Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CPP Genes in Tomato
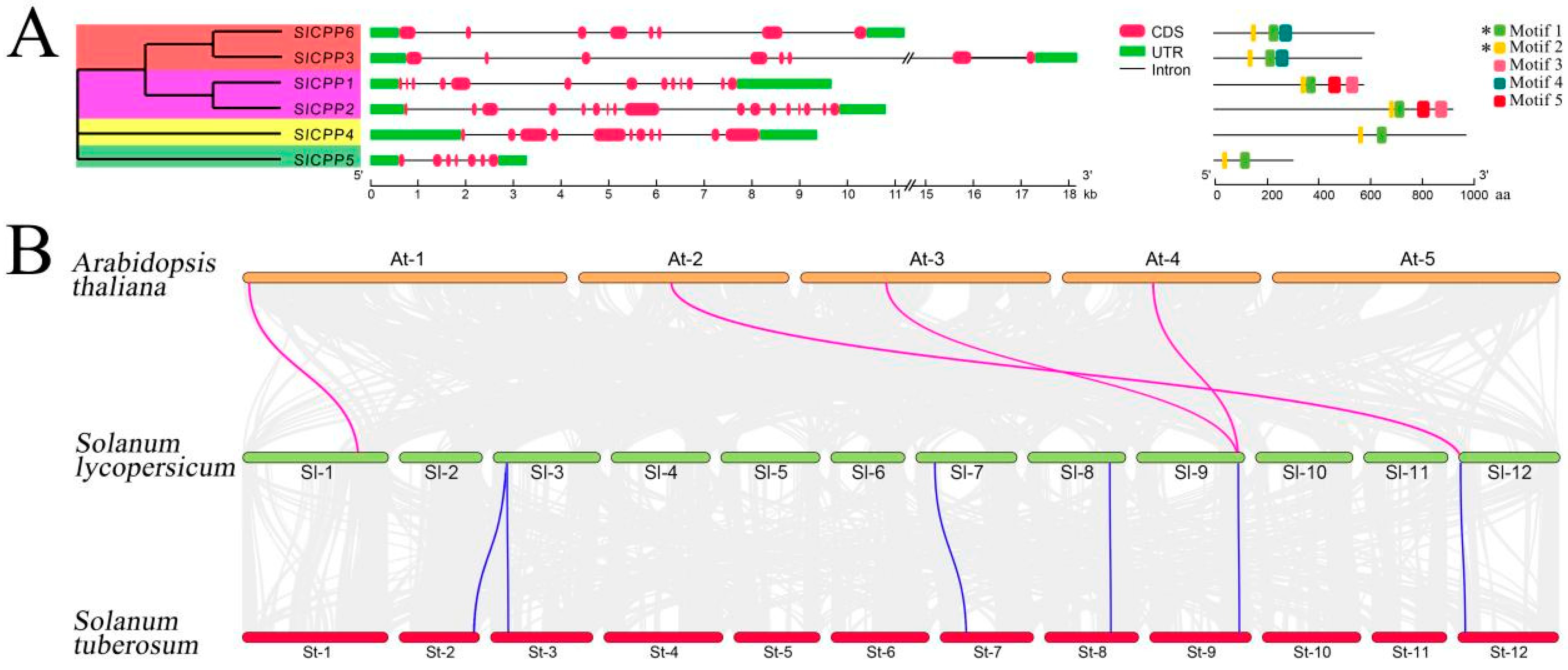
2.2. Phylogenetic Relationships and Gene Structure Analysis of SlCPPs
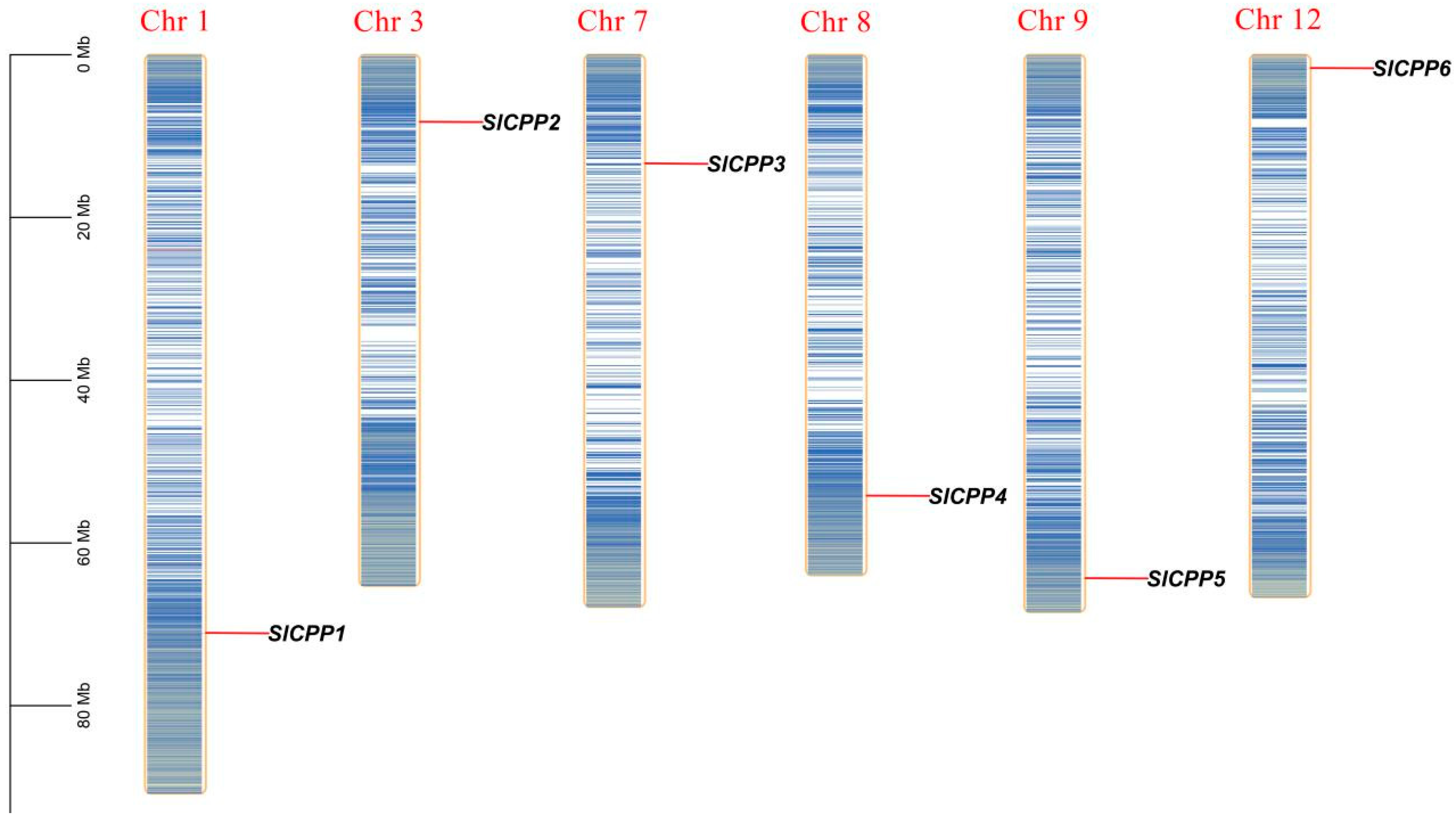
2.3. Synteny Analysis of SlCPP Genes
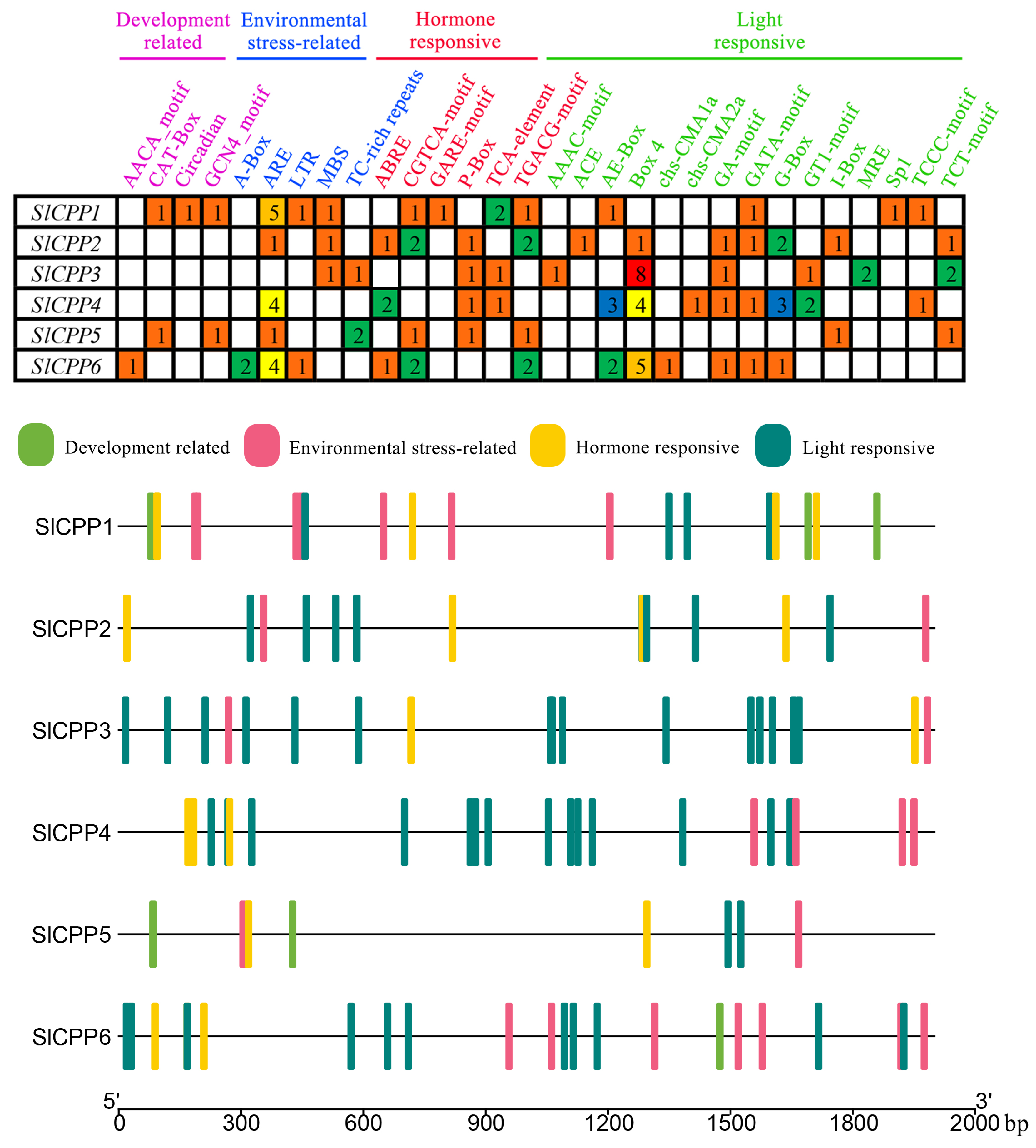
2.4. Detection of Cis-Acting Elements in the Promoter Regions of SlCPPs
2.5. Prediction of the Tertiary Structure of SlCPP Proteins
2.6. Expression Patterns of SlCPP Genes Revealed by Transcriptome Analysis
2.7. Expression Profiles of SlCPP Genes Analyzed by qRT–PCR
2.8. Gene Silencing of SlCPP3 Reduces Drought Resistance in Tomato
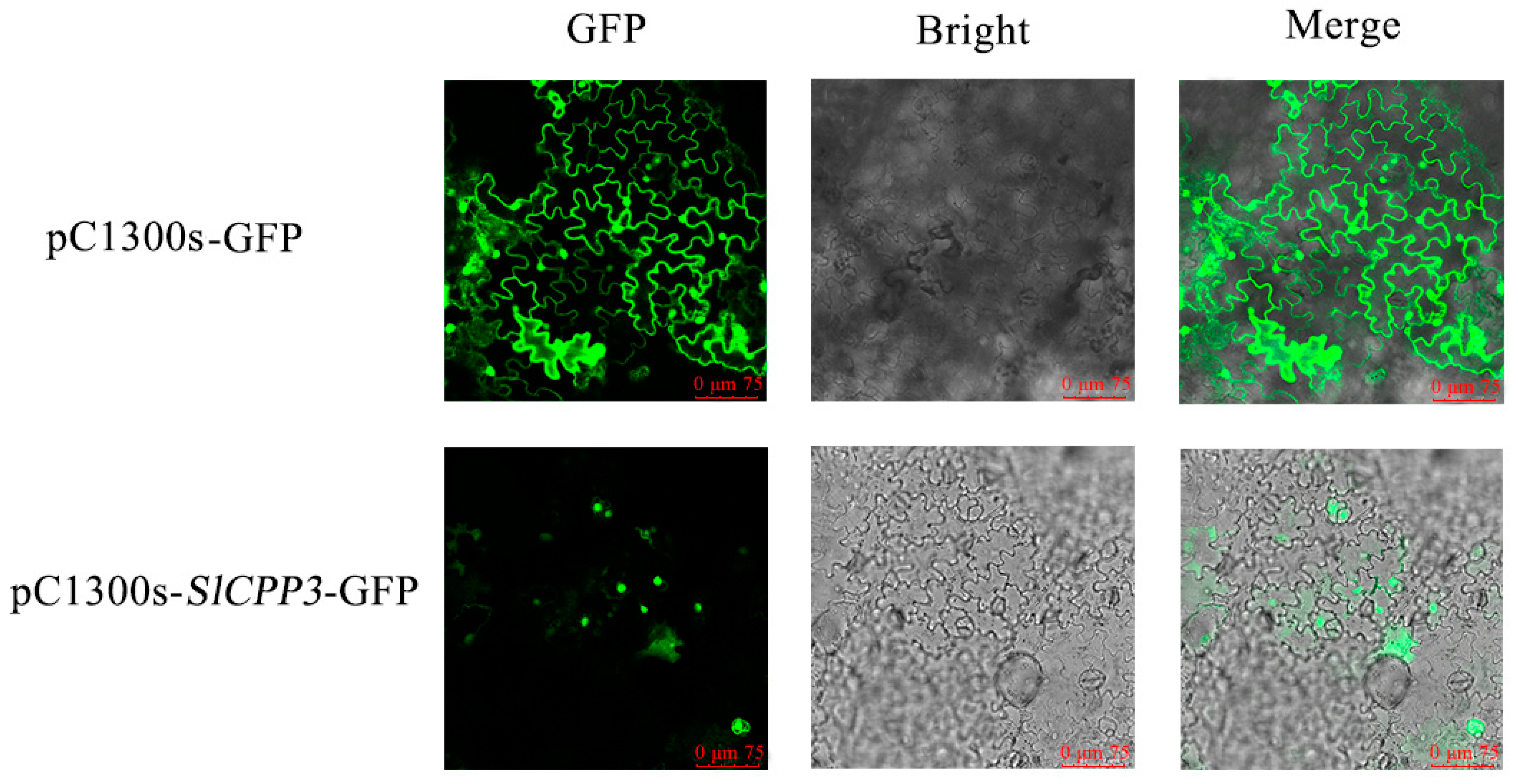
2.9. Expression of SlCPP3 in the Nucleus
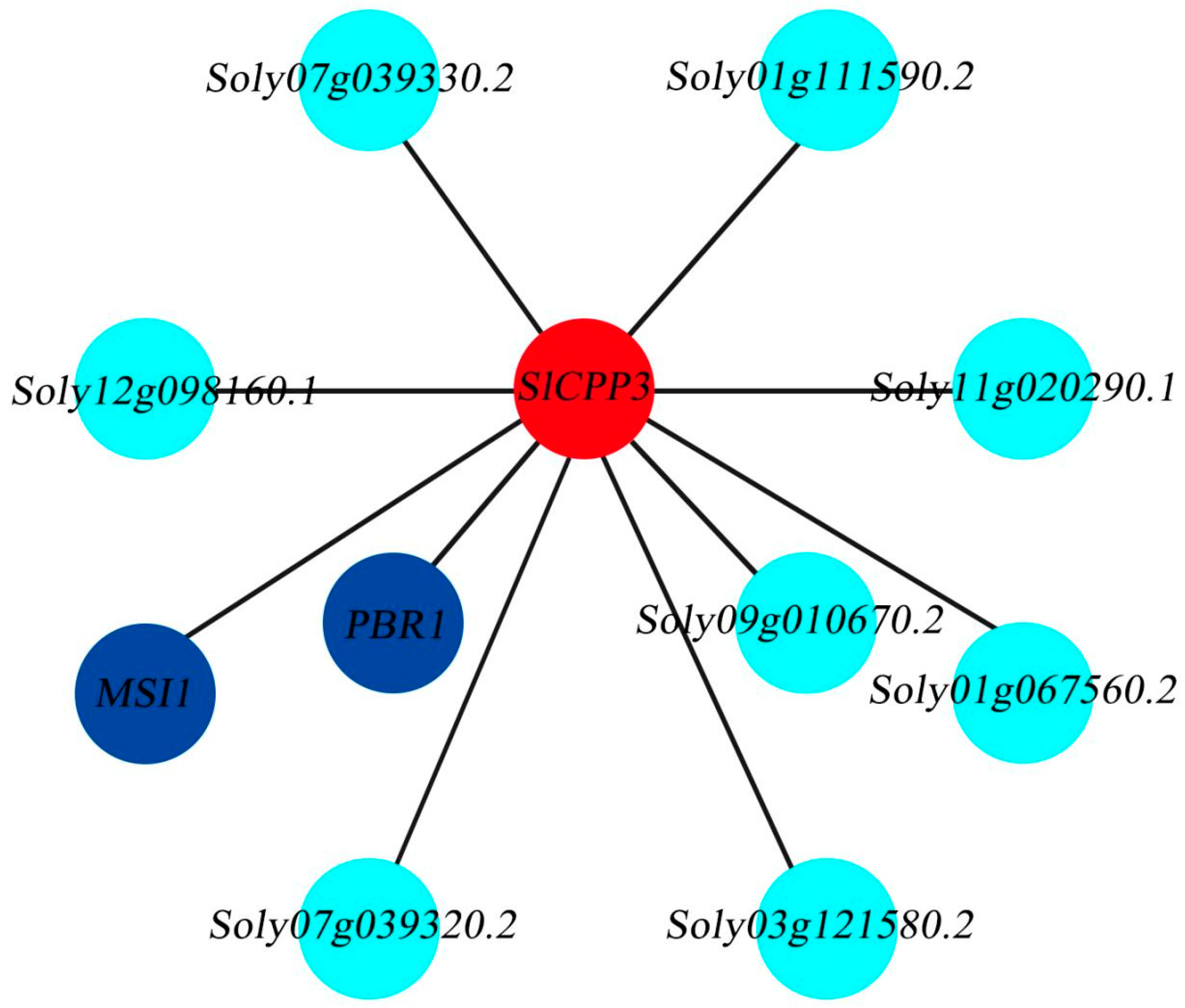
2.10. Analysis of SlCPP3 Gene Expression Network
3. Discussion
3.1. Identification and Physicochemical Properties of CPP Genes in Tomato
3.2. Distribution of the CPP Gene Family in Plants
3.3. Plant Evolutionary Relationships and Genetic Structure
3.4. Gene Duplication Events and Synteny Relationships
3.5. The Promoters of SlCPP Genes Contain Many Cis-Acting Elements
3.6. Tertiary Structure of SlCPP Proteins
3.7. Transcriptomics Combined with qRT–PCR Reveals the Expression Profile of SlCPP Genes in Tomato
3.8. The Role of SlCPP3 Gene in Abiotic and Biotic Stress
4. Materials and Methods
4.1. Plant Growth and Treatments
4.2. Identification of CPP Gene Family Members in Tomato
4.3. Bioinformatics Analysis of CPP Gene Family in Tomato
4.4. Tertiary Structure Prediction of SlCPP Proteins
4.5. Analysis of SlCPP Expression Patterns Based on Transcriptomic Data
4.6. qRT–PCR Analysis of SlCPPs Expression
4.7. VIGS Vector Construction and Agroinfiltration
4.8. Observation of Stained Tissue
