A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity †
Abstract
1. Introduction
2. UL Cellular Origin and Composition
3. Diversity of Endo-, Auto- and Paracrine Mechanisms Regulating UL Development
4. The Role of Woman’s Genotype in UL Aetiology
5. Spectrum of Somatic Genetic Aberrations in UL Cells
5.1. MED12 Mutations
5.2. Chromosomal Abnormalities
5.2.1. 12q14-15 Rearrangements
5.2.2. 6p21 Rearrangements
5.2.3. 10q22 Rearrangements
5.2.4. Deletions in the Long Arm of Chromosome 7
5.2.5. Chromosome 1 Rearrangements
5.2.6. Chromothripsis
5.2.7. Other Chromosomal Abnormalities
5.3. The Clinical Significance of Somatic Genetic Abnormalities in UL Cells
5.4. Speculations on the Origin of Somatic Genetic Abnormalities in UL Cells
6. Epigenetic Changes Associated with UL Development
6.1. Histone Post-Translational Modifications in ULs
6.2. DNA Methylation and Demethylation in ULs
6.3. Non-Coding RNAs
6.4. Speculations on the Role of Epigenetic Changes in UL Aetiology
7. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Day Baird, D.; Dunson, D.B.; Hill, M.C.; Cousins, D.; Schectman, J.M. High Cumulative Incidence of Uterine Leiomyoma in Black and White Women: Ultrasound Evidence. Am. J. Obstet. Gynecol. 2003, 188, 100–107. [Google Scholar] [CrossRef]
- Bulun, S.E. Uterine Fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef]
- Al-Hendy, A.; Myers, E.R.; Stewart, E. Uterine Fibroids: Burden and Unmet Medical Need. Semin. Reprod. Med. 2017, 35, 473–480. [Google Scholar] [CrossRef]
- Coutinho, L.M.; Assis, W.A.; Spagnuolo-Souza, A.; Reis, F.M. Uterine Fibroids and Pregnancy: How Do They Affect Each Other? Reprod. Sci. Thousand Oaks Calif. 2022, 29, 2145–2151. [Google Scholar] [CrossRef]
- Parazzini, F.; Tozzi, L.; Bianchi, S. Pregnancy Outcome and Uterine Fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 34, 74–84. [Google Scholar] [CrossRef]
- Sato, F.; Mori, M.; Nishi, M.; Kudo, R.; Miyake, H. Familial Aggregation of Uterine Myomas in Japanese Women. J. Epidemiol. 2002, 12, 249–253. [Google Scholar] [CrossRef]
- Day Baird, D.; Dunson, D.B. Why Is Parity Protective for Uterine Fibroids? Epidemiology 2003, 14, 247–250. [Google Scholar] [CrossRef]
- Stewart, E.; Cookson, C.; Gandolfo, R.; Schulze-Rath, R. Epidemiology of Uterine Fibroids: A Systematic Review. BJOG Int. J. Obstet. Gynaecol. 2017, 124, 1501–1512. [Google Scholar] [CrossRef]
- Yang, Q.; Ciebiera, M.; Bariani, M.V.; Ali, M.; Elkafas, H.; Boyer, T.G.; Al-Hendy, A. Comprehensive Review of Uterine Fibroids: Developmental Origin, Pathogenesis, and Treatment. Endocr. Rev. 2022, 43, 678–719. [Google Scholar] [CrossRef]
- Commandeur, A.E.; Styer, A.K.; Teixeira, J.M. Epidemiological and Genetic Clues for Molecular Mechanisms Involved in Uterine Leiomyoma Development and Growth. Hum. Reprod. Update 2015, 21, 593–615. [Google Scholar] [CrossRef]
- Pavone, D.; Clemenza, S.; Sorbi, F.; Fambrini, M.; Petraglia, F. Epidemiology and Risk Factors of Uterine Fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 46, 3–11. [Google Scholar] [CrossRef]
- Moravek, M.B.; Bulun, S.E. Endocrinology of Uterine Fibroids: Steroid Hormones, Stem Cells, and Genetic Contribution. Curr. Opin. Obstet. Gynecol. 2015, 27, 276–283. [Google Scholar] [CrossRef]
- Baranov, V.S.; Osinovskaya, N.S.; Yarmolinskaya, M.I. Pathogenomics of Uterine Fibroids Development. Int. J. Mol. Sci. 2019, 20, 6151. [Google Scholar] [CrossRef] [PubMed]
- Flake, G.P.; Moore, A.B.; Sutton, D.; Flagler, N.; Clayton, N.; Kissling, G.E.; Hall, B.W.; Horton, J.; Walmer, D.; Robboy, S.J.; et al. The Life Cycle of the Uterine Fibroid Myocyte. Curr. Obstet. Gynecol. Rep. 2018, 7, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Miura, S.; Khan, K.N.; Kitajima, M.; Hiraki, K.; Moriyama, S.; Masuzaki, H.; Samejima, T.; Fujishita, A.; Ishimaru, T. Differential Infiltration of Macrophages and Prostaglandin Production by Different Uterine Leiomyomas. Hum. Reprod. Oxf. Engl. 2006, 21, 2545–2554. [Google Scholar] [CrossRef]
- Holdsworth-Carson, S.J.; Zhao, D.; Cann, L.; Bittinger, S.; Nowell, C.J.; Rogers, P.A.W. Differences in the Cellular Composition of Small versus Large Uterine Fibroids. Reprod. Camb. Engl. 2016, 152, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Othman, E.R.; Elgamal, D.A.; Refaiy, A.M.; Abdelaal, I.I.; Abdel-Mola, A.F.; Al-Hendy, A. Identification and Potential Role of Telocytes in Human Uterine Leiomyoma. Contracept. Reprod. Med. 2016, 1, 12. [Google Scholar] [CrossRef]
- Giray, B.; Esim-Buyukbayrak, E.; Hallac-Keser, S.; Karageyim-Karsidag, A.Y.; Turkgeldi, A. Comparison of Nerve Fiber Density between Patients with Uterine Leiomyoma with and without Pain: A Prospective Clinical Study. Geburtshilfe Frauenheilkd. 2018, 78, 407–411. [Google Scholar] [CrossRef]
- Aleksandrovych, V.; Kurnik-Łucka, M.; Bereza, T.; Białas, M.; Pasternak, A.; Cretoiu, D.; Walocha, J.A.; Gil, K. The Autonomic Innervation and Uterine Telocyte Interplay in Leiomyoma Formation. Cell Transplant. 2019, 28, 619–629. [Google Scholar] [CrossRef]
- Li, Y.; Qiang, W.; Griffin, B.B.; Gao, T.; Chakravarti, D.; Bulun, S.; Kim, J.J.; Wei, J.-J. HMGA2-Mediated Tumorigenesis through Angiogenesis in Leiomyoma. Fertil. Steril. 2020, 114, 1085–1096. [Google Scholar] [CrossRef]
- Zannotti, A.; Greco, S.; Pellegrino, P.; Giantomassi, F.; Delli Carpini, G.; Goteri, G.; Ciavattini, A.; Ciarmela, P. Macrophages and Immune Responses in Uterine Fibroids. Cells 2021, 10, 982. [Google Scholar] [CrossRef]
- Goad, J.; Rudolph, J.; Zandigohar, M.; Tae, M.; Dai, Y.; Wei, J.-J.; Bulun, S.E.; Chakravarti, D.; Rajkovic, A. Single-Cell Sequencing Reveals Novel Cellular Heterogeneity in Uterine Leiomyomas. Hum. Reprod. Oxf. Engl. 2022, 37, 2334–2349. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth-Carson, S.J.; Zaitseva, M.; Vollenhoven, B.J.; Rogers, P.A.W. Clonality of Smooth Muscle and Fibroblast Cell Populations Isolated from Human Fibroid and Myometrial Tissues. Mol. Hum. Reprod. 2014, 20, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Maruyama, T.; Masuda, H.; Kajitani, T.; Nagashima, T.; Arase, T.; Ito, M.; Ohta, K.; Uchida, H.; Asada, H.; et al. Side Population in Human Uterine Myometrium Displays Phenotypic and Functional Characteristics of Myometrial Stem Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 18700–18705. [Google Scholar] [CrossRef]
- Gálvez, B.G.; Martín, N.S.; Salama-Cohen, P.; Lazcano, J.J.; Coronado, M.J.; Lamelas, M.L.; Alvarez-Barrientes, A.; Eiró, N.; Vizoso, F.; Rodríguez, C. An Adult Myometrial Pluripotential Precursor That Promotes Healing of Damaged Muscular Tissues. Vivo Athens Greece 2010, 24, 431–441. [Google Scholar]
- Mas, A.; Nair, S.; Laknaur, A.; Simón, C.; Diamond, M.P.; Al-Hendy, A. Stro-1/CD44 as Putative Human Myometrial and Fibroid Stem Cell Markers. Fertil. Steril. 2015, 104, 225–234. [Google Scholar] [CrossRef]
- Chang, H.L.; Senaratne, T.N.; Zhang, L.; Szotek, P.P.; Stewart, E.; Dombkowski, D.; Preffer, F.; Donahoe, P.K.; Teixeira, J. Uterine Leiomyomas Exhibit Fewer Stem/Progenitor Cell Characteristics When Compared with Corresponding Normal Myometrium. Reprod. Sci. Thousand Oaks Calif. 2010, 17, 158–167. [Google Scholar] [CrossRef]
