Genome-Wide Analysis of Long Non-Coding RNAs Related to UV-B Radiation in the Antarctic Moss Pohlia nutans
Abstract
1. Introduction
2. Results
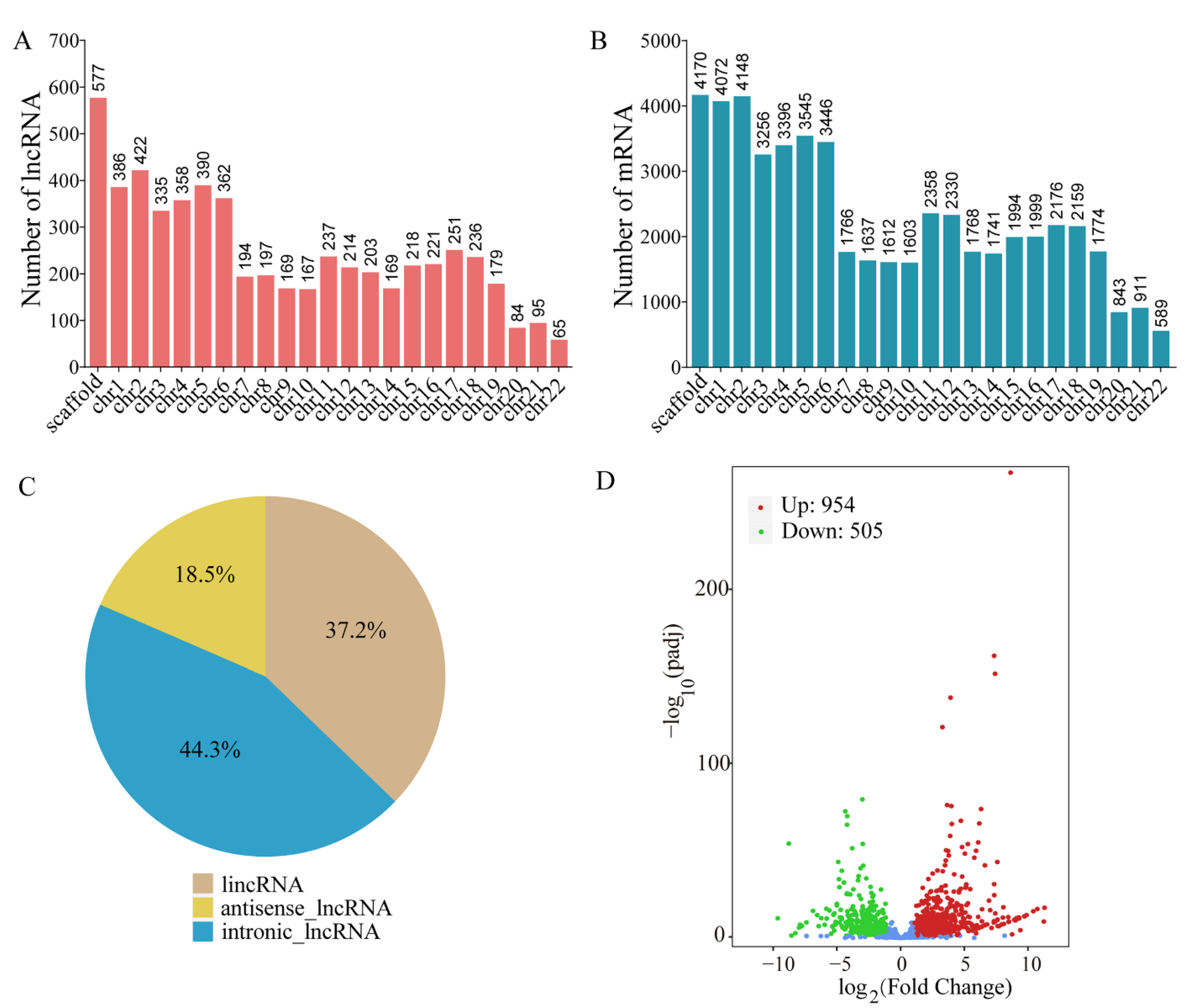
2.1. Identification and Characterization of lncRNAs in Pohlia nutans
2.2. Prediction, Classification and Expression Profiles of lncRNAs in Pohlia nutans
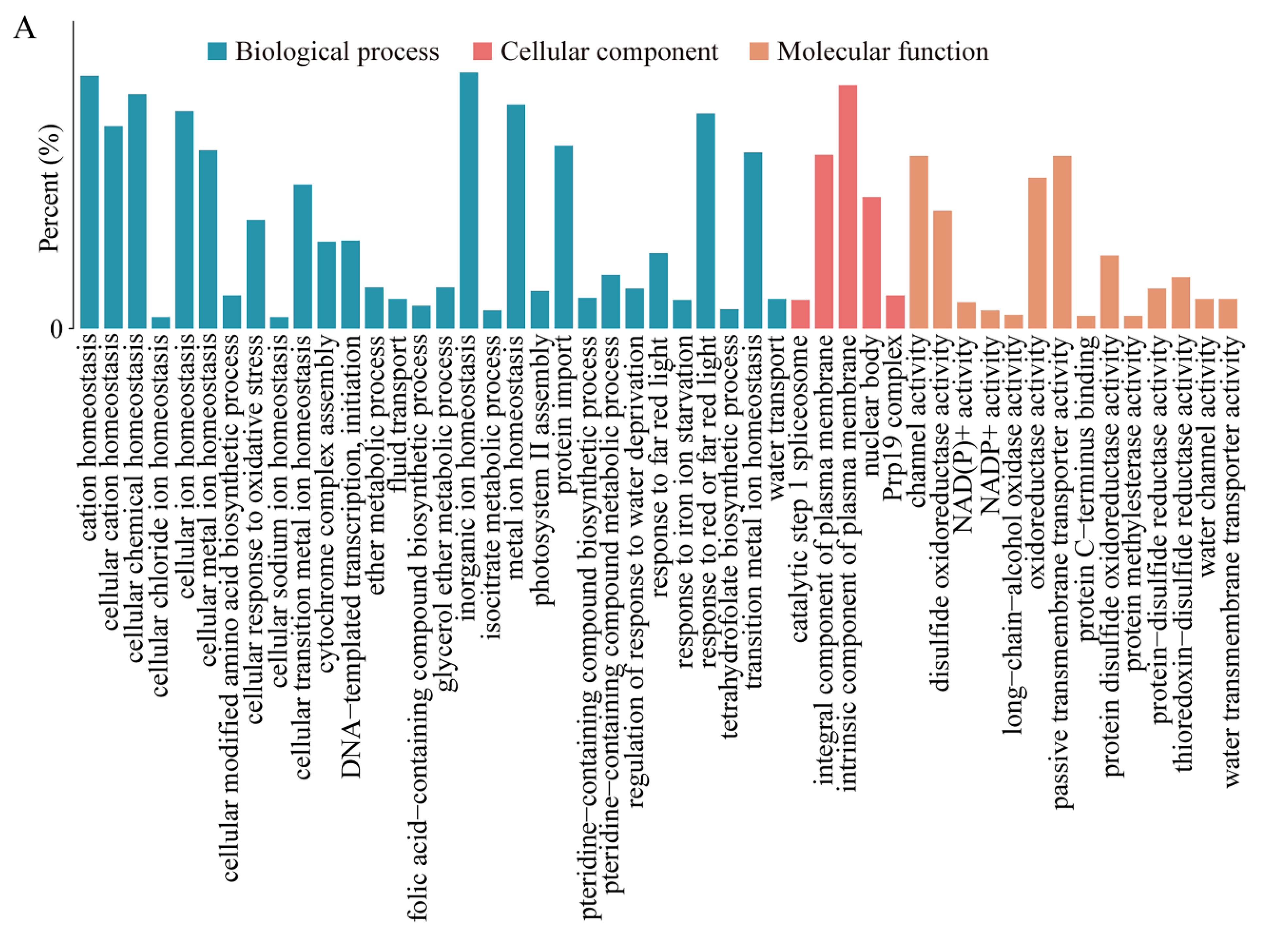
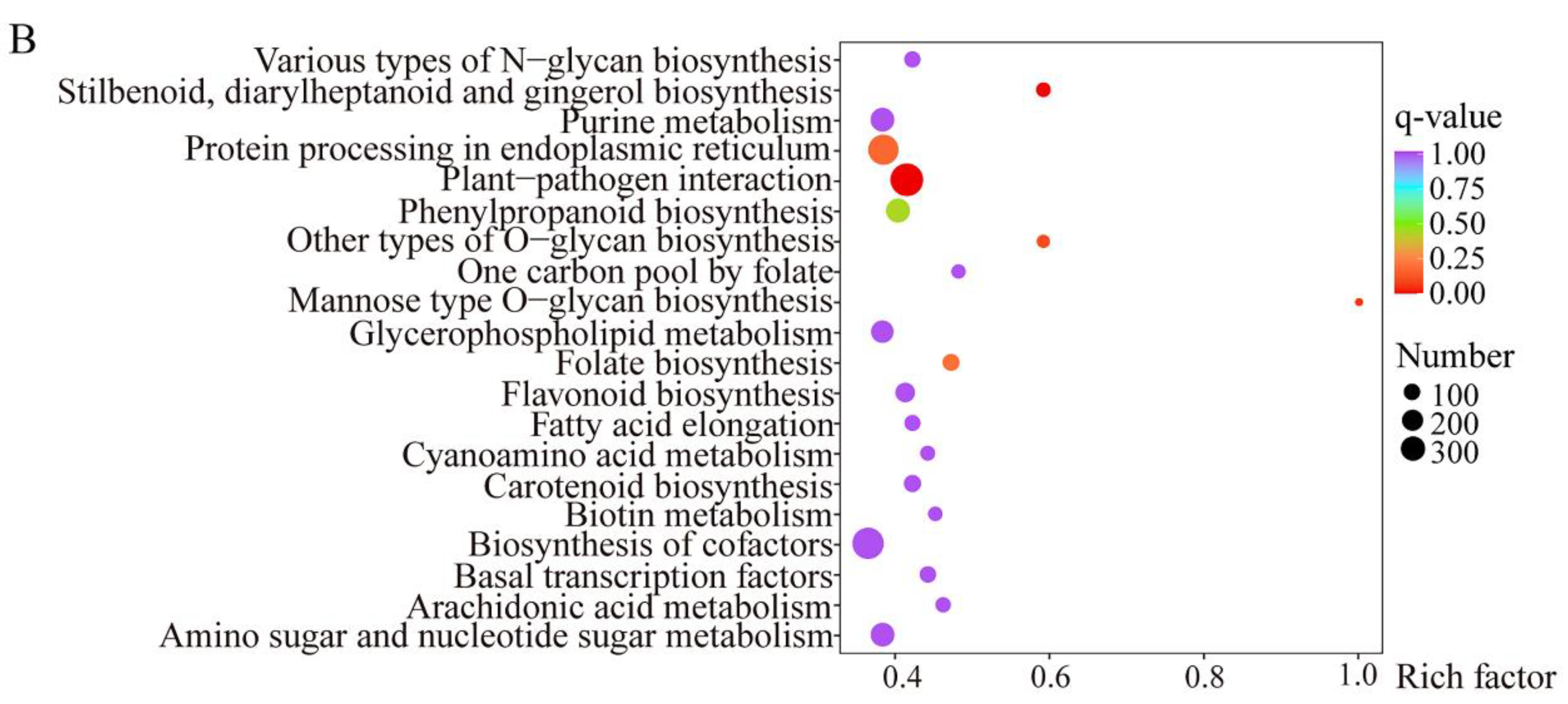
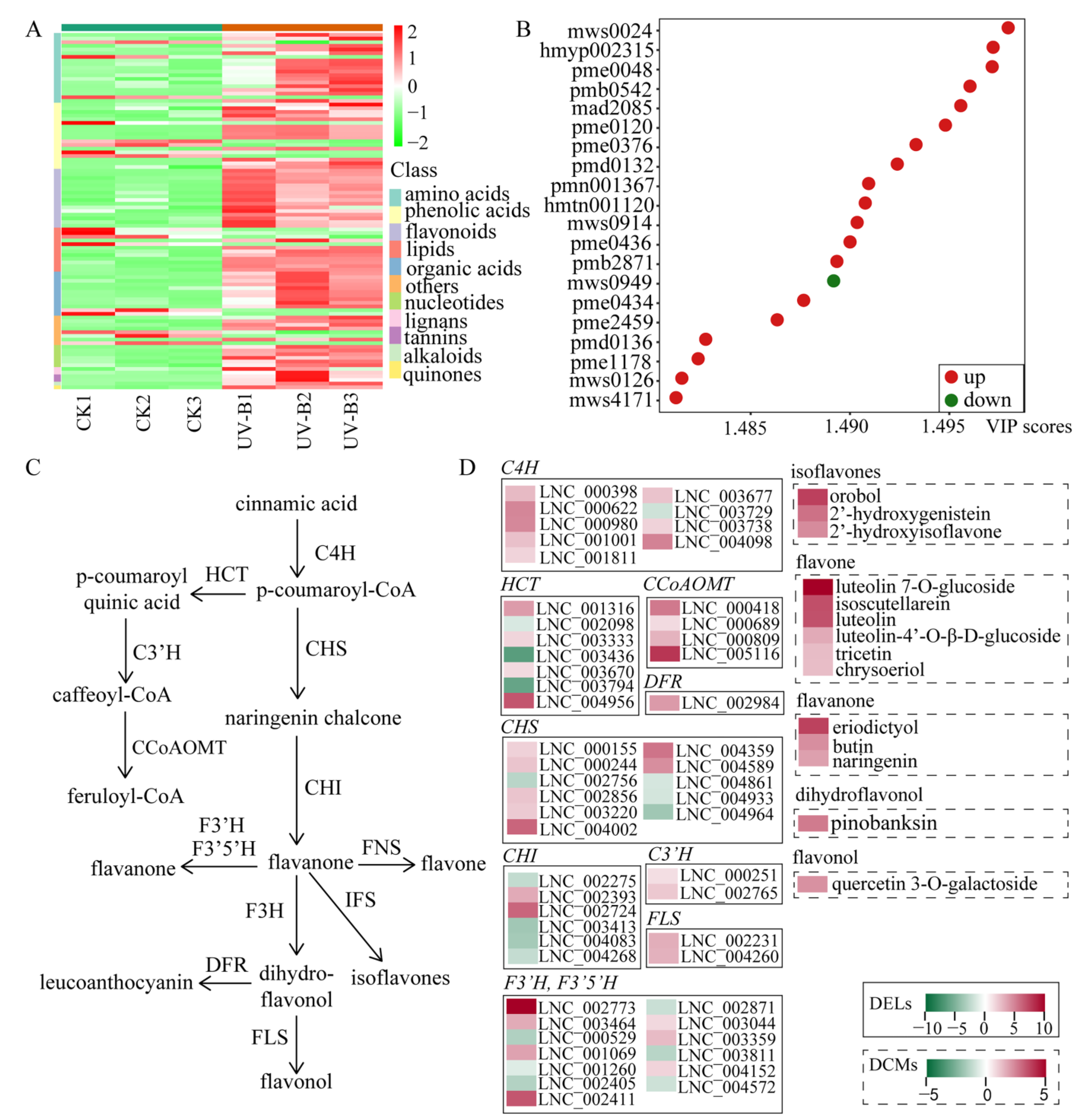
2.3. Functional Annotation of Cis- and Trans-Target Genes Involved in Flavonoid Biosynthesis and Plant-Pathogen Interaction Pathways
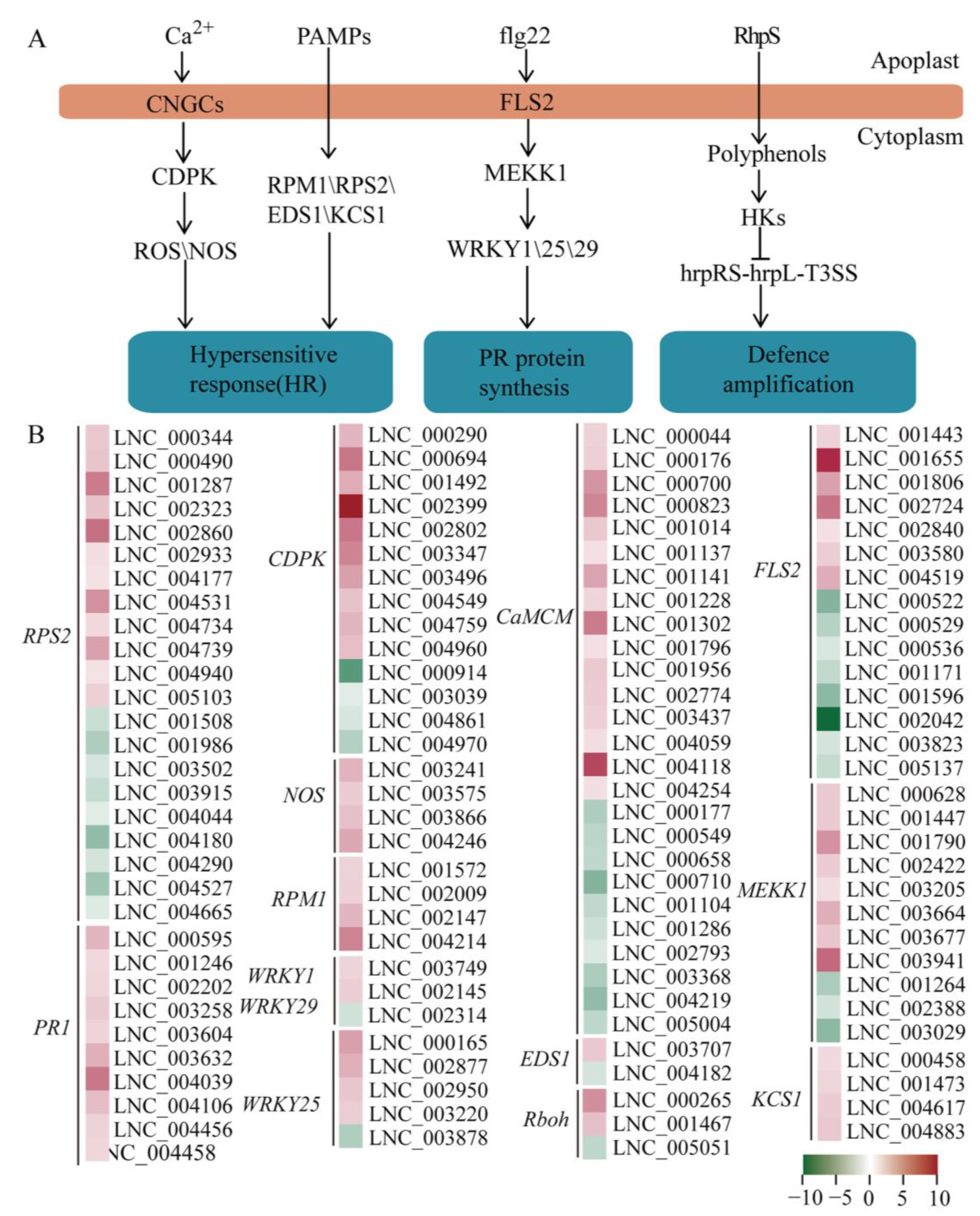
2.4. DELs in Plant-Pathogen Interaction
2.5. Integrated Multi-Omics Analyses Highlight the Role of the Flavonoid Biosynthesis Pathway under UV-B Radiation
3. Discussion
4. Experimental Procedures
4.1. Plant Samples and UV-B Radiation Treatments
4.2. RNA Extraction and Library Construction
4.3. Read Mapping to the Reference Genome
4.4. Differential Expression Analysis and Functional Annotation
4.5. Widely Targeted Metabolomics Analysis
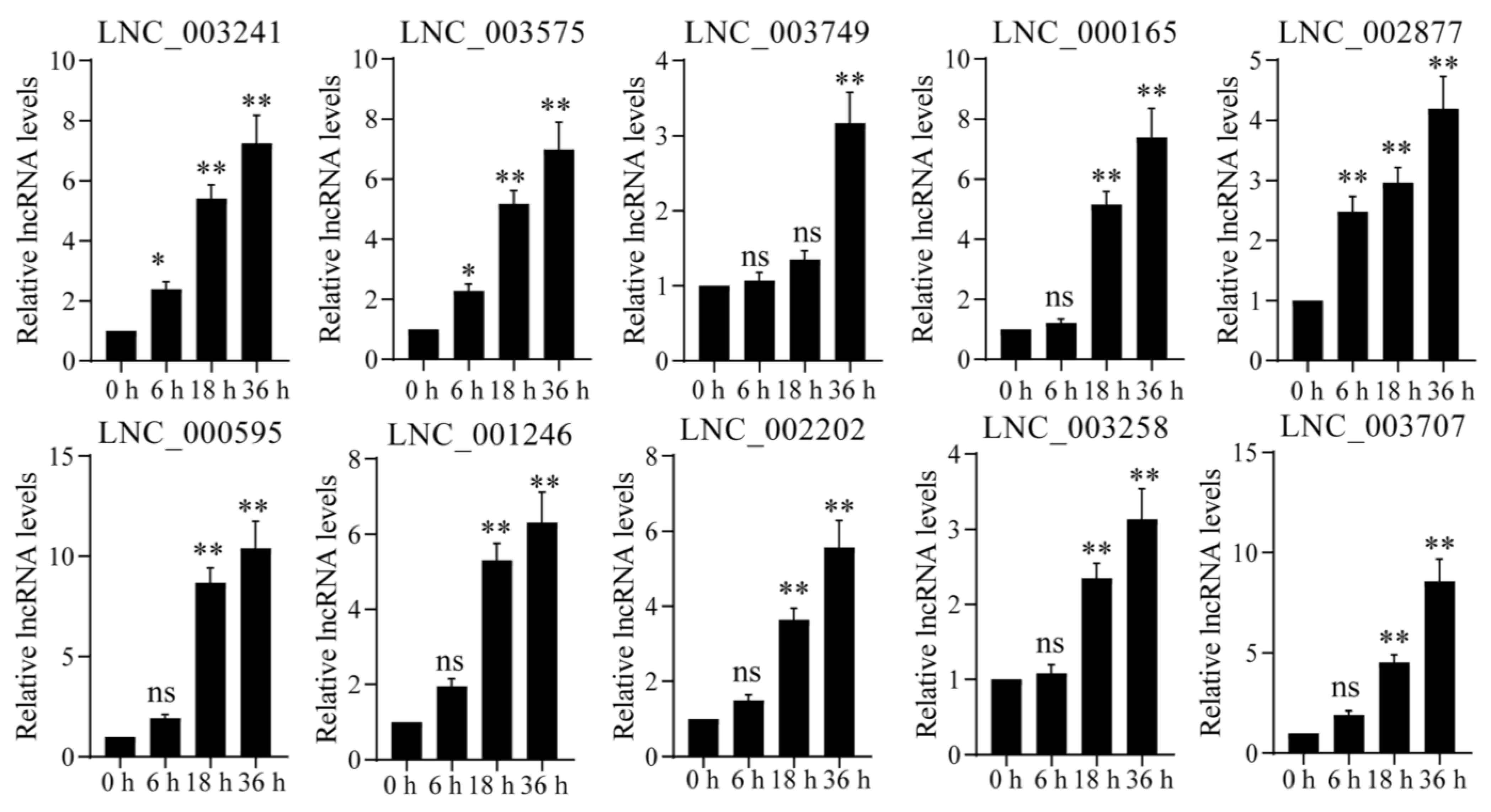
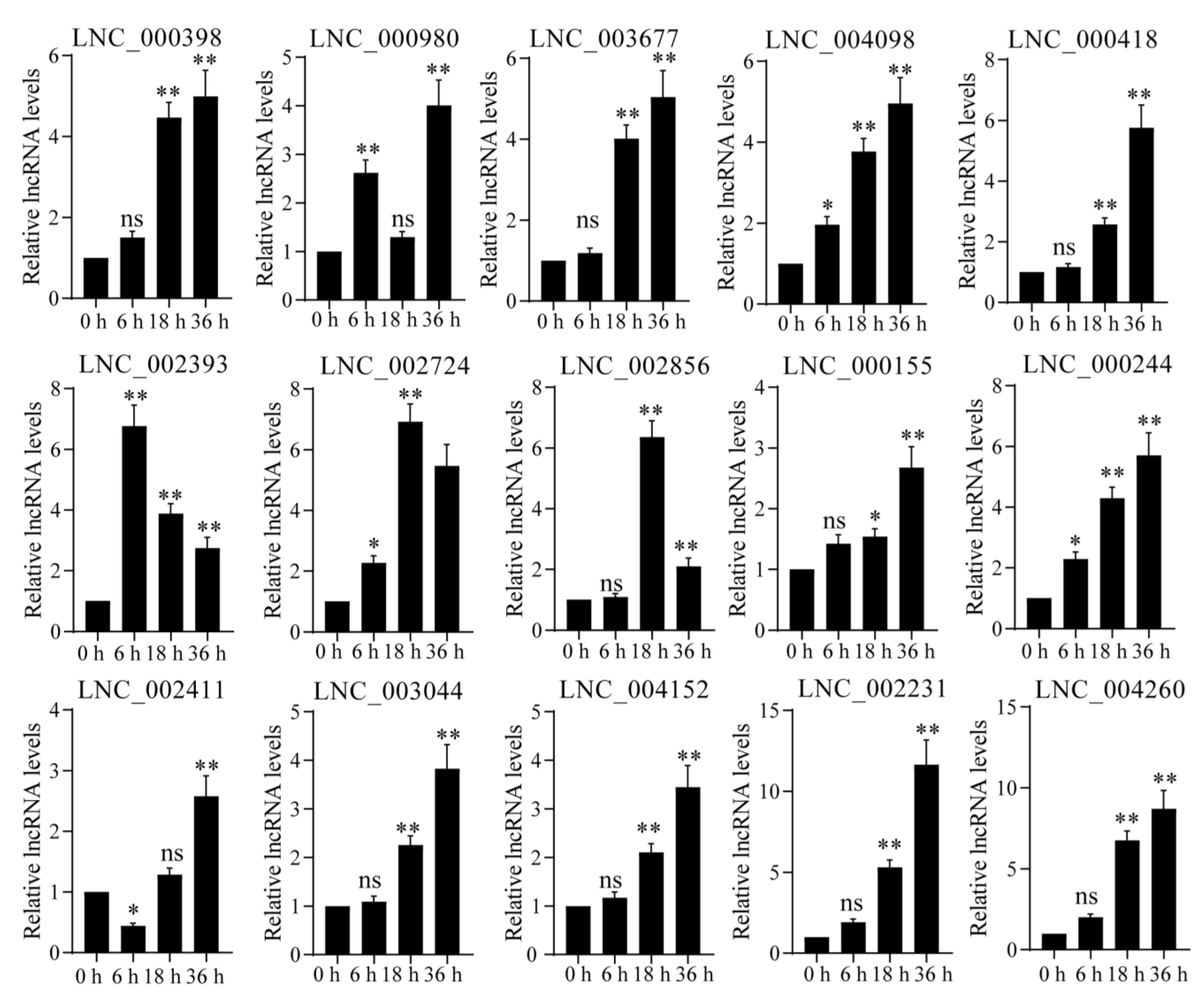
4.6. Quantitative Real-Time RT-PCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Escobar-Bravo, R.; Nederpel, C.; Naranjo, S.; Kim, H.K.; Rodríguez-López, M.J.; Chen, G.; Glauser, G.; Leiss, K.A.; Klinkhamer, P.G.L. Ultraviolet radiation modulates both constitutive and inducible plant defenses against thrips but is dose and plant genotype dependent. J. Pest Sci. 2021, 94, 69–81. [Google Scholar]
- Zedek, F.; Šmerda, J.; Veselý, P.; Horová, L.; Kocmanová, J.; Bureš, P. Elevation-dependent endopolyploid response suggests that plants with holocentric chromosomes are less stressed by UV-B. Bot. J. Linn. Soc. 2020, 195, 106–113. [Google Scholar] [CrossRef]
- Shi, C.; Liu, H. How plants protect themselves from ultraviolet-B radiation stress. Plant Physiol. 2021, 187, 1096–1103. [Google Scholar]
- Xu, C.; Sullivan, J.H. Reviewing the technical designs for experiments with ultraviolet-B radiation and impact on photosynthesis, DNA and secondary metabolism. J. Integr. Plant Biol. 2010, 52, 377–387. [Google Scholar]
- McKenzie, R.L.; Aucamp, P.J.; Bais, A.F.; Björn, L.O.; Ilyas, M. Changes in biologically-active ultraviolet radiation reaching the Earth’s surface. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2007, 6, 218–231. [Google Scholar] [CrossRef]
