical requirement for determining whether a long transcript is an lncRNA. The absence of RBS on lncWOX11a was detected by bioinformatics software, indicating that it could not be translated into a protein. lncRNAs have several major mechanisms of action, such as guide molecules [43] and scaffold molecules [44]. In addition, lncRNAs can act as decoy molecules that can bind to miRNAs [44]. miRNAs can target and repress the expression of protein-coding genes. When such lncRNAs bind to miRNAs, they are able to compete with the target genes of miRNAs, which reduces the quantity of miRNA-targeting genes and alleviates the repression of protein-coding genes by miRNAs. These lncRNAs are also regarded as competing endogenous RNAs (ceRNAs) [45]. The prediction of miRNA binding sites revealed that lncWOX11a does not function by acting as a decoy molecule. Furthermore, by conducting transient expression experiments in protoplasts, we concluded that lncWOX11a is a novel non-coding RNA.
The important role of the WOX11 gene in root organ development is evident from a large number of reports [16]. PagWOX11/12a can modulate ROS scavenging by regulating the expression of PagCYP736A12 to enhance salt tolerance in poplar [46], while ROS has been shown to be closely associated with the development of ARs in several species [47,48]. Overall, the way in which WOX11 promotes adventitious rooting in angiosperms is at least evolutionarily conserved, and it could be a vital molecular tool to promote rooting from stem or leaf cuttings. Therefore, functional studies and mechanistic analyses of the WOX11 gene in poplar still need to be continued. In plants and other eukaryotes, the expression level of lncRNA is usually much lower than that of mRNA [49]. We selected various poplar organs to measure the expression pattern of lncWOX11a. The expression of lncWOX11a was generally high in various organs, and no significant organ-specific expression pattern was found in the roots. In addition, the overexpression of lncWOX11a in poplar suppressed AR formation to some extent, which complemented the biological function of WOX11a.
Regulating the expression of downstream functional genes is a valuable mechanism through which lncRNAs participate in plant growth. lncRNAs can regulate the expression of nearby genes in cis and distant genes in trans [50,51]. By prediction, we found that lncWOX11a may regulate some protein-coding genes by cis-regulating action. Although the CRISPR/Cas9 editing system is gradually becoming a critical instrument for genetic breeding and gene function research, a stable genetic transformation system is still lacking for many forest trees. Therefore, we performed CRISPR/Cas9 knockout experiments using poplar protoplasts. Accordingly, we elaborated on the relationship between lncWOX11a and the potential protein-coding genes (Figure S2). Among these putative target genes, the gene annotations showed that POPTR_0013s06300 encodes a Class III peroxidase PRX70 protein; POPTR-0013s06240 belongs to glycosyltransferase family 14 (GT14). Glycosyltransferase (GT) is a key enzyme in the synthesis of plant cell wall polysaccharides and glycoproteins. To date, studies on the GT14 family have focused on poplar, A. thaliana, O. sativa [52,53], and others. Combined with the above-described experimental results, we speculate that lncWOX11a may have a regulatory relationship with PeWOX11a and POPTR_0013s06270. In addition, we found that the expression of the POPTR-0013s06240 gene increased after both lncWOX11a overexpression and knockdown. Based on this result, we speculate that a direct regulatory relationship may not exist between lncWOX11a and POPTR-0013s06240. In our previous study, the 35S::lncWOX11 plasmid was transiently expressed in protoplasts, after which the expression of PeWOX11a was found to be upregulated [42]. This experiment builds on previous studies to further validate the biological function of lncWOX11a in poplar through experiments such as genetic transformation and CRISPR/Cas9 gene knockdown. Finally, we proposed a hypothesis that lncWOX11a may exert a suppressive effect on PeWOX11a, thus inhibiting the growth and development of ARs in transgenic plants. However, the genetic evidence and the regulatory module between lncWOX11a and PeWOX11a must be better confirmed in subsequent experiments.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tissue-cultured seedlings of a cultivated variety (Populus deltoides × P. euramericana cv. ‘Nanlin 895’) were used as the materials. The seedlings were grown on Murashige and Skoog (MS) medium (pH 5.8) in a humid chamber at a temperature of 25/18 °C (day/night). The photoperiod was 16/8 h per day (light/dark), and the relative humidity was 60–80%. Tissue-cultured ‘Nanlin 895’ seedlings were used for gene cloning, and the leaves from 40-day-old seedlings were used for transient protoplast expression. Nine organs from 2-year-old ‘Nanlin 895’ poplars were used for RNA extraction to measure the transcript abundance of lncWOX11a. The samples were stored in a freezing chamber at −80 °C for further experiments. We used hybrid poplar (P. davidiana × P. bolleana cv. ‘Shanxin’) as the transformation acceptor plantlets in this study and cultivated it as described above.