4.9. Subcellular Localization of SlCPP3
4.10. Analysis of the Expression Network of SlCPP3 in Tomato
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergougnoux, V. The history of tomato: From domestication to biopharming. Biotechnol. Adv. 2014, 32, 170–189. [Google Scholar] [CrossRef]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.F.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef]
- Hoshikawa, K.; Pham, D.; Ezura, H.; Schafleitner, R.; Nakashima, K. Genetic and Molecular Mechanisms Conferring Heat Stress Tolerance in Tomato Plants. Front. Plant Sci. 2021, 12, e786688. [Google Scholar] [CrossRef]
- Yin, M.; Wang, Y.; Zhang, L.; Li, J.; Quan, W.; Yang, L.; Wang, Q.; Chan, Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017, 68, 2991–3005. [Google Scholar] [CrossRef]
- Hamann, E.; Denney, D.; Day, S.; Lombardi, E.; Jameel, M.I.; MacTavish, R.; Anderson, J.T. Review: Plant eco-evolutionary responses to climate change: Emerging directions. Plant Sci. 2021, 304, e110737. [Google Scholar] [CrossRef]
- Albacete, A.A.; Martínez-Andújar, C.; Pérez-Alfocea, F. Hormonal and metabolic regulation of source-sink relations under salinity and drought: From plant survival to crop yield stability. Biotechnol. Adv. 2014, 32, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Singh, G.; Bhattacharya, S.; Singh, A. Comparative transcriptome meta-analysis of Arabidopsis thaliana under drought and cold stress. PLoS ONE 2018, 13, e0203266. [Google Scholar] [CrossRef]
- Muhammad, A.M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Xue, M.; Zhang, H.; Zhao, F.; Zhao, T.; Li, H.; Jiang, D. The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A.Z eviction and active transcription in Arabidopsis. Mol. Plant 2021, 14, 1799–1813. [Google Scholar] [CrossRef]