- Bais, A.F.; Bernhard, G.; McKenzie, R.L.; Aucamp, P.J.; Young, P.J.; Ilyas, M.; Jöckel, P.; Deushi, M. Ozone-climate interactions and effects on solar ultraviolet radiation. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2019, 18, 602–640. [Google Scholar]
- Damany-Pearce, L.; Johnson, B.; Wells, A.; Osborne, M.; Allan, J.; Belcher, C.; Jones, A.; Haywood, J. Australian wildfires cause the largest stratospheric warming since Pinatubo and extends the lifetime of the Antarctic ozone hole. Sci. Rep. 2022, 12, 12665. [Google Scholar]
- Barnes, P.W.; Robson, T.M.; Neale, P.J.; Williamson, C.E.; Zepp, R.G.; Madronich, S.; Wilson, S.R.; Andrady, A.L.; Heikkilä, A.M.; Bernhard, G.H.; et al. Environmental effects of stratospheric ozone depletion, UV radiation, and interactions with climate change: UNEP Environmental Effects Assessment Panel, Update 2021. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2022, 21, 275–301. [Google Scholar]
- Wierzbicki, A.T.; Blevins, T.; Swiezewski, S. Long Noncoding RNAs in Plants. Annu. Rev. Plant Biol. 2021, 72, 245–271. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Q.H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Lim, D.A. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018, 19, e46955. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.V.; Chekanova, J.A. Long Noncoding RNAs in Plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar] [PubMed]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen, J.; Mysore, K.S.; Zhang, W.H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Singh, M.B.; Bhalla, P.L. The Long Intergenic Noncoding RNA (LincRNA) Landscape of the Soybean Genome. Plant Physiol. 2018, 176, 2133–2147. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, X.; Traore, S.M.; Xin, Z.; Ning, L.; Li, K.; Zhao, K.; Li, Z.; He, G.; Yin, D. Genome-wide identification and analysis of long noncoding RNAs (lncRNAs) during seed development in peanut (Arachis hypogaea L.). BMC Plant Biol. 2020, 20, 192. [Google Scholar] [CrossRef]
- Liu, H.; Wang, R.; Mao, B.; Zhao, B.; Wang, J. Identification of lncRNAs involved in rice ovule development and female gametophyte abortion by genome-wide screening and functional analysis. BMC Genom. 2019, 20, 90. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, X.; Lin, F.; He, G.; Terzaghi, W.; Zhu, D.; Deng, X.W. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 2014, 111, 10359–10364. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution expression map of the arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Lu, Q.; Xu, Q.; Guo, F.; Lv, Y.; Song, C.; Feng, M.; Yu, J.; Zhang, D.; Cang, J. Identification and characterization of long non-coding RNAs as competing endogenous RNAs in the cold stress response of Triticum aestivum. Plant Biol. 2020, 22, 635–645. [Google Scholar] [CrossRef]
- Huo, C.; Zhang, B.; Wang, R. Research progress on plant noncoding RNAs in response to low-temperature stress. Plant Signal. Behav. 2022, 17, 2004035. [Google Scholar] [CrossRef] [PubMed]
- Gelaw, T.A.; Sanan-Mishra, N. Non-Coding RNAs in response to drought stress. Int. J. Mol. Sci. 2021, 22, 12519. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Zhu, R.; Wang, P.; Ye, W.; Ma, D.; Xu, H. Potential effects of ultraviolet radiation reduction on tundra nitrous oxide and methane fluxes in maritime Antarctica. Sci. Rep. 2018, 8, 3716. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Gautam, S.; Bhushan Pant, A. Effect of UV-B radiation on UV absorbing compounds and pigments of moss and lichen of Schirmacher oasis region, East Antarctica. Cell. Mol. Biol. 2012, 58, 80–84. [Google Scholar]
- Singh, J.; Singh, R.P. Adverse effects of UV-B radiation on plants growing at Schirmacher Oasis, East Antarctica. Toxicol. Int. 2014, 21, 101–106. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Chollet-Krugler, M.; Gouault, N.; Tomasi, S. UV-protectant metabolites from lichens and their symbiotic partners. Nat. Prod. Rep. 2013, 30, 1490–1508. [Google Scholar] [CrossRef]
- Liu, S.; Fang, S.; Liu, C.; Zhao, L.; Cong, B.; Zhang, Z. Transcriptomics integrated with metabolomics reveal the effects of Ultraviolet-B radiation on flavonoid biosynthesis in Antarctic moss. Front. Plant Sci. 2021, 12, 788377. [Google Scholar] [CrossRef] [PubMed]
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 5, eaaz0888. [Google Scholar] [CrossRef] [PubMed]
- Day, T.A.; Ruhland, C.T.; Xiong, F.S. Influence of solar ultraviolet-B radiation on Antarctic terrestrial plants: Results from a 4-year field study. J. Photochem. Photobiol. B Biol. 2001, 62, 78–87. [Google Scholar] [CrossRef]