4.2. Cloning and Sequence Analysis
We designed amplification-specific primers (Table S1) for cloning based on the lncWOX11a sequences obtained in our previous study [42]. First, RNA and DNA were extracted separately from 1-week-old roots of ‘Nanlin 895’ tissue-cultured seedlings. Genomic DNA was obtained using a DNeasy Plant Mini Kit (Tiangen, China). Total RNA was separated using the RNAprep Pure Plant Plus kit (Tiangen, China), and cDNA was then synthesized from RNA using the PrimeScript RT Master Mix Kit (Takara, China). To explore whether lncRNAs homologous to lncWOX11a exist in other species, cDNA sequence similarity comparisons were performed with NCBI BLASTN (https://blast.ncbi.nlm.nih.gov/ (accessed on 10 March 2022)), the Arabidopsis TAIR database (https://www.arabidopsis.org/ (accessed on 10 March 2022)), and the NONCODE database (http://www.noncode.org/ (accessed on 10 March 2022)) with the threshold set to E < 0.005; the subcellular localization was predicted using lncLocator; the miRNA binding sites were predicted using psRNATarget (https://www.zhaolab.org/ (accessed on 15 July 2022)); RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (accessed on 15 July 2022)) was used to predict the secondary structure; CPC2 software (http://cpc2.cbi.pku.edu.cn/ (accessed on 16 July 2022)) was used for the assessment of the coding ability; the ORF was searched using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/ (accessed on 16 July 2022)); and IRESite software (http://www.iresite.org/ (accessed on 20 August 2022)) was aimed at determining RBSs.
4.3. Transient Expression in Poplar Protoplasts
Some studies have shown that carrying the 5’UTR of a gene at the 5’ end of the ORF can facilitate the translation of the gene. To experimentally validate the coding ability of lncWOX11a, we designed primers (Table S1) targeting lncWOX11a 5’UTR+sORF-GFP. PCR amplification was first performed using DNA polymerase, and the sequence was obtained by sequence verification. The correct sequence was transferred to the p2GWF7.0 vector carrying GFP using Gateway technology (Invitrogen) following the manufacturer’s protocol. We used a method that optimized the type and concentration of enzymes and the digestion time for the extraction of poplar protoplasts [54]. Finally, the plasmids were introduced into poplar protoplasts using the polyethylene glycol (PEG)-mediated transient expression system [54]. The fluorescence of the GFP tags was monitored under a fluorescence microscope (Scope A1 Carl Zesiss, Jena, Germany).
4.4. Quantitative Real-Time Quantification
Quantitative real-time polymerase chain reaction (qRT–PCR) aimed to assess the expression pattern of lncWOX11a in diverse organs from ‘Nanlin 895’. Nine organs, including the xylem, phloem, petiole, leaf, root, stem, and bud organs, were taken from 2-year-old ‘Nanlin 895’ poplar. First, total RNA was isolated from nine different organs, and cDNA was then synthesized from RNA. qRT–PCR was performed with an ABI ViiA™ 7 real-time PCR system using the PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, Carlsbad, CA, USA) following the manufacturer’s manual. The 2−ΔΔCT value between the target gene and the reference, 18S ribosomal RNA (18S) [55], was used to compute the relative expression [56]. One-way ANOVA was used to examine statistically significant differences. The primers for qRT–PCR were designed using oligo6 and are provided in Supplementary Table S1.