- Kahlon, P.S.; Stam, R. Polymorphisms in plants to restrict losses to pathogens: From gene family expansions to complex network evolution. Curr. Opin. Plant Biol. 2021, 62, e102040. [Google Scholar] [CrossRef]
- Dong, L.; Huo, N.; Wang, Y.; Deal, K.; Wang, D.; Hu, T.; Dvorak, J.; Anderson, O.D.; Luo, M.C.; Gu, Y.Q. Rapid evolutionary dynamics in a 2.8-Mb chromosomal region containing multiple prolamin and resistance gene families in Aegilops tauschii. Plant J. 2016, 87, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hu, L.; Ye, S.; Jiang, L.; Liu, S. Genome-wide identification and characterization of cysteine-rich polycomb-like protein (cpp) family genes in cucumber (cucumis sativus) and their roles in stress responses. Biologia 2018, 73, 425–435. [Google Scholar] [CrossRef]
- Wang, K. Bioinformatic Analysis of the CPP Transcription Factors Family in Arabidopsis and Rice. Biotechnol. Bull. 2010, 20, 76–84. [Google Scholar]
- Pan, R.R.; Wei, M.M.; Wang, Y.J.; Hu, X.X.; Li, W.G. Cloning and expression analysis of HbCCP1 in rubber tree(Hevea brasiliensis). Plant Physiol. J. 2018, 54, 763–772. [Google Scholar]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Yang, Z.; Gu, S.; Wang, X.; Li, W.; Tang, Z.; Xu, C. Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. J. Mol. Evol. 2008, 67, 266–277. [Google Scholar] [CrossRef]
- Song, X.Y.; Zhang, Y.Y.; Wu, F.C.; Zhang, L. Genome-wide analysis of the maize (Zea may L.) CPP-like gene family and expression profiling under abiotic stress. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, H.K.; Wang, Y.M.; Yuan, C.P.; Zhang, Y.Y.; Li, H.Y.; Yan, X.F.; Li, Q.Y.; Dong, Y.S. Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genet. Mol. Res. 2015, 14, 1260–1268. [Google Scholar] [CrossRef]