- Waterman, M.J.; Bramley-Alves, J.; Miller, R.E.; Keller, P.A.; Robinson, S.A. Photoprotection enhanced by red cell wall pigments in three East Antarctic mosses. Biol. Res. 2018, 51, 49. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Mewis, I.; Schreiner, M.; Nguyen, C.N.; Krumbein, A.; Ulrichs, C.; Lohse, M.; Zrenner, R. UV-B irradiation changes specifically the secondary metabolite profile in broccoli sprouts: Induced signaling overlaps with defense response to biotic stressors. Plant Cell Physiol. 2012, 53, 1546–1560. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Takshak, S.; Agrawal, S.B. Defense potential of secondary metabolites in medicinal plants under UV-B stress. J. Photochem. Photobiol. B Biol. 2019, 193, 51–88. [Google Scholar] [CrossRef]
- Xie, Y.; Ding, Y.; Shao, X.; Yao, C.; Li, J.; Liu, J.; Deng, X. Pseudomonas syringae senses polyphenols via phosphorelay crosstalk to inhibit virulence. EMBO Rep. 2021, 22, e52805. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.R.; Feron, S.; Damiani, A.; Redondas, A.; Carrasco, J.; Sepúlveda, E.; Jorquera, J.; Fernandoy, F.; Llanillo, P.; Rowe, P.M.; et al. Persistent extreme ultraviolet irradiance in Antarctica despite the ozone recovery onset. Sci. Rep. 2022, 12, 1266. [Google Scholar] [CrossRef] [PubMed]
- Heijde, M.; Ulm, R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012, 17, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Singh, D.; Lingwan, M.; Yadukrishnan, P.; Masakapalli, S.K.; Datta, S. Light signaling and UV-B-mediated plant growth regulation. J. Integr. Plant Biol. 2020, 62, 1270–1292. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long non-coding RNA in plants in the era of reference sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Alptekin, B.; Budak, H. RNA Sequencing and Co-expressed Long Non-coding RNA in Modern and Wild Wheats. Sci. Rep. 2017, 7, 10670. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Muslu, T.; Reddy, G.V.P.; Dogramaci, M.; Budak, H. Wheat long noncoding RNAs from organelle and nuclear genomes carry conserved microRNA precursors which may together comprise intricate networks in insect responses. Int. J. Mol. Sci. 2023, 24, 2226. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Chen, J.; Zhong, Y.; Qi, X. LncRNA TCONS_00021861 is functionally associated with drought tolerance in rice (Oryza sativa L.) via competing endogenous RNA regulation. BMC Plant Biol. 2021, 21, 410. [Google Scholar] [CrossRef]
- Xu, Y.C.; Zhang, J.; Zhang, D.Y.; Nan, Y.H.; Ge, S.; Guo, Y.L. Identification of long noncoding natural antisense transcripts (lncNATs) correlated with drought stress response in wild rice (Oryza nivara). BMC Genom. 2021, 22, 424. [Google Scholar] [CrossRef]