- Mas, A.; Cervelló, I.; Gil-Sanchis, C.; Faus, A.; Ferro, J.; Pellicer, A.; Simón, C. Identification and Characterization of the Human Leiomyoma Side Population as Putative Tumor-Initiating Cells. Fertil. Steril. 2012, 98, 741–751. [Google Scholar] [CrossRef]
- Ono, M.; Qiang, W.; Serna, V.A.; Yin, P.; Coon, J.S.; Navarro, A.; Monsivais, D.; Kakinuma, T.; Dyson, M.; Druschitz, S.; et al. Role of Stem Cells in Human Uterine Leiomyoma Growth. PLoS ONE 2012, 7, e36935. [Google Scholar] [CrossRef]
- Orciani, M.; Caffarini, M.; Biagini, A.; Lucarini, G.; Delli Carpini, G.; Berretta, A.; Di Primio, R.; Ciavattini, A. Chronic Inflammation May Enhance Leiomyoma Development by the Involvement of Progenitor Cells. Stem Cells Int. 2018, 2018, 1716246. [Google Scholar] [CrossRef]
- Prusinski Fernung, L.E.; Al-Hendy, A.; Yang, Q. A Preliminary Study: Human Fibroid Stro-1+/CD44+ Stem Cells Isolated From Uterine Fibroids Demonstrate Decreased DNA Repair and Genomic Integrity Compared to Adjacent Myometrial Stro-1+/CD44+ Cells. Reprod. Sci. Thousand Oaks Calif. 2019, 26, 619–638. [Google Scholar] [CrossRef]
- Ono, M.; Yin, P.; Navarro, A.; Moravek, M.B.; Coon, J.S.; Druschitz, S.A.; Serna, V.A.; Qiang, W.; Brooks, D.C.; Malpani, S.S.; et al. Paracrine Activation of WNT/β-Catenin Pathway in Uterine Leiomyoma Stem Cells Promotes Tumor Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 17053–17058. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Kajitani, T.; Uchida, H.; Arase, T.; Oda, H.; Uchida, S.; Ota, K.; Nagashima, T.; Masuda, H.; Miyazaki, K.; et al. CD34 and CD49f Double-Positive and Lineage Marker-Negative Cells Isolated from Human Myometrium Exhibit Stem Cell-Like Properties Involved in Pregnancy-Induced Uterine Remodeling. Biol. Reprod. 2015, 93, 37. [Google Scholar] [CrossRef] [PubMed]
- Popescu, L.M.; Faussone-Pellegrini, M.-S. TELOCYTES—A Case of Serendipity: The Winding Way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J. Cell. Mol. Med. 2010, 14, 729–740. [Google Scholar] [CrossRef]
- Varga, I.; Klein, M.; Urban, L.; Danihel, L.; Polak, S.; Danihel, L. Recently Discovered Interstitial Cells “Telocytes” as Players in the Pathogenesis of Uterine Leiomyomas. Med. Hypotheses 2018, 110, 64–67. [Google Scholar] [CrossRef]
- Aleksandrovych, V.; Gil, A.; Wrona, A. Sex Steroid Hormone Receptors of Telocytes—Potential Key Role in Leiomyoma Development. Folia Med. Cracov. 2020, 60, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Moravek, M.B.; Yin, P.; Ono, M.; Coon, J.S.; Dyson, M.T.; Navarro, A.; Marsh, E.E.; Chakravarti, D.; Kim, J.J.; Wei, J.-J.; et al. Ovarian Steroids, Stem Cells and Uterine Leiomyoma: Therapeutic Implications. Hum. Reprod. Update 2015, 21, 1–12. [Google Scholar] [CrossRef]
- Sumitani, H.; Shozu, M.; Segawa, T.; Murakami, K.; Yang, H.J.; Shimada, K.; Inoue, M. In Situ Estrogen Synthesized by Aromatase P450 in Uterine Leiomyoma Cells Promotes Cell Growth Probably via an Autocrine/Intracrine Mechanism. Endocrinology 2000, 141, 3852–3861. [Google Scholar] [CrossRef]
- Nelson, L.R.; Bulun, S.E. Estrogen Production and Action. J. Am. Acad. Dermatol. 2001, 45, S116–S124. [Google Scholar] [CrossRef]
- Yang, Y.; He, Y.; Zeng, Q.; Li, S. Association of Body Size and Body Fat Distribution with Uterine Fibroids among Chinese Women. J. Womens Health 2002 2014, 23, 619–626. [Google Scholar] [CrossRef]
- Ciebiera, M.; Włodarczyk, M.; Słabuszewska-Jóźwiak, A.; Nowicka, G.; Jakiel, G. Influence of Vitamin D and Transforming Growth Factor Β3 Serum Concentrations, Obesity, and Family History on the Risk for Uterine Fibroids. Fertil. Steril. 2016, 106, 1787–1792. [Google Scholar] [CrossRef]
- Sun, K.; Xie, Y.; Zhao, N.; Li, Z. A Case-Control Study of the Relationship between Visceral Fat and Development of Uterine Fibroids. Exp. Ther. Med. 2019, 18, 404–410. [Google Scholar] [CrossRef]
- Qin, H.; Lin, Z.; Vásquez, E.; Luan, X.; Guo, F.; Xu, L. Association between Obesity and the Risk of Uterine Fibroids: A Systematic Review and Meta-Analysis. J. Epidemiol. Community Health 2021, 75, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Diamond, M.P.; Al-Hendy, A. Early Life Adverse Environmental Exposures Increase the Risk of Uterine Fibroid Development: Role of Epigenetic Regulation. Front. Pharmacol. 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Lamminen, S.; Rantala, I.; Helin, H.; Rorarius, M.; Tuimala, R. Proliferative Activity of Human Uterine Leiomyoma Cells as Measured by Automatic Image Analysis. Gynecol. Obstet. Investig. 1992, 34, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ishi, K.; Serna, V.A.; Kakazu, R.; Bulun, S.E.; Kurita, T. Progesterone Is Essential for Maintenance and Growth of Uterine Leiomyoma. Endocrinology 2010, 151, 2433–2442. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Fujii, S.; Konishi, I.; Nanbu, Y.; Nonogaki, H.; Mori, T. Mitotic Activity in Uterine Leiomyomas during the Menstrual Cycle. Am. J. Obstet. Gynecol. 1989, 160, 637–641. [Google Scholar] [CrossRef]
- Wu, X.; Blanck, A.; Olovsson, M.; Möller, B.; Favini, R.; Lindblom, B. Apoptosis, Cellular Proliferation and Expression of P53 in Human Uterine Leiomyomas and Myometrium during the Menstrual Cycle and after Menopause. Acta Obstet. Gynecol. Scand. 2000, 79, 397–404. [Google Scholar]
- Palomba, S.; Sena, T.; Morelli, M.; Noia, R.; Zullo, F.; Mastrantonio, P. Effect of Different Doses of Progestin on Uterine Leiomyomas in Postmenopausal Women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2002, 102, 199–201. [Google Scholar] [CrossRef]
- Patel, A.; Malik, M.; Britten, J.; Cox, J.; Catherino, W.H. Mifepristone Inhibits Extracellular Matrix Formation in Uterine Leiomyoma. Fertil. Steril. 2016, 105, 1102–1110. [Google Scholar] [CrossRef]
- Voronin, D.; Sotnikova, N.; Rukavishnikov, K.; Malyshkina, A.; Nagornii, S.; Antsiferova, Y. Differential Regulatory Effect of Progesterone on the Proliferation and Apoptosis of Uterine Leiomyoma Tissue Explants and Primary Leiomyoma Cell Cultures. JBRA Assist. Reprod. 2021, 25, 540–548. [Google Scholar] [CrossRef]
- Patel, B.; Elguero, S.; Thakore, S.; Dahoud, W.; Bedaiwy, M.; Mesiano, S. Role of Nuclear Progesterone Receptor Isoforms in Uterine Pathophysiology. Hum. Reprod. Update 2015, 21, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Sefton, E.C. The Role of Progesterone Signaling in the Pathogenesis of Uterine Leiomyoma. Mol. Cell. Endocrinol. 2012, 358, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Elkafas, H.; Qiwei, Y.; Al-Hendy, A. Origin of Uterine Fibroids: Conversion of Myometrial Stem Cells to Tumor-Initiating Cells. Semin. Reprod. Med. 2017, 35, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, A.V.; Sefton, E.C.; Berry, E.; Lu, Z.; Hardt, J.; Marsh, E.; Yin, P.; Clardy, J.; Chakravarti, D.; Bulun, S.; et al. Progestins Activate the AKT Pathway in Leiomyoma Cells and Promote Survival. J. Clin. Endocrinol. Metab. 2009, 94, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Al-Hendy, A.; Laknaur, A.; Diamond, M.P.; Ismail, N.; Boyer, T.G.; Halder, S.K. Silencing Med12 Gene Reduces Proliferation of Human Leiomyoma Cells Mediated via Wnt/β-Catenin Signaling Pathway. Endocrinology 2017, 158, 592–603. [Google Scholar] [CrossRef]