- Hauser, B.A.; Villanueva, J.M.; Gasser, C.S. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics 1998, 150, 411–423. [Google Scholar] [CrossRef]
- Hauser, B.A.; He, J.Q.; Park, S.O.; Gasser, C.S. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 2000, 127, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.U.; Algreen-Petersen, R.G.; Hoedl, M.; Jurkiewicz, A.; Cvitanich, C.; Braunschweig, U.; Schauser, L.; Oh, S.A.; Twell, D.; Jensen, E.O. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3657–3670. [Google Scholar] [CrossRef]
- Sijacic, P.; Wang, W.; Liu, Z. Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 2011, 7, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sijacic, P.; Xu, P.; Lian, H.; Liu, Z. Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc. Natl. Acad. Sci. USA 2018, 115, E3045–E3054. [Google Scholar] [CrossRef]
- Cvitanich, C.; Pallisgaard, N.; Nielsen, K.A.; Hansen, A.C.; Larsen, K.; Pihakaski-Maunsbach, K.; Marcker, K.A.; Jensen, E.O. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc. Natl. Acad. Sci. USA 2000, 97, 8163–8168. [Google Scholar] [CrossRef]
- Zhao, T.; Schranz, M.E. Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 2017, 36, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Barthelson, R.; McFarlin, A.J.; Rounsley, S.D.; Young, S. Plantagora: Modeling whole genome sequencing and assembly of plant genomes. PLoS ONE 2011, 6, e28436. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; Geest, H.V.D.; Maumus, F.; Saha, S. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. BioRxiv 2019, 767764. [Google Scholar]
- Xiong, Q.; Jiang, X.; Liu, X.; Zhou, P.; Ding, K. Prediction of IER5 structure and function using a bioinformatics approach. Mol. Med. Rep. 2019, 19, 4631–4636. [Google Scholar] [CrossRef]