- Wang, R.; Shu, P.; Zhang, C.; Zhang, J.; Chen, Y.; Zhang, Y.; Du, K.; Xie, Y.; Li, M.; Ma, T.; et al. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 2022, 233, 373–389. [Google Scholar] [CrossRef]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 8249. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Rozema, J.; Björn, L.O.; Bornman, J.F.; Gaberscik, A.; Häder, D.P.; Trost, T.; Germ, M.; Klisch, M.; Gröniger, A.; Sinha, R.P.; et al. The role of UV-B radiation in aquatic and terrestrial ecosystems—An experimental and functional analysis of the evolution of UV-absorbing compounds. J. Photochem. Photobiol. B Biol. 2002, 66, 2–12. [Google Scholar] [CrossRef]
- Brosché, M.; Strid, Å.J.P.P. Molecular events following perception of ultraviolet-B radiation by plants. Physiol. Plant. 2003, 117, 1–10. [Google Scholar] [CrossRef]
- Jenkins, G.I. Signal transduction in responses to UV-B radiation. Annu. Rev. Plant Biol. 2009, 60, 407–431. [Google Scholar] [CrossRef] [PubMed]
- Palma, C.F.F.; Castro-Alves, V.; Morales, L.O.; Rosenqvist, E.; Ottosen, C.O.; Strid, Å. Spectral composition of light affects sensitivity to UV-B and photoinhibition in Cucumber. Front. Plant Sci. 2020, 11, 610011. [Google Scholar] [CrossRef] [PubMed]
- Podolec, R.; Lau, K.; Wagnon, T.B.; Hothorn, M.; Ulm, R. A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2017284118. [Google Scholar] [CrossRef] [PubMed]
- Podolec, R.; Demarsy, E.; Ulm, R. Perception and signaling of Ultraviolet-B radiation in plants. Annu. Rev. Plant Biol. 2021, 72, 793–822. [Google Scholar] [CrossRef] [PubMed]
- Tossi, V.E.; Regalado, J.J.; Iannicelli, J.; Laino, L.E.; Burrieza, H.P.; Escandón, A.S.; Pitta-Álvarez, S.I. Beyond arabidopsis: Differential UV-B response mediated by UVR8 in diverse species. Front. Plant Sci. 2019, 10, 780. [Google Scholar] [CrossRef]
- Hollósy, F. Effects of ultraviolet radiation on plant cells. Micron 2002, 33, 179–197. [Google Scholar] [CrossRef] [PubMed]
- Nawkar, G.M.; Maibam, P.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. UV-Induced cell death in plants. Int. J. Mol. Sci. 2013, 14, 1608–1628. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Van de Poel, B.; De Coninck, B. UV-B light and its application potential to reduce disease and pest incidence in crops. Hortic. Res. 2021, 8, 194. [Google Scholar] [CrossRef]