- El Andaloussi, A.; Al-Hendy, A.; Ismail, N.; Boyer, T.G.; Halder, S.K. Introduction of Somatic Mutation in MED12 Induces Wnt4/β-Catenin and Disrupts Autophagy in Human Uterine Myometrial Cell. Reprod. Sci. Thousand Oaks Calif 2020, 27, 823–832. [Google Scholar] [CrossRef]
- Ono, M.; Yin, P.; Navarro, A.; Moravek, M.B.; Coon V, J.S.; Druschitz, S.A.; Gottardi, C.J.; Bulun, S.E. Inhibition of Canonical WNT Signaling Attenuates Human Leiomyoma Cell Growth. Fertil. Steril. 2014, 101, 1441–1449. [Google Scholar] [CrossRef]
- Al-Hendy, A.; Diamond, M.P.; El-Sohemy, A.; Halder, S.K. 1,25-Dihydroxyvitamin D3 Regulates Expression of Sex Steroid Receptors in Human Uterine Fibroid Cells. J. Clin. Endocrinol. Metab. 2015, 100, E572–E582. [Google Scholar] [CrossRef]
- Corachán, A.; Ferrero, H.; Aguilar, A.; Garcia, N.; Monleon, J.; Faus, A.; Cervelló, I.; Pellicer, A. Inhibition of Tumor Cell Proliferation in Human Uterine Leiomyomas by Vitamin D via Wnt/β-Catenin Pathway. Fertil. Steril. 2019, 111, 397–407. [Google Scholar] [CrossRef]
- El Sabeh, M.; Saha, S.K.; Afrin, S.; Borahay, M.A. Simvastatin Inhibits Wnt/β-Catenin Pathway in Uterine Leiomyoma. Endocrinology 2021, 162, bqab211. [Google Scholar] [CrossRef] [PubMed]
- Borahay, M.A.; Al-Hendy, A.; Kilic, G.S.; Boehning, D. Signaling Pathways in Leiomyoma: Understanding Pathobiology and Implications for Therapy. Mol. Med. Camb. Mass 2015, 21, 242–256. [Google Scholar] [CrossRef]
- Islam, M.S.; Afrin, S.; Singh, B.; Jayes, F.L.; Brennan, J.T.; Borahay, M.A.; Leppert, P.C.; Segars, J.H. Extracellular Matrix and Hippo Signaling as Therapeutic Targets of Antifibrotic Compounds for Uterine Fibroids. Clin. Transl. Med. 2021, 11, e475. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.-A.; Jamaluddin, M.F.B.; Adebayo, M.; Bajwa, P.; Scott, R.J.; Dharmarajan, A.M.; Nahar, P.; Tanwar, P.S. Extracellular Matrix (ECM) Activates β-Catenin Signaling in Uterine Fibroids. Reprod. Camb. Engl. 2018, 155, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Wear, L.E. Uterine Myoma as a Hereditary Disease. Lancet Lond. Engl. 1957, 272, 25–26. [Google Scholar] [CrossRef] [PubMed]
- Vikhlyaeva, E.M.; Khodzhaeva, Z.S.; Fantschenko, N.D. Familial Predisposition to Uterine Leiomyomas. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 1995, 51, 127–131. [Google Scholar] [CrossRef]
- Luoto, R.; Kaprio, J.; Rutanen, E.M.; Taipale, P.; Perola, M.; Koskenvuo, M. Heritability and Risk Factors of Uterine Fibroids--the Finnish Twin Cohort Study. Maturitas 2000, 37, 15–26. [Google Scholar] [CrossRef]
- Marshall, L.M.; Spiegelman, D.; Barbieri, R.L.; Goldman, M.B.; Manson, J.E.; Colditz, G.A.; Willett, W.C.; Hunter, D.J. Variation in the Incidence of Uterine Leiomyoma among Premenopausal Women by Age and Race. Obstet. Gynecol. 1997, 90, 967–973. [Google Scholar] [CrossRef]
- Keaton, J.M.; Jasper, E.A.; Hellwege, J.N.; Jones, S.H.; Torstenson, E.S.; Edwards, T.L.; Velez Edwards, D.R. Evidence That Geographic Variation in Genetic Ancestry Associates with Uterine Fibroids. Hum. Genet. 2021, 140, 1433–1440. [Google Scholar] [CrossRef]
- Stewart, E.A.; Nicholson, W.K.; Bradley, L.; Borah, B.J. The Burden of Uterine Fibroids for African-American Women: Results of a National Survey. J. Womens Health 2002 2013, 22, 807–816. [Google Scholar] [CrossRef]
- Cha, P.-C.; Takahashi, A.; Hosono, N.; Low, S.-K.; Kamatani, N.; Kubo, M.; Nakamura, Y. A Genome-Wide Association Study Identifies Three Loci Associated with Susceptibility to Uterine Fibroids. Nat. Genet. 2011, 43, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhao, X.; Zhou, C.; Yang, L.; Liu, Y.; Bian, C.; Gou, J.; Lin, X.; Wang, Z.; Zhao, X. The Associations between the Val158Met in the Catechol-O-Methyltransferase (COMT) Gene and the Risk of Uterine Leiomyoma (ULM). Gene 2013, 529, 296–299. [Google Scholar] [CrossRef]
- Hellwege, J.N.; Jeff, J.M.; Wise, L.A.; Gallagher, C.S.; Wellons, M.; Hartmann, K.E.; Jones, S.F.; Torstenson, E.S.; Dickinson, S.; Ruiz-Narváez, E.A.; et al. A Multi-Stage Genome-Wide Association Study of Uterine Fibroids in African Americans. Hum. Genet. 2017, 136, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Rafnar, T.; Gunnarsson, B.; Stefansson, O.A.; Sulem, P.; Ingason, A.; Frigge, M.L.; Stefansdottir, L.; Sigurdsson, J.K.; Tragante, V.; Steinthorsdottir, V.; et al. Variants Associating with Uterine Leiomyoma Highlight Genetic Background Shared by Various Cancers and Hormone-Related Traits. Nat. Commun. 2018, 9, 3636. [Google Scholar] [CrossRef] [PubMed]
- Välimäki, N.; Kuisma, H.; Pasanen, A.; Heikinheimo, O.; Sjöberg, J.; Bützow, R.; Sarvilinna, N.; Heinonen, H.-R.; Tolvanen, J.; Bramante, S.; et al. Genetic Predisposition to Uterine Leiomyoma Is Determined by Loci for Genitourinary Development and Genome Stability. eLife 2018, 7, e37110. [Google Scholar] [CrossRef]
- Edwards, T.L.; Giri, A.; Hellwege, J.N.; Hartmann, K.E.; Stewart, E.A.; Jeff, J.M.; Bray, M.J.; Pendergrass, S.A.; Torstenson, E.S.; Keaton, J.M.; et al. A Trans-Ethnic Genome-Wide Association Study of Uterine Fibroids. Front. Genet. 2019, 10, 511. [Google Scholar] [CrossRef]
- Gallagher, C.S.; Mäkinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-Wide Association and Epidemiological Analyses Reveal Common Genetic Origins between Uterine Leiomyomata and Endometriosis. Nat. Commun. 2019, 10, 4857. [Google Scholar] [CrossRef]
- Masuda, T.; Low, S.-K.; Akiyama, M.; Hirata, M.; Ueda, Y.; Matsuda, K.; Kimura, T.; Murakami, Y.; Kubo, M.; Kamatani, Y.; et al. GWAS of Five Gynecologic Diseases and Cross-Trait Analysis in Japanese. Eur. J. Hum. Genet. EJHG 2020, 28, 95–107. [Google Scholar] [CrossRef]
- Sakai, K.; Tanikawa, C.; Hirasawa, A.; Chiyoda, T.; Yamagami, W.; Kataoka, F.; Susumu, N.; Terao, C.; Kamatani, Y.; Takahashi, A.; et al. Identification of a Novel Uterine Leiomyoma GWAS Locus in a Japanese Population. Sci. Rep. 2020, 10, 1197. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Reshetnikov, E.; Polonikov, A.; Verzilina, I.; Sorokina, I.; Yermachenko, A.; Dvornyk, V.; Churnosov, M. Candidate Genes for Age at Menarche Are Associated With Uterine Leiomyoma. Front. Genet. 2020, 11, 512940. [Google Scholar] [CrossRef]
- Alset, D.; Pokudina, I.O.; Butenko, E.V.; Shkurat, T.P. The Effect of Estrogen-Related Genetic Variants on the Development of Uterine Leiomyoma: Meta-Analysis. Reprod. Sci. Thousand Oaks Calif. 2022, 29, 1921–1929. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, M.V.; Sogoyan, N.S.; Donnikov, A.J.; Trofimov, D.Y.; Adamyan, L.V.; Mishina, N.D.; Shubina, J.; Zelensky, D.V.; Sukhikh, G.T. Familial Predisposition to Leiomyomata: Searching for Protective Genetic Factors. Biomedicines 2022, 10, 508. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.L.; Hartmann, K.E.; Velez Edwards, D.R. Variants in BET1L and TNRC6B Associate with Increasing Fibroid Volume and Fibroid Type among European Americans. Hum. Genet. 2013, 132, 1361–1369. [Google Scholar] [CrossRef]