- Nan, H.; Lin, Y.; Wang, X.; Gao, L. Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Hortic 2021, 7, 469–478. [Google Scholar] [CrossRef]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2012, 2, 119. [Google Scholar] [CrossRef]
- Janies, D.; DeSalle, R. Development, evolution, and corroboration. Anat. Rec. 1999, 257, 6–14. [Google Scholar] [CrossRef]
- Jeong, Y.M.; Mun, J.H.; Lee, I.; Woo, J.C.; Hong, C.B.; Kim, S.G. Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol. 2006, 140, 196–209. [Google Scholar] [CrossRef]
- Keidar, D.; Doron, C.; Kashkush, K. Genome-wide analysis of a recently active retrotransposon, Au SINE, in wheat: Content, distribution within subgenomes and chromosomes, and gene associations. Plant Cell Rep. 2018, 37, 193–208. [Google Scholar] [CrossRef]
- Bandyopadhyay, D.; Huan, J.; Liu, J.; Prins, J.; Snoeyink, J.; Wang, W.; Tropsha, A. Functional neighbors: Inferring relationships between nonhomologous protein families using family-specific packing motifs. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Feng, G.; Yang, Z.; Xu, X.; Huang, L.; Yang, Q.; Zhang, X. Genome-wide AP2/ERF gene family analysis reveals the classification, structure, expression profiles and potential function in orchardgrass (Dactylis glomerata). Mol. Biol. Rep. 2020, 47, 5225–5241. [Google Scholar] [CrossRef] [PubMed]
- Tsitsekian, D.; Daras, G.; Alatzas, A.; Templalexis, D.; Hatzopoulos, P.; Rigas, S. Comprehensive analysis of Lon proteases in plants highlights independent gene duplication events. J. Exp. Bot. 2019, 70, 2185–2197. [Google Scholar] [CrossRef]
- Soltis, D.E.; Ma, H.; Frohlich, M.W.; Soltis, P.S.; Albert, V.A.; Oppenheimer, D.G.; Altman, N.S.; dePamphilis, C.; Leebens-Mack, J. The floral genome: An evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 2007, 12, 358–367. [Google Scholar] [CrossRef]