4.5. Generation of Transgenic Poplar Lines
The lncWOX11a sequence was cloned into pH35GS using the Gateway System to construct the overexpression vector. The leaves from 40-day-old tissue-cultured ‘shanxin’ seedlings were used as the transformation acceptor plantlets. The 35S::lncWOX11a plasmid was introduced into EHA105 and used for the genetic transformation of hybrid poplar ‘Shanxin’ using the Agrobacterium-based leaf disc method [57,58]. The OE lines were obtained after screening in MS medium with kanamycin. Afterward, amplification reactions at the DNA and RNA levels were performed on all putative OE lines and WT controls. The DNA of the transgenic plants was extracted for PCR amplification using p35SF on the promoter as the forward primer and the downstream primer of lncWOX11a as the reverse primer.
4.6. Target Gene Prediction
Based on genome annotation, genes located within 100 kb upstream or downstream of lncWOX11a were chosen as candidate target genes in the cis-regulatory mechanism [42]. The cis-regulatory module of lncWOX11a was mapped using Cytoscape software.
4.7. Construction of the pYLCRISPR/Cas9-lncWOX11a Vector
The CRISPR/Cas9 system [59] was used to confirm the regulatory module of lncWOX11a. We designed four sgRNA sequences that targeted lncWOX11a using the online website CRISPR-GE [59] (http://skl.scau.edu.cn/ (accessed on 20 March 2022)). The four target sites were ligated to the four sgRNA intermediate vectors by overlapping PCR, and the sgRNAs of the four target sites were actuated by the AtU3b, AtU3d, AtU6-1, and AtU6-29 promoters from A. thaliana. The four correct sgRNA expression cassettes were cloned into the pYLCRISPR/Cas9pUbi-N and pYLCRISPR/Cas9p35S-N vectors to complete the constructs by Golden Gate ligation [60,61]. The control and knockout vectors were transferred into poplar protoplasts, and the genomic DNA was then isolated from the plasmid-transferred protoplasts. The primers that contained the four target sites were used to perform PCR amplification and were then sequenced by the Sanger method. The data were analyzed using Synthego software to identify base insertions or deletions near the target sites, and additional experiments were subsequently conducted. All the primers needed to construct the knockout vectors are shown in Supplementary Table S2.
5. Conclusions
In conclusion, we isolated and cloned lncWOX11a from ‘Nanlin 895’, which is 215 bp in length. Both bioinformatic analysis and transient expression confirmed that lncWOX11a has no ability to encode proteins. Through genetic transformation in poplar, we elucidated the role of lncWOX11a in the regulation of AR development. The regulatory relationship between lncWOX11a and its target genes was explained by CRISPR/Cas9 knockout experiments in poplar protoplasts. Finally, we hypothesized that lncWOX11a could repress the expression of PeWOX11a to negatively regulate AR development.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24065766/s1.