- Aissani, B.; Zhang, K.; Wiener, H. Follow-up to Genome-Wide Linkage and Admixture Mapping Studies Implicates Components of the Extracellular Matrix in Susceptibility to and Size of Uterine Fibroids. Fertil. Steril. 2015, 103, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.J.; Edwards, T.L.; Wellons, M.F.; Jones, S.H.; Hartmann, K.E.; Velez Edwards, D.R. Admixture Mapping of Uterine Fibroid Size and Number in African American Women. Fertil. Steril. 2017, 108, 1034–1042. [Google Scholar] [CrossRef]
- Dzhemlikhanova, L.K.; Efimova, O.A.; Osinovskaya, N.S.; Parfenyev, S.E.; Niauri, D.A.; Sultanov, I.Y.; Malysheva, O.V.; Pendina, A.A.; Shved, N.Y.; Ivashchenko, T.E.; et al. Catechol-O-Methyltransferase Val158Met Polymorphism Is Associated with Increased Risk of Multiple Uterine Leiomyomas Either Positive or Negative for MED12 Exon 2 Mutations. J. Clin. Pathol. 2017, 70, 233–236. [Google Scholar] [CrossRef]
- Bray, M.J.; Wellons, M.F.; Jones, S.H.; Torstenson, E.S.; Edwards, T.L.; Velez Edwards, D.R. Transethnic and Race-Stratified Genome-Wide Association Study of Fibroid Characteristics in African American and European American Women. Fertil. Steril. 2018, 110, 737–745. [Google Scholar] [CrossRef]
- Launonen, V.; Vierimaa, O.; Kiuru, M.; Isola, J.; Roth, S.; Pukkala, E.; Sistonen, P.; Herva, R.; Aaltonen, L.A. Inherited Susceptibility to Uterine Leiomyomas and Renal Cell Cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 3387–3392. [Google Scholar] [CrossRef]
- Tomlinson, I.P.M.; Alam, N.A.; Rowan, A.J.; Barclay, E.; Jaeger, E.E.M.; Kelsell, D.; Leigh, I.; Gorman, P.; Lamlum, H.; Rahman, S.; et al. Germline Mutations in FH Predispose to Dominantly Inherited Uterine Fibroids, Skin Leiomyomata and Papillary Renal Cell Cancer. Nat. Genet. 2002, 30, 406–410. [Google Scholar] [CrossRef]
- Lehtonen, R.; Kiuru, M.; Vanharanta, S.; Sjöberg, J.; Aaltonen, L.-M.; Aittomäki, K.; Arola, J.; Butzow, R.; Eng, C.; Husgafvel-Pursiainen, K.; et al. Biallelic Inactivation of Fumarate Hydratase (FH) Occurs in Nonsyndromic Uterine Leiomyomas but Is Rare in Other Tumors. Am. J. Pathol. 2004, 164, 17–22. [Google Scholar] [CrossRef]
- Miettinen, M.; Felisiak-Golabek, A.; Wasag, B.; Chmara, M.; Wang, Z.; Butzow, R.; Lasota, J. Fumarase-Deficient Uterine Leiomyomas: An Immunohistochemical, Molecular Genetic, and Clinicopathologic Study of 86 Cases. Am. J. Surg. Pathol. 2016, 40, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Harrison, W.J.; Andrici, J.; Maclean, F.; Madadi-Ghahan, R.; Farzin, M.; Sioson, L.; Toon, C.W.; Clarkson, A.; Watson, N.; Pickett, J.; et al. Fumarate Hydratase-Deficient Uterine Leiomyomas Occur in Both the Syndromic and Sporadic Settings. Am. J. Surg. Pathol. 2016, 40, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Popp, B.; Erber, R.; Kraus, C.; Vasileiou, G.; Hoyer, J.; Burghaus, S.; Hartmann, A.; Beckmann, M.W.; Reis, A.; Agaimy, A. Targeted Sequencing of FH-Deficient Uterine Leiomyomas Reveals Biallelic Inactivating Somatic Fumarase Variants and Allows Characterization of Missense Variants. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2020, 33, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Siegler, L.; Erber, R.; Burghaus, S.; Brodkorb, T.; Wachter, D.; Wilkinson, N.; Bolton, J.; Stringfellow, H.; Haller, F.; Beckmann, M.W.; et al. Fumarate Hydratase (FH) Deficiency in Uterine Leiomyomas: Recognition by Histological Features versus Blind Immunoscreening. Virchows Arch. Int. J. Pathol. 2018, 472, 789–796. [Google Scholar] [CrossRef]
- Kashtan, C.E. Alport Syndrome. An Inherited Disorder of Renal, Ocular, and Cochlear Basement Membranes. Medicine (Baltimore) 1999, 78, 338–360. [Google Scholar] [CrossRef]
- Liu, C.; Dillon, J.; Beavis, A.L.; Liu, Y.; Lombardo, K.; Fader, A.N.; Hung, C.-F.; Wu, T.-C.; Vang, R.; Garcia, J.E.; et al. Prevalence of Somatic and Germline Mutations of Fumarate Hydratase in Uterine Leiomyomas from Young Patients. Histopathology 2020, 76, 354–365. [Google Scholar] [CrossRef]
- Mehine, M.; Mäkinen, N.; Heinonen, H.-R.; Aaltonen, L.A.; Vahteristo, P. Genomics of Uterine Leiomyomas: Insights from High-Throughput Sequencing. Fertil. Steril. 2014, 102, 621–629. [Google Scholar] [CrossRef]
- Yatsenko, S.A.; Mittal, P.; Wood-Trageser, M.A.; Jones, M.W.; Surti, U.; Edwards, R.P.; Sood, A.K.; Rajkovic, A. Highly Heterogeneous Genomic Landscape of Uterine Leiomyomas by Whole Exome Sequencing and Genome-Wide Arrays. Fertil. Steril. 2017, 107, 457–466. [Google Scholar] [CrossRef]
- Shaik, N.A.; Lone, W.G.; Khan, I.A.; Vaidya, S.; Rao, K.P.; Kodati, V.L.; Hasan, Q. Detection of Somatic Mutations and Germline Polymorphisms in Mitochondrial DNA of Uterine Fibroids Patients. Genet. Test. Mol. Biomark. 2011, 15, 537–541. [Google Scholar] [CrossRef]
- Mäkinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the Mediator Complex Subunit 12 Gene, Is Mutated at High Frequency in Uterine Leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef]
- McGuire, M.M.; Yatsenko, A.; Hoffner, L.; Jones, M.; Surti, U.; Rajkovic, A. Whole Exome Sequencing in a Random Sample of North American Women with Leiomyomas Identifies MED12 Mutations in Majority of Uterine Leiomyomas. PLoS ONE 2012, 7, e33251. [Google Scholar] [CrossRef]
- Heinonen, H.-R.; Sarvilinna, N.S.; Sjöberg, J.; Kämpjärvi, K.; Pitkänen, E.; Vahteristo, P.; Mäkinen, N.; Aaltonen, L.A. MED12 Mutation Frequency in Unselected Sporadic Uterine Leiomyomas. Fertil. Steril. 2014, 102, 1137–1142. [Google Scholar] [CrossRef]
- Osinovskaya, N.S.; Malysheva, O.V.; Shved, N.Y.; Ivashchenko, T.E.; Sultanov, I.Y.; Efimova, O.A.; Yarmolinskaya, M.I.; Bezhenar, V.F.; Baranov, V.S. Frequency and Spectrum of MED12 Exon 2 Mutations in Multiple Versus Solitary Uterine Leiomyomas From Russian Patients. Int. J. Gynecol. Pathol. Off. J. Int. Soc. Gynecol. Pathol. 2016, 35, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Cheon, K.; Chae, B.; Hwang, H.; Kim, H.-K.; Chung, Y.-J.; Song, J.-Y.; Cho, H.-H.; Kim, J.-H.; Kim, M.-R. Analysis of MED12 Mutation in Multiple Uterine Leiomyomas in South Korean Patients. Int. J. Med. Sci. 2018, 15, 124–128. [Google Scholar] [CrossRef]
- He, C.; Nelson, W.; Li, H.; Xu, Y.-D.; Dai, X.-J.; Wang, Y.-X.; Ding, Y.-B.; Li, Y.-P.; Li, T. Frequency of MED12 Mutation in Relation to Tumor and Patient’s Clinical Characteristics: A Meta-Analysis. Reprod. Sci. Thousand Oaks Calif. 2022, 29, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, H.-R.; Pasanen, A.; Heikinheimo, O.; Tanskanen, T.; Palin, K.; Tolvanen, J.; Vahteristo, P.; Sjöberg, J.; Pitkänen, E.; Bützow, R.; et al. Multiple Clinical Characteristics Separate MED12-Mutation-Positive and -Negative Uterine Leiomyomas. Sci. Rep. 2017, 7, 1015. [Google Scholar] [CrossRef]
- Mäkinen, N.; Vahteristo, P.; Kämpjärvi, K.; Arola, J.; Bützow, R.; Aaltonen, L.A. MED12 Exon 2 Mutations in Histopathological Uterine Leiomyoma Variants. Eur. J. Hum. Genet. EJHG 2013, 21, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, N.; Kämpjärvi, K.; Frizzell, N.; Bützow, R.; Vahteristo, P. Characterization of MED12, HMGA2, and FH Alterations Reveals Molecular Variability in Uterine Smooth Muscle Tumors. Mol. Cancer 2017, 16, 101. [Google Scholar] [CrossRef]