- Bornman, J.F.; Barnes, P.W.; Robinson, S.A.; Ballaré, C.L.; Flint, S.D.; Caldwell, M.M. Solar ultraviolet radiation and ozone depletion-driven climate change: Effects on terrestrial ecosystems. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2015, 14, 88–107. [Google Scholar] [CrossRef]
- Schenke, D.; Utami, H.P.; Zhou, Z.; Gallegos, M.T.; Cai, D. Suppression of UV-B stress induced flavonoids by biotic stress: Is there reciprocal crosstalk? Plant Physiol. Biochem. PPB 2019, 134, 53–63. [Google Scholar] [CrossRef]
- Demkura, P.V.; Ballaré, C.L. UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 2012, 5, 642–652. [Google Scholar] [CrossRef]
- Brunetti, C.; Sebastiani, F.; Tattini, M. Review: ABA, flavonols, and the evolvability of land plants. Plant Sci. Int. J. Exp. Plant Biol. 2019, 280, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Yuan, H.; Dong, X.; Peng, M.; Jing, X.; Xu, Q.; Tang, T.; Wang, Y.; Zha, S.; Gao, M.; et al. Genome-wide Dissection of Co-selected UV-B Responsive Pathways in the UV-B Adaptation of Qingke. Mol. Plant 2020, 13, 112–127. [Google Scholar] [CrossRef]
- Zhou, Z.; Schenke, D.; Miao, Y.; Cai, D. Investigation of the crosstalk between the flg22 and the UV-B-induced flavonol pathway in Arabidopsis thaliana seedlings. Plant Cell Environ. 2017, 40, 453–458. [Google Scholar] [CrossRef]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef] [PubMed]
- Neugart, S.; Tobler, M.A.; Barnes, P.W. Different irradiances of UV and PAR in the same ratios alter the flavonoid profiles of Arabidopsis thaliana wild types and UV-signalling pathway mutants. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2019, 18, 1685–1699. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Silvestre, K.E.; Santiz-Gómez, J.A.; Luján-Hidalgo, M.C.; Ruiz-Lau, N.; Sánchez-Roque, Y.; Gutiérrez-Miceli, F.A. Effect of UV-B Radiation on Flavonoids and Phenols Accumulation in Tempisque (Sideroxylon capiri Pittier) Callus. Plants 2022, 11, 473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Huang, J.; Guo, H.; Yang, C.; Li, Y.; Shen, S.; Zhan, C.; Qu, L.; Liu, X.; Wang, S.; et al. OsRLCK160 contributes to flavonoid accumulation and UV-B tolerance by regulating OsbZIP48 in rice. Sci. China Life Sci. 2022, 65, 1380–1394. [Google Scholar] [CrossRef] [PubMed]