Author Contributions
M.X. conceived the project; N.R., S.L., H.Q. and J.W. performed the experiments; T.S. and W.X. analyzed the data. N.R. drafted the manuscript. M.X. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Key R & D Program of China during the 14th Five-year Plan Period (2021YFD2200103), the National Natural Science Foundation of China (31971679; 31570671), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (19KJB180001), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data are shown in the main manuscript and in the Supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Legué, V.; Rigal, A.; Bhalerao, R. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant. 2014, 151, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Dickmann, D.; Kuzovkina, J. Poplars and willows of the world, with emphasis on silviculturally important species. In Poplars and Willows: Trees for Society and the Environment; CABI: Wallingford, UK, 2014; pp. 8–91. [Google Scholar]
- Lin, Y.C.; Wang, J.; Delhomme, N.; Schiffthaler, B.; Sundstrom, G.; Zuccolo, A.; Nystedt, B.; Hvidsten, T.R.; de la Torre, A.; Cossu, R.M.; et al. Functional and evolutionary genomic inferences in Populus through genome and population sequencing of American and European aspen. Proc. Natl. Acad. Sci. USA 2018, 115, E10970–E10978. [Google Scholar] [CrossRef]
- Stettler, R.F.; Bradshaw, H.D.; Heilman, P.E.; Hinckley, T.M. Biology of Populus and Its Implications for Management and Conservation; NRC Research Press: Ottawa, ON, Canada, 1996. [Google Scholar]
- Zhai, S.; Cai, W.; Xiang, Z.-X.; Chen, C.-Y.; Lu, Y.-T.; Yuan, T.-T. PIN3-mediated auxin transport contributes to blue light-induced adventitious root formation in Arabidopsis. Plant Sci. 2021, 312, 111044. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Tayengwa, R.; Cheng, Z.M.; Peer, W.A.; Murphy, A.S.; Zhao, M. Auxin regulates adventitious root formation in tomato cuttings. BMC Plant Biol. 2019, 19, 435. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plant. 2019, 166, 663–676. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, C.; Wang, N.; Wei, L.; Li, W.; Yao, Y.; Liao, W. Roles of Small-Molecule Compounds in Plant Adventitious Root Development. Biomolecules 2019, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, S.; Ruiz-Cano, H.; Fernandez, M.A.; Sanchez-Garcia, A.B.; Villanova, J.; Micol, J.L.; Perez-Perez, J.M. A Network-Guided Genetic Approach to Identify Novel Regulators of Adventitious Root Formation in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 461. [Google Scholar] [CrossRef]
- Lin, C.; Sauter, M. Polar Auxin Transport Determines Adventitious Root Emergence and Growth in Rice. Front. Plant Sci. 2019, 10, 444. [Google Scholar] [CrossRef]
- Singh, Z.; Singh, H.; Garg, T.; Mushahary, K.K.K.; Yadav, S.R. Genetic and Hormonal Blueprint of Shoot-Borne Adventitious Root Development in Rice and Maize. Plant Cell Physiol. 2022, 63, 1806–1813. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The Physiology of Adventitious Roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Lorbiecke, R.; Sauter, M. Adventitious Root Growth and Cell-Cycle Induction in Deepwater Rice1. Plant Physiol. 1999, 119, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Hilo, A.; Perez-Perez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, N.; Xie, D.; Liu, W.; Xu, L. WOX11: The founder of plant organ regeneration. Cell. Regen. 2023, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.K.; Luijten, M.; Miyashima, S.; Lenhard, M.; Hashimoto, T.; Nakajima, K.; Scheres, B.; Heidstra, R.; Laux, T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 2007, 446, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Baesso, B.; Chiatante, D.; Terzaghi, M.; Zenga, D.; Nieminen, K.; Mahonen, A.P.; Siligato, R.; Helariutta, Y.; Scippa, G.S.; Montagnoli, A. Transcription factors PRE3 and WOX11 are involved in the formation of new lateral roots from secondary growth taproot in A. thaliana. Plant Biol. 2018, 20, 426–432. [Google Scholar] [CrossRef]