- Van-de-Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Moghe, G.D.; Shiu, S.H. The causes and molecular consequences of polyploidy in flowering plants. Ann. N. Y. Acad. Sci. 2014, 1320, 16–34. [Google Scholar] [CrossRef]
- Huang, Z.; Van-Houten, J.; Gonzalez, G.; Xiao, H.; van-der-Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013, 288, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Kucho, K.; Yoshioka, S.; Taniguchi, F.; Ohyama, K.; Fukuzawa, H. Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 2003, 133, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhong, Y.; Zhou, W. Molecular mechanisms of mesocotyl elongation induced by brassinosteroid in maize under deep-seeding stress by RNA-sequencing, microstructure observation, and physiological metabolism. Genomics 2021, 113, 3565–3581. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.Y.; Ji, X.R.; Guo, D.L. Genome-wide identification, characterization, and expression analysis of GDSL-type esterases/lipases gene family in relation to grape berry ripening. Sci. Hortic. 2020, 264, e109162. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Skolnick, J.; Gao, M.; Zhou, H.; Singh, S. AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function. J. Chem. Inf. Model. 2021, 61, 4827–4831. [Google Scholar] [CrossRef]
- David, A.; Islam, S.; Tankhilevich, E.; Sternberg, M.J.E. The AlphaFold Database of Protein Structures: A Biologist’s Guide. J. Mol. Biol. 2022, 434, e167336. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Zia, R.; Nawaz, M.S.; Siddique, M.J.; Hakim, S.; Imran, A. Plant survival under drought stress: Implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiol. Res. 2021, 242, e126626. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of Stomatal Closure in Plants Exposed to Drought and Cold Stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar] [PubMed]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Grüttner, C.; Schnittger, A. Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet. 2018, 14, e1007797. [Google Scholar] [CrossRef]
- Alexandre, C.; Möller-Steinbach, Y.; Schönrock, N.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant 2009, 2, 675–687. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zhao, S.; Liu, J.F.; Zhao, H.Y.; Sun, X.Y.; Wu, T.R.; Pei, T.; Wang, Y.; Liu, Q.F.; Yang, H.H.; et al. Genome-wide identification of Tomato Golden 2-Like transcription factors and abiotic stress related members screening. BMC Plant Biol. 2022, 22, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wu, T.; Zhang, J.; Wang, Z.; Pei, T.; Yang, H.; Li, J.; Xu, X. Genome-Wide Analyses of the Genetic Screening of C2H2-Type Zinc Finger Transcription Factors and Abiotic and Biotic Stress Responses in Tomato (Solanum lycopersicum) Based on RNA-Seq Data. Front. Genet. 2020, 11, 540. [Google Scholar] [CrossRef]
- Zhang, D.; Bao, Y.; Sun, Y.; Yang, H.; Zhao, T.; Li, H.; Du, C.; Jiang, J.; Li, J.; Xie, L.; et al. Comparative transcriptome analysis reveals the response mechanism of Cf-16-mediated resistance to Cladosporium fulvum infection in tomato. BMC Plant Biol. 2020, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhao, T.; Jiang, J.; Chen, X.; Zhang, H.; Liu, G.; Zhang, D.; Du, C.; Wang, S.; Xu, X.; et al. Transcriptome Analysis of the Sm-Mediated Hypersensitive Response to Stemphylium lycopersici in Tomato. Front. Plant Sci. 2017, 8, 1257. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef]