- Vanhaelewyn, L.; Van Der Straeten, D.; De Coninck, B.; Vandenbussche, F. Ultraviolet Radiation From a Plant Perspective: The Plant-Microorganism Context. Front. Plant Sci. 2020, 11, 597642. [Google Scholar] [CrossRef]
- Li, C.; Liu, S.; Zhang, W.; Chen, K.; Zhang, P. Transcriptional profiling and physiological analysis reveal the critical roles of ROS-scavenging system in the Antarctic moss Pohlia nutans under Ultraviolet-B radiation. Plant Physiol. Biochem. PPB 2019, 134, 113–122. [Google Scholar] [CrossRef]
- Kim, Y.J. Antimelanogenic and antioxidant properties of gallic acid. Biol. Pharm. Bull. 2007, 30, 1052–1055. [Google Scholar] [CrossRef]
- Kubo, I.; Fujita, K.; Nihei, K.; Masuoka, N. Non-antibiotic antibacterial activity of dodecyl gallate. Bioorganic Med. Chem. 2003, 11, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kubo, I.; Xiao, P.; Fujita, K. Antifungal activity of octyl gallate: Structural criteria and mode of action. Bioorganic Med. Chem. Lett. 2001, 11, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Gatto, M.A.; Ippolito, A.; Linsalata, V.; Cascarano, N.A.; Nigro, F.; Vanadia, S.; Di Venere, D. Activity of extracts from wild edible herbs against postharvest fungal diseases of fruit and vegetables. Postharvest Biol. Technol. 2011, 61, 72–82. [Google Scholar] [CrossRef]
- Fraser, D.P.; Sharma, A.; Fletcher, T.; Budge, S.; Moncrieff, C.; Dodd, A.N.; Franklin, K.A. UV-B antagonises shade avoidance and increases levels of the flavonoid quercetin in coriander (Coriandrum sativum). Sci. Rep. 2017, 7, 17758. [Google Scholar] [CrossRef] [PubMed]
- Hectors, K.; van Oevelen, S.; Guisez, Y.; Prinsen, E.; Jansen, M.A. The phytohormone auxin is a component of the regulatory system that controls UV-mediated accumulation of flavonoids and UV-induced morphogenesis. Physiol. Plant. 2012, 145, 594–603. [Google Scholar] [CrossRef]
- Wolf, L.; Rizzini, L.; Stracke, R.; Ulm, R.; Rensing, S.A. The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 2010, 153, 1123–1134. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 24, 7–1338. [Google Scholar]
- Liu, S.; Fang, S.; Cong, B.; Li, T.; Yi, D.; Zhang, Z.; Zhao, L.; Zhang, P. The Antarctic moss Pohlia nutans genome provides insights into the evolution of bryophytes and the adaptation to extreme terrestrial habitats. Front. Plant Sci. 2022, 13, 920138. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Galvez, S.; Sen, T.Z.; Budak, H. LncMachine: A machine learning algorithm for long noncoding RNA annotation in plants. Funct. Integr. Genom. 