- Zhang, T.; Li, R.; Xing, J.; Yan, L.; Wang, R.; Zhao, Y. The YUCCA-Auxin-WOX11 Module Controls Crown Root Development in Rice. Front. Plant Sci. 2018, 9, 523. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Chen, Z.; Yue, Y.; Huang, H.; Wu, B.; Liu, Y.; Zhou, D.X.; Zhao, Y. ROS-stimulated Protein Lysine Acetylation Is Required for Crown Root Development in Rice. J. Adv. Res. 2022, in press. [Google Scholar] [CrossRef]
- Xu, M.; Xie, W.; Huang, M. Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant 2015, 155, 446–456. [Google Scholar] [CrossRef]
- Liu, R.; Wen, S.-S.; Sun, T.-T.; Wang, R.; Zuo, W.-T.; Yang, T.; Wang, C.; Hu, J.-J.; Lu, M.-Z.; Wang, L.-Q. PagWOX11/12a positively regulates the PagSAUR36 gene that enhances adventitious root development in poplar. J. Exp. Bot. 2022, 73, 7298–7311. [Google Scholar] [CrossRef]
- Baesso, B.; Terzaghi, M.; Chiatante, D.; Scippa, G.S.; Montagnoli, A. WOX genes expression during the formation of new lateral roots from secondary structures in Populus nigra (L.) taproot. Sci. Rep. 2020, 10, 18890. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, K.; Yu, R.; Zhou, B.; Huang, P.; Cao, Z.; Zhou, Y.; Wang, J. From “Dark Matter” to “Star”: Insight Into the Regulation Mechanisms of Plant Functional Long Non-Coding RNAs. Front. Plant Sci. 2021, 12, 650926. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef]
- Herr, A.J.; Jensen, M.B.; Dalmay, T.; Baulcombe, D.C. RNA polymerase IV directs silencing of endogenous DNA. Science 2005, 308, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Zou, X.; Ali, F.; Jin, S.; Li, F.; Wang, Z. RNA-Seq with a novel glabrous-ZM24fl reveals some key lncRNAs and the associated targets in fiber initiation of cotton. BMC Plant Biol. 2022, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, J.; Wang, C.; Xu, Y.; Li, X.; Yang, J.; Chen, K.; Kang, Y.; Wang, Y.; Cao, P.; et al. Comprehensive Analysis of Long Non-coding RNA Modulates Axillary Bud Development in Tobacco (Nicotiana tabacum L.). Front. Plant Sci. 2022, 13, 809435. [Google Scholar] [CrossRef]
- Wang, H.; Chu, Z.; Chang, S.; Jia, S.; Pang, L.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Han, S. Transcriptomic identification of long noncoding RNAs and their hormone-associated nearby coding genes involved in the differential development of caryopses localized on different branches in rice. J. Plant Physiol. 2022, 271, 153663. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, G.; Zhang, F.; Ma, J.; Wen, C.; Li, H. Genome-Wide Identification of Powdery Mildew Responsive Long Non-Coding RNAs in Cucurbita pepo. Front. Genet. 2022, 13, 933022. [Google Scholar] [CrossRef]
- Liu, J.; Chen, S.; Liu, M.; Chen, Y.; Fan, W.; Lee, S.; Xiao, H.; Kudrna, D.; Li, Z.; Chen, X.; et al. Full-Length Transcriptome Sequencing Reveals Alternative Splicing and lncRNA Regulation during Nodule Development in Glycine max. Int. J. Mol. Sci. 2022, 23, 7371. [Google Scholar] [CrossRef]
- Xuhui, L.; Weiwei, C.; Siqi, L.; Junteng, F.; Hang, Z.; Xiangbo, Z.; Yongwen, Q. Full-length transcriptome analysis of maize root tips reveals the molecular mechanism of cold stress during the seedling stage. BMC Plant Biol. 2022, 22, 398. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Fang, M.; Li, Z.; Zhang, M.; Liu, X.; Peng, Y.; Wan, Y.; Chen, J. Third-Generation Sequencing Reveals LncRNA-Regulated HSP Genes in the Populus × canadensis Moench Heat Stress Response. Front. Genet. 2020, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, P.; Liu, P.; Bu, C.; Zhang, D. High-Temperature-Responsive Poplar lncRNAs Modulate Target Gene Expression via RNA Interference and Act as RNA Scaffolds to Enhance Heat Tolerance. Int. J. Mol. Sci. 2020, 21, 6808. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.Y.; Chen, X.; Shi, W.G.; Deng, S.R.; Luo, Z.B. Genome-Wide Identification and Characterization of Long Noncoding RNAs in Populus × canescens Roots Treated With Different Nitrogen Fertilizers. Front. Plant Sci. 2022, 13, 890453. [Google Scholar] [CrossRef]