- Yang, H.; Sun, Y.; Wang, H.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against Stemphylium lycopersici in tomato. BMC Plant Biol. 2021, 21, 72. [Google Scholar] [CrossRef]
- Sun, Y.; He, Y.; Wang, H.; Jiang, J.; Yang, H.; Xu, X. Genome-wide identification and expression analysis of GDSL esterase/lipase genes in tomato. J. Integr. Agric. 2022, 21, 389–406. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Chakravarthy, S.; Martin, G.B. Virus-induced Gene Silencing (VIGS) in Nicotiana benthamiana and Tomato. J. Vis. Exp. 2009, 28, e1292. [Google Scholar] [CrossRef]
- Kumar, D.; Yusuf, M.A.; Singh, P.; Sardar, M.; Sarin, N.B. Histochemical detection of superoxide and H2O2 accumulation in brassica juncea seedlings. Bio-Protoc. 2014, 4, e1108. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Jia, X.; Chen, D.; Fu, Q.; Chen, J.; Yang, W.; Yang, H.; Xu, X. Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato. Int. J. Mol. Sci. 2023, 24, 5762. https://doi.org/10.3390/ijms24065762
Sun Y, Jia X, Chen D, Fu Q, Chen J, Yang W, Yang H, Xu X. Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato. International Journal of Molecular Sciences. 2023; 24(6):5762. https://doi.org/10.3390/ijms24065762
Chicago/Turabian StyleSun, Yaoguang, Xinyi Jia, Dexia Chen, Qingjun Fu, Jinxiu Chen, Wenhui Yang, Huanhuan Yang, and Xiangyang Xu. 2023. "Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato" International Journal of Molecular Sciences 24, no. 6: 5762. https://doi.org/10.3390/ijms24065762
APA StyleSun, Y., Jia, X., Chen, D., Fu, Q., Chen, J., Yang, W., Yang, H., & Xu, X. (2023). Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato. International Journal of Molecular Sciences, 24(6), 5762. https://doi.org/10.3390/ijms24065762