- Wu, X.; Serna, V.A.; Thomas, J.; Qiang, W.; Blumenfeld, M.L.; Kurita, T. Subtype-Specific Tumor-Associated Fibroblasts Contribute to the Pathogenesis of Uterine Leiomyoma. Cancer Res. 2017, 77, 6891–6901. [Google Scholar] [CrossRef]
- Äyräväinen, A.; Pasanen, A.; Ahvenainen, T.; Heikkinen, T.; Pakarinen, P.; Härkki, P.; Vahteristo, P. Systematic Molecular and Clinical Analysis of Uterine Leiomyomas from Fertile-Aged Women Undergoing Myomectomy. Hum. Reprod. Oxf. Engl. 2020, 35, 2237–2244. [Google Scholar] [CrossRef]
- Maekawa, R.; Sato, S.; Tamehisa, T.; Sakai, T.; Kajimura, T.; Sueoka, K.; Sugino, N. Different DNA Methylome, Transcriptome and Histological Features in Uterine Fibroids with and without MED12 Mutations. Sci. Rep. 2022, 12, 8912. [Google Scholar] [CrossRef]
- Kämpjärvi, K.; Park, M.J.; Mehine, M.; Kim, N.H.; Clark, A.D.; Bützow, R.; Böhling, T.; Böhm, J.; Mecklin, J.-P.; Järvinen, H.; et al. Mutations in Exon 1 Highlight the Role of MED12 in Uterine Leiomyomas. Hum. Mutat. 2014, 35, 1136–1141. [Google Scholar] [CrossRef]
- Di Tommaso, S.; Tinelli, A.; Malvasi, A.; Massari, S. Missense Mutations in Exon 2 of the MED12 Gene Are Involved in IGF-2 Overexpression in Uterine Leiomyoma. Mol. Hum. Reprod. 2014, 20, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Shin, Y.-H.; Yatsenko, S.A.; Castro, C.A.; Surti, U.; Rajkovic, A. Med12 Gain-of-Function Mutation Causes Leiomyomas and Genomic Instability. J. Clin. Investig. 2015, 125, 3280–3284. [Google Scholar] [CrossRef]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Mäkinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine Leiomyoma-Linked MED12 Mutations Disrupt Mediator-Associated CDK Activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef]
- Park, M.J.; Shen, H.; Kim, N.H.; Gao, F.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Heikkinen, T.A.; Vahteristo, P.; et al. Mediator Kinase Disruption in MED12-Mutant Uterine Fibroids From Hispanic Women of South Texas. J. Clin. Endocrinol. Metab. 2018, 103, 4283–4292. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Shen, H.; Spaeth, J.M.; Tolvanen, J.H.; Failor, C.; Knudtson, J.F.; McLaughlin, J.; Halder, S.K.; Yang, Q.; Bulun, S.E.; et al. Oncogenic Exon 2 Mutations in Mediator Subunit MED12 Disrupt Allosteric Activation of Cyclin C-CDK8/19. J. Biol. Chem. 2018, 293, 4870–4882. [Google Scholar] [CrossRef] [PubMed]
- Mehine, M.; Kaasinen, E.; Heinonen, H.-R.; Mäkinen, N.; Kämpjärvi, K.; Sarvilinna, N.; Aavikko, M.; Vähärautio, A.; Pasanen, A.; Bützow, R.; et al. Integrated Data Analysis Reveals Uterine Leiomyoma Subtypes with Distinct Driver Pathways and Biomarkers. Proc. Natl. Acad. Sci. USA 2016, 113, 1315–1320. [Google Scholar] [CrossRef]
- Liu, S.; Yin, P.; Kujawa, S.A.; Coon, J.S.; Okeigwe, I.; Bulun, S.E. Progesterone Receptor Integrates the Effects of Mutated MED12 and Altered DNA Methylation to Stimulate RANKL Expression and Stem Cell Proliferation in Uterine Leiomyoma. Oncogene 2019, 38, 2722–2735. [Google Scholar] [CrossRef]
- Hutchinson, A.P.; Yin, P.; Neale, I.; Coon, J.S.; Kujawa, S.A.; Liu, S.; Bulun, S.E. Tryptophan 2,3-Dioxygenase-2 in Uterine Leiomyoma: Dysregulation by MED12 Mutation Status. Reprod. Sci. Thousand Oaks Calif. 2022, 29, 743–749. [Google Scholar] [CrossRef]
- Moyo, M.B.; Parker, J.B.; Chakravarti, D. Altered Chromatin Landscape and Enhancer Engagement Underlie Transcriptional Dysregulation in MED12 Mutant Uterine Leiomyomas. Nat. Commun. 2020, 11, 1019. [Google Scholar] [CrossRef] [PubMed]
- Kol’tsova, A.S.; Pendina, A.A.; Efimova, O.A.; Kaminskaya, A.N.; Tikhonov, A.V.; Osinovskaya, N.S.; Sultanov, I.Y.; Shved, N.Y.; Kakhiani, M.I.; Baranov, V.S. Differential DNA Hydroxymethylation in Human Uterine Leiomyoma Cells Depending on the Phase of Menstrual Cycle and Presence of MED12 Gene Mutations. Bull. Exp. Biol. Med. 2017, 163, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Muralimanoharan, S.; Shamby, R.; Stansbury, N.; Schenken, R.; de la Pena Avalos, B.; Javanmardi, S.; Dray, E.; Sung, P.; Boyer, T.G. Aberrant R-Loop-Induced Replication Stress in MED12-Mutant Uterine Fibroids. Sci. Rep. 2022, 12, 6169. [Google Scholar] [CrossRef] [PubMed]
- Nadine Markowski, D.; Tadayyon, M.; Bartnitzke, S.; Belge, G.; Maria Helmke, B.; Bullerdiek, J. Cell Cultures in Uterine Leiomyomas: Rapid Disappearance of Cells Carrying MED12 Mutations. Genes Chromosomes Cancer 2014, 53, 317–323. [Google Scholar] [CrossRef]
- Luo, N.; Guan, Q.; Zheng, L.; Qu, X.; Dai, H.; Cheng, Z. Estrogen-Mediated Activation of Fibroblasts and Its Effects on the Fibroid Cell Proliferation. Transl. Res. J. Lab. Clin. Med. 2014, 163, 232–241. [Google Scholar] [CrossRef]
- Okamoto, T.; Sato, J.D.; Barnes, D.W.; Sato, G.H. Biomedical Advances from Tissue Culture. Cytotechnology 2013, 65, 967–971. [Google Scholar] [CrossRef]
- Malik, M.; Britten, J.; Catherino, W.H. Development and Validation of Hormonal Impact of a Mouse Xenograft Model for Human Uterine Leiomyoma. Reprod. Sci. Thousand Oaks Calif. 2020, 27, 1304–1317. [Google Scholar] [CrossRef]
- Salas, A.; López, J.; Reyes, R.; Évora, C.; de Oca, F.M.; Báez, D.; Delgado, A.; Almeida, T.A. Organotypic Culture as a Research and Preclinical Model to Study Uterine Leiomyomas. Sci. Rep. 2020, 10, 5212. [Google Scholar] [CrossRef]
- Shved, N.; Egorova, A.; Osinovskaya, N.; Kiselev, A. Development of Primary Monolayer Cell Model and Organotypic Model of Uterine Leiomyoma. Methods Protoc. 2022, 5, 16. [Google Scholar] [CrossRef]
- Markowski, D.N.; Bartnitzke, S.; Löning, T.; Drieschner, N.; Helmke, B.M.; Bullerdiek, J. MED12 Mutations in Uterine Fibroids--Their Relationship to Cytogenetic Subgroups. Int. J. Cancer 2012, 131, 1528–1536. [Google Scholar] [CrossRef]
- Kämpjärvi, K.; Mäkinen, N.; Mehine, M.; Välipakka, S.; Uimari, O.; Pitkänen, E.; Heinonen, H.-R.; Heikkinen, T.; Tolvanen, J.; Ahtikoski, A.; et al. MED12 Mutations and FH Inactivation Are Mutually Exclusive in Uterine Leiomyomas. Br. J. Cancer 2016, 114, 1405–1411. [Google Scholar] [CrossRef]
- Galindo, L.J.; Hernández-Beeftink, T.; Salas, A.; Jung, Y.; Reyes, R.; de Oca, F.M.; Hernández, M.; Almeida, T.A. HMGA2 and MED12 Alterations Frequently Co-Occur in Uterine Leiomyomas. Gynecol. Oncol. 2018, 150, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Mello, J.B.H.; Barros-Filho, M.C.; Abreu, F.B.; Cirilo, P.D.R.; Domingues, M.a.C.; Pontes, A.; Rogatto, S.R. MicroRNAs Involved in the HMGA2 Deregulation and Its Co-Occurrence with MED12 Mutation in Uterine Leiomyoma. Mol. Hum. Reprod. 2018, 24, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Koltsova, A.S.; Efimova, O.A.; Malysheva, O.V.; Osinovskaya, N.S.; Liehr, T.; Al-Rikabi, A.; Shved, N.Y.; Sultanov, I.Y.; Chiryaeva, O.G.; Yarmolinskaya, M.I.; et al. Cytogenomic Profile of Uterine Leiomyoma: In Vivo vs. In Vitro Comparison. Biomedicines 2021, 9, 1777. [Google Scholar] [CrossRef]