- Lu, Y.; Deng, S.; Li, Z.; Wu, J.; Liu, Q.; Liu, W.; Yu, W.J.; Zhang, Y.; Shi, W.; Zhou, J.; et al. Competing Endogenous RNA Networks Underlying Anatomical and Physiological Characteristics of Poplar Wood in Acclimation to Low Nitrogen Availability. Plant Cell Physiol. 2019, 60, 2478–2495. [Google Scholar] [CrossRef]
- Xu, H.; Chen, B.; Zhao, Y.; Guo, Y.; Liu, G.; Li, R.; Zeisler-Diehl, V.V.; Chen, Y.; He, X.; Schreiber, L.; et al. Non-Coding RNA Analyses of Seasonal Cambium Activity in Populus tomentosa. Cells 2022, 11, 640. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, H.; Chen, B.; Grünhofer, P.; Schreiber, L.; Lin, J.; Zhao, Y. Genome-wide analysis of long non-coding RNAs in shoot apical meristem and vascular cambium in Populus tomentosa. J. Plant Physiol. 2022, 275, 153759. [Google Scholar] [CrossRef]
- Chen, P.; Song, Y.; Liu, X.; Xiao, L.; Bu, C.; Liu, P.; Zhao, L.; Ingvarsson, P.K.; Wu, H.X.; El-Kassaby, Y.A.; et al. LncRNA PMAT-PtoMYB46 module represses PtoMATE and PtoARF2 promoting Pb(2+) uptake and plant growth in poplar. J. Hazard Mater. 2022, 433, 128769. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Z.; Xu, M. Identification and characterization of long non-coding RNAs involved in the formation and development of poplar adventitious roots. Ind. Crops Prod. 2018, 118, 334–346. [Google Scholar] [CrossRef]
- Heo Jae, B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wen, S.S.; Wang, R.; Wang, C.; Gao, B.; Lu, M.Z. PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 2021, 19, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Wang, Y.; Liu, Y.; Wang, G.; She, X. Reactive oxygen species regulate auxin levels to mediate adventitious root induction in Arabidopsis hypocotyl cuttings. J. Integr. Plant Biol. 2020, 62, 912–926. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, B.K.; Singh, S.; Tripathi, D.K.; Sharma, S.; Prasad, S.M.; Chauhan, D.K.; Kumar, V.; Singh, V.P. New adventitious root formation and primary root biomass accumulation are regulated by nitric oxide and reactive oxygen species in rice seedlings under arsenate stress. J. Hazard. Mater. 2019, 361, 134–140. [Google Scholar] [CrossRef]
- Chekanova, J.A.; Gregory, B.D.; Reverdatto, S.V.; Chen, H.; Kumar, R.; Hooker, T.; Yazaki, J.; Li, P.; Skiba, N.; Peng, Q.; et al. Genome-Wide High-Resolution Mapping of Exosome Substrates Reveals Hidden Features in the Arabidopsis Transcriptome. Cell 2007, 131, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem. Sci. 2014, 39, 35–43. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Dilokpimol, A.; Geshi, N. Arabidopsis thaliana glucuronosyltransferase in family GT14. Plant Signal Behav. 2014, 9, e28891. [Google Scholar] [CrossRef]
- Ye, C.-Y.; Li, T.; Tuskan, G.A.; Tschaplinski, T.J.; Yang, X. Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci. 2011, 181, 688–695. [Google Scholar] [CrossRef]
- Tan, B.; Xu, M.; Chen, Y.; Huang, M. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 2013, 114, 11–18. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bruegmann, T.; Polak, O.; Deecke, K.; Nietsch, J.; Fladung, M. Poplar Transformation. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Springer: New York, NY, USA, 2019; pp. 165–177. [Google Scholar]
- Eck, J.V.; Kirk, D.D.; Walmsley, A.M. Tomato (Lycopersicum esculentum). In Agrobacterium Protocols; Wang, K., Ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 459–474. [Google Scholar]
- Zhu, Q.; Liu, Y.-G.; Ma, X.; Zeng, D.; Xie, X. A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants. Sci. Sin. Vitae 2018, 48, 783–794. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.-G. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.36.1–31.36.21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).