2021, 21, 195–204. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261.e212. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Cao, G.; Hou, X.; Huang, M.; Du, P.; Tan, T.; Zhang, Y.; Zhou, H.; Liu, X.; Liu, L.; et al. Development of a widely targeted volatilomics method for profiling volatilomes in plants. Mol. Plant 2022, 15, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, T.; Zhang, P.; Zhao, L.; Yi, D.; Zhang, Z.; Cong, B. Insights into the Jasmonate Signaling in Basal Land Plant Revealed by the Multi-Omics Analysis of an Antarctic Moss Pohlia nutans Treated with OPDA. Int. J. Mol. Sci. 2022, 23, 13507. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2(−ΔΔCt) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]


| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| CK1 | 79,930,786 | 79,469,444 | 11.92 | 0.03 | 96.3 | 90.14 | 44.02 |
| CK2 | 99,222,926 | 98,500,292 | 14.78 | 0.03 | 96.21 | 89.96 | 43.84 |
| CK3 | 90,289,934 | 88,589,850 | 13.29 | 0.03 | 96.74 | 91 | 44.94 |
| UV-B1 | 87,048,974 | 86,334,606 | 12.95 | 0.03 | 96.43 | 90.38 | 45 |
| UV-B2 | 84,624,622 | 83,986,726 | 12.6 | 0.03 | 96.77 | 91.04 | 44.34 |
| UV-B3 | 96,313,384 | 95,201,204 | 14.28 | 0.03 | 96.95 | 91.4 | 45.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, S.; Cong, B.; Zhao, L.; Liu, C.; Zhang, Z.; Liu, S. Genome-Wide Analysis of Long Non-Coding RNAs Related to UV-B Radiation in the Antarctic Moss Pohlia nutans. Int. J. Mol. Sci. 2023, 24, 5757. https://doi.org/10.3390/ijms24065757
Fang S, Cong B, Zhao L, Liu C, Zhang Z, Liu S. Genome-Wide Analysis of Long Non-Coding RNAs Related to UV-B Radiation in the Antarctic Moss Pohlia nutans. International Journal of Molecular Sciences. 2023; 24(6):5757. https://doi.org/10.3390/ijms24065757
Chicago/Turabian StyleFang, Shuo, Bailin Cong, Linlin Zhao, Chenlin Liu, Zhaohui Zhang, and Shenghao Liu. 2023. "Genome-Wide Analysis of Long Non-Coding RNAs Related to UV-B Radiation in the Antarctic Moss Pohlia nutans" International Journal of Molecular Sciences 24, no. 6: 5757. https://doi.org/10.3390/ijms24065757
APA StyleFang, S., Cong, B., Zhao, L., Liu, C., Zhang, Z., & Liu, S. (2023). Genome-Wide Analysis of Long Non-Coding RNAs Related to UV-B Radiation in the Antarctic Moss Pohlia nutans. International Journal of Molecular Sciences, 24(6), 5757. https://doi.org/10.3390/ijms24065757