- Koltsova, A.S.; Efimova, O.A.; Pendina, A.A.; Chiryaeva, O.G.; Osinovskaya, N.S.; Shved, N.Y.; Yarmolinskaya, M.I.; Polenov, N.I.; Kunitsa, V.V.; Sagurova, Y.M.; et al. Uterine Leiomyomas with an Apparently Normal Karyotype Comprise Minor Heteroploid Subpopulations Differently Represented in Vivo and in Vitro. Cytogenet. Genome Res. 2021, 161, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.A. Updates on the Cytogenetics and Molecular Genetics of Bone and Soft Tissue Tumors: Leiomyoma. Cancer Genet. Cytogenet. 2005, 158, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Nilbert, M.; Heim, S.; Mandahl, N.; Flodérus, U.M.; Willén, H.; Mitelman, F. Characteristic Chromosome Abnormalities, Including Rearrangements of 6p, Del(7q), +12, and t(12;14), in 44 Uterine Leiomyomas. Hum. Genet. 1990, 85, 605–611. [Google Scholar] [CrossRef]
- Hu, J.; Surti, U. Subgroups of Uterine Leiomyomas Based on Cytogenetic Analysis. Hum. Pathol. 1991, 22, 1009–1016. [Google Scholar] [CrossRef]
- Mashal, R.D.; Fejzo, M.L.; Friedman, A.J.; Mitchner, N.; Nowak, R.A.; Rein, M.S.; Morton, C.C.; Sklar, J. Analysis of Androgen Receptor DNA Reveals the Independent Clonal Origins of Uterine Leiomyomata and the Secondary Nature of Cytogenetic Aberrations in the Development of Leiomyomata. Genes Chromosomes Cancer 1994, 11, 1–6. [Google Scholar] [CrossRef]
- Hayashi, S.; Miharu, N.; Okamoto, E.; Samura, O.; Hara, T.; Ohama, K. Detection of Chromosomal Abnormalities of Chromosome 12 in Uterine Leiomyoma Using Fluorescence in Situ Hybridization. Jpn. J. Hum. Genet. 1996, 41, 193–202. [Google Scholar] [CrossRef]
- Schoenmakers, E.F.; Mols, R.; Wanschura, S.; Kools, P.F.; Geurts, J.M.; Bartnitzke, S.; Bullerdiek, J.; van den Berghe, H.; Van de Ven, W.J. Identification, Molecular Cloning, and Characterization of the Chromosome 12 Breakpoint Cluster Region of Uterine Leiomyomas. Genes Chromosomes Cancer 1994, 11, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Mehine, M.; Kaasinen, E.; Mäkinen, N.; Katainen, R.; Kämpjärvi, K.; Pitkänen, E.; Heinonen, H.-R.; Bützow, R.; Kilpivaara, O.; Kuosmanen, A.; et al. Characterization of Uterine Leiomyomas by Whole-Genome Sequencing. N. Engl. J. Med. 2013, 369, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, C.; Markowski, D.N.; Koczan, D.; Küpker, W.; Helmke, B.M.; Bullerdiek, J. Cytogenetically Normal Uterine Leiomyomas without MED12-Mutations—A Source to Identify Unknown Mechanisms of the Development of Uterine Smooth Muscle Tumors. Mol. Cytogenet. 2014, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Heim, S.; Nilbert, M.; Vanni, R.; Floderus, U.M.; Mandahl, N.; Liedgren, S.; Lecca, U.; Mitelman, F. A Specific Translocation, t(12;14)(Q14-15;Q23-24), Characterizes a Subgroup of Uterine Leiomyomas. Cancer Genet. Cytogenet. 1988, 32, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Hennig, Y.; Deichert, U.; Bonk, U.; Thode, B.; Bartnitzke, S.; Bullerdiek, J. Chromosomal Translocations Affecting 12q14-15 but Not Deletions of the Long Arm of Chromosome 7 Associated with a Growth Advantage of Uterine Smooth Muscle Cells. Mol. Hum. Reprod. 1999, 5, 1150–1154. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg Fejzo, M.; Ashar, H.R.; Krauter, K.S.; Powell, W.L.; Rein, M.S.; Weremowicz, S.; Yoon, S.J.; Kucherlapati, R.S.; Chada, K.; Morton, C.C. Translocation Breakpoints Upstream of the HMGIC Gene in Uterine Leiomyomata Suggest Dysregulation of This Gene by a Mechanism Different from That in Lipomas. Genes Chromosomes Cancer 1996, 17, 1–6. [Google Scholar] [CrossRef]
- Gattas, G.J.; Quade, B.J.; Nowak, R.A.; Morton, C.C. HMGIC Expression in Human Adult and Fetal Tissues and in Uterine Leiomyomata. Genes Chromosomes Cancer 1999, 25, 316–322. [Google Scholar] [CrossRef]
- Parisi, S.; Piscitelli, S.; Passaro, F.; Russo, T. HMGA Proteins in Stemness and Differentiation of Embryonic and Adult Stem Cells. Int. J. Mol. Sci. 2020, 21, 362. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Ditzel, H.J.; Duijf, P.H.G.; Khaze, V.; Gjerstorff, M.F.; Baradaran, B. HMGA2 as a Critical Regulator in Cancer Development. Genes 2021, 12, 269. [Google Scholar] [CrossRef]
- Quade, B.J.; Weremowicz, S.; Neskey, D.M.; Vanni, R.; Ladd, C.; Dal Cin, P.; Morton, C.C. Fusion Transcripts Involving HMGA2 Are Not a Common Molecular Mechanism in Uterine Leiomyomata with Rearrangements in 12q15. Cancer Res. 2003, 63, 1351–1358. [Google Scholar]
- Pradhan, B.; Sarvilinna, N.; Matilainen, J.; Aska, E.; Sjöberg, J.; Kauppi, L. Detection and Screening of Chromosomal Rearrangements in Uterine Leiomyomas by Long-Distance Inverse PCR. Genes Chromosomes Cancer 2016, 55, 215–226. [Google Scholar] [CrossRef]
- Peng, Y.; Laser, J.; Shi, G.; Mittal, K.; Melamed, J.; Lee, P.; Wei, J.-J. Antiproliferative Effects by Let-7 Repression of High-Mobility Group A2 in Uterine Leiomyoma. Mol. Cancer Res. MCR 2008, 6, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Klemke, M.; Meyer, A.; Hashemi Nezhad, M.; Belge, G.; Bartnitzke, S.; Bullerdiek, J. Loss of Let-7 Binding Sites Resulting from Truncations of the 3’ Untranslated Region of HMGA2 MRNA in Uterine Leiomyomas. Cancer Genet. Cytogenet. 2010, 196, 119–123. [Google Scholar] [CrossRef]
- Klemke, M.; Meyer, A.; Nezhad, M.H.; Bartnitzke, S.; Drieschner, N.; Frantzen, C.; Schmidt, E.H.; Belge, G.; Bullerdiek, J. Overexpression of HMGA2 in Uterine Leiomyomas Points to Its General Role for the Pathogenesis of the Disease. Genes Chromosomes Cancer 2009, 48, 171–178. [Google Scholar] [CrossRef] [PubMed]
- George, J.W.; Fan, H.; Johnson, B.; Carpenter, T.J.; Foy, K.K.; Chatterjee, A.; Patterson, A.L.; Koeman, J.; Adams, M.; Madaj, Z.B.; et al. Integrated Epigenome, Exome, and Transcriptome Analyses Reveal Molecular Subtypes and Homeotic Transformation in Uterine Fibroids. Cell Rep. 2019, 29, 4069–4085. [Google Scholar] [CrossRef]
- Takahashi, K.; Kawamura, N.; Ishiko, O.; Ogita, S. Shrinkage Effect of Gonadotropin Releasing Hormone Agonist Treatment on Uterine Leiomyomas and t(12;14). Int. J. Oncol. 2002, 20, 279–283. [Google Scholar] [CrossRef]
- Markowski, D.N.; Bartnitzke, S.; Belge, G.; Drieschner, N.; Helmke, B.M.; Bullerdiek, J. Cell Culture and Senescence in Uterine Fibroids. Cancer Genet. Cytogenet. 2010, 202, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Markowski, D.N.; von Ahsen, I.; Nezhad, M.H.; Wosniok, W.; Helmke, B.M.; Bullerdiek, J. HMGA2 and the P19Arf-TP53-CDKN1A Axis: A Delicate Balance in the Growth of Uterine Leiomyomas. Genes Chromosomes Cancer 2010, 49, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Markowski, D.N.; Helmke, B.M.; Belge, G.; Nimzyk, R.; Bartnitzke, S.; Deichert, U.; Bullerdiek, J. HMGA2 and P14Arf: Major Roles in Cellular Senescence of Fibroids and Therapeutic Implications. Anticancer Res. 2011, 31, 753–761. [Google Scholar]
- Mas, A.; Cervelló, I.; Fernández-Álvarez, A.; Faus, A.; Díaz, A.; Burgués, O.; Casado, M.; Simón, C. Overexpression of the Truncated Form of High Mobility Group A Proteins (HMGA2) in Human Myometrial Cells Induces Leiomyoma-like Tissue Formation. Mol. Hum. Reprod. 2015, 21, 330–338. [Google Scholar] [CrossRef]
- Helmke, B.M.; Markowski, D.N.; Müller, M.H.; Sommer, A.; Müller, J.; Möller, C.; Bullerdiek, J. HMGA Proteins Regulate the Expression of FGF2 in Uterine Fibroids. Mol. Hum. Reprod. 2011, 17, 135–142. [Google Scholar] [CrossRef]
- Hodge, J.C.; Kim, T.-M.; Dreyfuss, J.M.; Somasundaram, P.; Christacos, N.C.; Rousselle, M.; Quade, B.J.; Park, P.J.; Stewart, E.A.; Morton, C.C. Expression Profiling of Uterine Leiomyomata Cytogenetic Subgroups Reveals Distinct Signatures in Matched Myometrium: Transcriptional Profilingof the t(12;14) and Evidence in Support of Predisposing Genetic Heterogeneity. Hum. Mol. Genet. 2012, 21, 2312–2329. [Google Scholar] [CrossRef] [PubMed]
- Klemke, M.; Müller, M.H.; Wosniok, W.; Markowski, D.N.; Nimzyk, R.; Helmke, B.M.; Bullerdiek, J. Correlated Expression of HMGA2 and PLAG1 in Thyroid Tumors, Uterine Leiomyomas and Experimental Models. PLoS ONE 2014, 9, e88126. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ubango, J.; Ban, Y.; Chakravarti, D.; Kim, J.J.; Wei, J.-J. Comparative Analysis of AKT and the Related Biomarkers in Uterine Leiomyomas with MED12, HMGA2, and FH Mutations. Genes Chromosomes Cancer 2018, 57, 485–494. [Google Scholar] [CrossRef]
- Liu, B.; Chen, G.; He, Q.; Liu, M.; Gao, K.; Cai, B.; Qu, J.; Lin, S.; Geng, A.; Li, S.; et al. An HMGA2-P62-ERα Axis Regulates Uterine Leiomyomas Proliferation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 10966–10983. [Google Scholar] [CrossRef]
- Ingraham, S.E.; Lynch, R.A.; Kathiresan, S.; Buckler, A.J.; Menon, A.G. HREC2, a RAD51-like Gene, Is Disrupted by t(12;14) (Q15;Q24.1) in a Uterine Leiomyoma. Cancer Genet. Cytogenet. 1999, 115, 56–61. [Google Scholar] [CrossRef]
- Schoenmakers, E.F.; Huysmans, C.; Van de Ven, W.J. Allelic Knockout of Novel Splice Variants of Human Recombination Repair Gene RAD51B in t(12;14) Uterine Leiomyomas. Cancer Res. 1999, 59, 19–23. [Google Scholar] [PubMed]
- Kurose, K.; Mine, N.; Doi, D.; Ota, Y.; Yoneyama, K.; Konishi, H.; Araki, T.; Emi, M. Novel Gene Fusion of COX6C at 8q22-23 to HMGIC at 12q15 in a Uterine Leiomyoma. Genes Chromosomes Cancer 2000, 27, 303–307. [Google Scholar] [CrossRef]
- Mine, N.; Kurose, K.; Konishi, H.; Araki, T.; Nagai, H.; Emi, M. Fusion of a Sequence from HEI10 (14q11) to the HMGIC Gene at 12q15 in a Uterine Leiomyoma. Jpn. J. Cancer Res. Gann 2001, 92, 135–139. [Google Scholar] [CrossRef]
- Mine, N.; Kurose, K.; Nagai, H.; Doi, D.; Ota, Y.; Yoneyama, K.; Konishi, H.; Araki, T.; Emi, M. Gene Fusion Involving HMGIC Is a Frequent Aberration in Uterine Leiomyomas. J. Hum. Genet. 2001, 46, 408–412. [Google Scholar] [CrossRef]
- Velagaleti, G.V.N.; Tonk, V.S.; Hakim, N.M.; Wang, X.; Zhang, H.; Erickson-Johnson, M.R.; Medeiros, F.; Oliveira, A.M. Fusion of HMGA2 to COG5 in Uterine Leiomyoma. Cancer Genet. Cytogenet. 2010, 202, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.; Swanson, D.; Tang, S.; Dickson, B.C.; Turashvili, G. Gene Fusions Characterize a Subset of Uterine Cellular Leiomyomas. Genes Chromosomes Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, B.; Bol, S.; Wanschura, S.; Bartnitzke, S.; Bullerdiek, J. PAC Clone Containing the HMGI(Y) Gene Spans the Breakpoint of a 6p21 Translocation in a Uterine Leiomyoma Cell Line. Genes Chromosomes Cancer 1996, 17, 191–193. [Google Scholar] [CrossRef]
- Nezhad, M.H.; Drieschner, N.; Helms, S.; Meyer, A.; Tadayyon, M.; Klemke, M.; Belge, G.; Bartnitzke, S.; Burchardt, K.; Frantzen, C.; et al. 6p21 Rearrangements in Uterine Leiomyomas Targeting HMGA1. Cancer Genet. Cytogenet. 2010, 203, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.J.; Powell, W.L.; Collins, T.; Morton, C.C. HMGI(Y) Expression in Human Uterine Leiomyomata. Involvement of Another High-Mobility Group Architectural Factor in a Benign Neoplasm. Am. J. Pathol. 1997, 150, 911–918. [Google Scholar] [PubMed]
- Tallini, G.; Vanni, R.; Manfioletti, G.; Kazmierczak, B.; Faa, G.; Pauwels, P.; Bullerdiek, J.; Giancotti, V.; Van Den Berghe, H.; Dal Cin, P. HMGI-C and HMGI(Y) Immunoreactivity Correlates with Cytogenetic Abnormalities in Lipomas, Pulmonary Chondroid Hamartomas, Endometrial Polyps, and Uterine Leiomyomas and Is Compatible with Rearrangement of the HMGI-C and HMGI(Y) Genes. Lab. Investig. J. Tech. Methods Pathol. 2000, 80, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Ozisik, Y.Y.; Meloni, A.M.; Surti, U.; Sandberg, A.A. Involvement of 10q22 in Leiomyoma. Cancer Genet. Cytogenet. 1993, 69, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.D.P.; Herrick, S.R.; Ince, T.A.; Kleinman, M.S.; Dal Cin, P.; Morton, C.C.; Quade, B.J. Uterine Leiomyomata with t(10;17) Disrupt the Histone Acetyltransferase MORF. Cancer Res. 2004, 64, 5570–5577. [Google Scholar] [CrossRef]
- Ainsworth, A.J.; Dashti, N.K.; Mounajjed, T.; Fritchie, K.J.; Davila, J.; Mopuri, R.; Jackson, R.A.; Halling, K.C.; Bakkum-Gamez, J.N.; Schoolmeester, J.K. Leiomyoma with KAT6B-KANSL1 Fusion: Case Report of a Rapidly Enlarging Uterine Mass in a Postmenopausal Woman. Diagn. Pathol. 2019, 14, 32. [Google Scholar] [CrossRef]
- Agaimy, A.; Clarke, B.A.; Kolin, D.L.; Lee, C.-H.; Lee, J.-C.; McCluggage, W.G.; Pöschke, P.; Stoehr, R.; Swanson, D.; Turashvili, G.; et al. Recurrent KAT6B/A::KANSL1 Fusions Characterize a Potentially Aggressive Uterine Sarcoma Morphologically Overlapping With Low-Grade Endometrial Stromal Sarcoma. Am. J. Surg. Pathol. 2022, 46, 1298–1308. [Google Scholar] [CrossRef]
- Xing, Y.P.; Powell, W.L.; Morton, C.C. The Del(7q) Subgroup in Uterine Leiomyomata: Genetic and Biologic Characteristics. Further Evidence for the Secondary Nature of Cytogenetic Abnormalities in the Pathobiology of Uterine Leiomyomata. Cancer Genet. Cytogenet. 1997, 98, 69–74. [Google Scholar] [CrossRef]
- Ozisik, Y.Y.; Meloni, A.M.; Powell, M.; Surti, U.; Sandberg, A.A. Chromosome 7 Biclonality in Uterine Leiomyoma. Cancer Genet. Cytogenet. 1993, 67, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Medikare, V.; Kandukuri, L.R.; Ananthapur, V.; Deenadayal, M.; Nallari, P. The Genetic Bases of Uterine Fibroids; a Review. J. Reprod. Infertil. 2011, 12, 181–191. [Google Scholar] [PubMed]
- Kingsley, K.L.; Meloni, A.M.; Peier, A.M.; Sandberg, A.A.; Surti, U. Deletion of Both Chromosome 7 Homologues in Leiomyoma. Cancer Genet. Cytogenet. 1995, 81, 99–100. [Google Scholar] [CrossRef]
- Sargent, M.S.; Weremowicz, S.; Rein, M.S.; Morton, C.C. Translocations in 7q22 Define a Critical Region in Uterine Leiomyomata. Cancer Genet. Cytogenet. 1994, 77, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Ishwad, C.S.; Ferrell, R.E.; Davare, J.; Meloni, A.M.; Sandberg, A.A.; Surti, U. Molecular and Cytogenetic Analysis of Chromosome 7 in Uterine Leiomyomas. Genes Chromosomes Cancer 1995, 14, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Ishwad, C.S.; Ferrell, R.E.; Hanley, K.; Davare, J.; Meloni, A.M.; Sandberg, A.A.; Surti, U. Two Discrete Regions of Deletion at 7q in Uterine Leiomyomas. Genes Chromosomes Cancer 1997, 19, 156–160. [Google Scholar] [CrossRef]
- van der Heijden, O.; Chiu, H.C.; Park, T.C.; Takahashi, H.; LiVolsi, V.A.; Risinger, J.I.; Barrett, J.C.; Berchuck, A.; Evans, A.C.; Behbakht, K.; et al. Allelotype Analysis of Uterine Leiomyoma: Localization of a Potential Tumor Suppressor Gene to a 4-CM Region of Chromosome 7q. Mol. Carcinog. 1998, 23, 243–247. [Google Scholar] [CrossRef]
- Vanharanta, S.; Wortham, N.C.; Langford, C.; El-Bahrawy, M.; van der Spuy, Z.; Sjöberg, J.; Lehtonen, R.; Karhu, A.; Tomlinson, I.P.M.; Aaltonen, L.A. Definition of a Minimal Region of Deletion of Chromosome 7 in Uterine Leiomyomas by Tiling-Path Microarray CGH and Mutation Analysis of Known Genes in This Region. Genes Chromosomes Cancer 2007, 46, 451–458. [Google Scholar] [CrossRef]
- Zeng, W.R.; Scherer, S.W.; Koutsilieris, M.; Huizenga, J.J.; Filteau, F.; Tsui, L.C.; Nepveu, A. Loss of Heterozygosity and Reduced Expression of the CUTL1 Gene in Uterine Leiomyomas. Oncogene 1997, 14, 2355–2365. [Google Scholar] [CrossRef]
- Schoenmakers, E.F.P.M.; Bunt, J.; Hermers, L.; Schepens, M.; Merkx, G.; Janssen, B.; Kersten, M.; Huys, E.; Pauwels, P.; Debiec-Rychter, M.; et al. Identification of CUX1 as the Recurrent Chromosomal Band 7q22 Target Gene in Human Uterine Leiomyoma. Genes Chromosomes Cancer 2013, 52, 11–23. [Google Scholar] [CrossRef]
- Quintana, D.G.; Thome, K.C.; Hou, Z.H.; Ligon, A.H.; Morton, C.C.; Dutta, A. ORC5L, a New Member of the Human Origin Recognition Complex, Is Deleted in Uterine Leiomyomas and Malignant Myeloid Diseases. J. Biol. Chem. 1998, 273, 27137–27145. [Google Scholar] [CrossRef]
- Sell, S.M.; Tullis, C.; Stracner, D.; Song, C.-Y.; Gewin, J. Minimal Interval Defined on 7q in Uterine Leiomyoma. Cancer Genet. Cytogenet. 2005, 157, 67–69. [Google Scholar] [CrossRef]
- Ptacek, T.; Song, C.; Walker, C.L.; Sell, S.M. Physical Mapping of Distinct 7q22 Deletions in Uterine Leiomyoma and Analysis of a Recently Annotated 7q22 Candidate Gene. Cancer Genet. Cytogenet. 2007, 174, 116–120. [Google Scholar] [CrossRef]
- Saito, E.; Okamoto, A.; Saito, M.; Shinozaki, H.; Takakura, S.; Yanaihara, N.; Ochiai, K.; Tanaka, T. Genes Associated with the Genesis of Leiomyoma of the Uterus in a Commonly Deleted Chromosomal Region at 7q22. Oncol. Rep. 2005, 13, 469–472. [Google Scholar] [CrossRef]
- Hodge, J.C.; Park, P.J.; Dreyfuss, J.M.; Assil-Kishawi, I.; Somasundaram, P.; Semere, L.G.; Quade, B.J.; Lynch, A.M.; Stewart, E.A.; Morton, C.C. Identifying the Molecular Signature of the Interstitial Deletion 7q Subgroup of Uterine Leiomyomata Using a Paired Analysis. Genes Chromosomes Cancer 2009, 48, 865–885. [Google Scholar] [CrossRef]
- Takahashi, K.; Kawamura, N.; Tsujimura, A.; Ichimura, T.; Ito, F.; Ishiko, O.; Ogita, S. Association of the Shrinkage of Uterine Leiomyoma Treated with GnRH Agonist and Deletion of Long Arm of Chromosome 7. Int. J. Oncol. 2001, 18, 1259–1263. [Google Scholar] [CrossRef]
- Nilbert, M.; Heim, S.; Mandahl, N.; Flodérus, U.M.; Willén, H.; Mitelman, F. Karyotypic Rearrangements in 20 Uterine Leiomyomas. Cytogenet. Cell Genet. 1988, 49, 300–304. [Google Scholar] [CrossRef]
- Nilbert, M.; Heim, S.; Mandahl, N.; Flodérus, U.M.; Willén, H.; Akerman, M.; Mitelman, F. Ring Formation and Structural Rearrangements of Chromosome 1 as Secondary Changes in Uterine Leiomyomas with t(12;14)(Q14-15;Q23-24). Cancer Genet. Cytogenet. 1988, 36, 183–190. [Google Scholar] [CrossRef]
- Pandis, N.; Heim, S.; Bardi, G.; Flodérus, U.M.; Willén, H.; Mandahl, N.; Mitelman, F. Parallel Karyotypic Evolution and Tumor Progression in Uterine Leiomyoma. Genes Chromosomes Cancer 1990, 2, 311–317. [Google Scholar] [CrossRef]
- Pandis, N.; Heim, S.; Bardi, G.; Flodérus, U.M.; Willén, H.; Mandahl, N.; Mitelman, F. Chromosome Analysis of 96 Uterine Leiomyomas. Cancer Genet. Cytogenet. 1991, 55, 11–18. [Google Scholar] [CrossRef]
- Levy, B.; Mukherjee, T.; Hirschhorn, K. Molecular Cytogenetic Analysis of Uterine Leiomyoma and Leiomyosarcoma by Comparative Genomic Hybridization. Cancer Genet. Cytogenet. 2000, 121, 1–8. [Google Scholar] [CrossRef]
- Christacos, N.C.; Quade, B.J.; Dal Cin, P.; Morton, C.C. Uterine Leiomyomata with Deletions of Ip Represent a Distinct Cytogenetic Subgroup Associated with Unusual Histologic Features. Genes Chromosomes Cancer 2006, 45, 304–312. [Google Scholar] [CrossRef]
- Hodge, J.C.; Pearce, K.E.; Clayton, A.C.; Taran, F.A.; Stewart, E.A. Uterine Cellular Leiomyomata with Chromosome 1p Deletions Represent a Distinct Entity. Am. J. Obstet. Gynecol. 2014, 210, 572.e1–572.e7. [Google Scholar] [CrossRef]
- Korbel, J.O.; Campbell, P.J. Criteria for Inference of Chromothripsis in Cancer Genomes. Cell 2013, 152, 1226–1236. [Google Scholar] [CrossRef]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Koster, J.; Molenaar, J.J. Prevalence and Clinical Implications of Chromothripsis in Cancer Genomes. Curr. Opin. Oncol. 2014, 26, 64–72. [Google Scholar] [CrossRef]
- Koltsova, A.S.; Pendina, A.A.; Efimova, O.A.; Chiryaeva, O.G.; Kuznetzova, T.V.; Baranov, V.S. On the Complexity of Mechanisms and Consequences of Chromothripsis: An Update. Front. Genet. 2019, 10, 393. [Google Scholar] [CrossRef]
- Pendina, A.A.; Koltsova, A.S.; Efimova, O.A.; Malysheva, O.V.; Osinovskaya, N.S.; Sultanov, I.Y.; Tikhonov, A.V.; Shved, N.Y.; Chiryaeva, O.G.; Simareva, A.D.; et al. Case of Chromothripsis in a Large Solitary Non-Recurrent Uterine Leiomyoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017, 219, 134–136. [Google Scholar] [CrossRef]
- Bowden, W.; Skorupski, J.; Kovanci, E.; Rajkovic, A. Detection of Novel Copy Number Variants in Uterine Leiomyomas Using High-Resolution SNP Arrays. Mol. Hum. Reprod. 2009, 15, 563–568. [Google Scholar] [CrossRef]
- Nagai, K.; Asano, R.; Sekiguchi, F.; Asai-Sato, M.; Miyagi, Y.; Miyagi, E. MED12 Mutations in Uterine Leiomyomas: Prediction of Volume Reduction by Gonadotropin-Releasing Hormone Agonists. Am. J. Obstet. Gynecol. 2022, S0002-9378(22)00748-7. [Google Scholar] [CrossRef] [PubMed]
- Kolterud, Å.; Välimäki, N.; Kuisma, H.; Patomo, J.; Ilves, S.T.; Mäkinen, N.; Kaukomaa, J.; Palin, K.; Kaasinen, E.; Karhu, A.; et al. Molecular Subclass of Uterine Fibroids Predicts Tumor Shrinkage in Response to Ulipristal Acetate. Hum. Mol. Genet. 2022, ddac217. [Google Scholar] [CrossRef] [PubMed]
- Nibert, M.; Heim, S. Uterine Leiomyoma Cytogenetics. Genes Chromosomes Cancer 1990, 2, 3–13. [Google Scholar] [CrossRef]
- Canevari, R.A.; Pontes, A.; Rosa, F.E.; Rainho, C.A.; Rogatto, S.R. Independent Clonal Origin of Multiple Uterine Leiomyomas That Was Determined by X Chromosome Inactivation and Microsatellite Analysis. Am. J. Obstet. Gynecol. 2005, 193, 1395–1403. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, C.; Hao, J.; Sung, C.J.; Quddus, M.R.; Steinhoff, M.M.; Lawrence, W.D. Use of X-Chromosome Inactivation Pattern to Determine the Clonal Origins of Uterine Leiomyoma and Leiomyosarcoma. Hum. Pathol. 2006, 37, 1350–1356. [Google Scholar] [CrossRef]
- Cai, Y.-R.; Diao, X.-L.; Wang, S.-F.; Zhang, W.; Zhang, H.-T.; Su, Q. X-Chromosomal Inactivation Analysis of Uterine Leiomyomas Reveals a Common Clonal Origin of Different Tumor Nodules in Some Multiple Leiomyomas. Int. J. Oncol. 2007, 31, 1379–1389. [Google Scholar] [CrossRef]
- Mehine, M.; Heinonen, H.-R.; Sarvilinna, N.; Pitkänen, E.; Mäkinen, N.; Katainen, R.; Tuupanen, S.; Bützow, R.; Sjöberg, J.; Aaltonen, L.A. Clonally Related Uterine Leiomyomas Are Common and Display Branched Tumor Evolution. Hum. Mol. Genet. 2015, 24, 4407–4416. [Google Scholar] [CrossRef]
- Fasih, N.; Prasad Shanbhogue, A.K.; Macdonald, D.B.; Fraser-Hill, M.A.; Papadatos, D.; Kielar, A.Z.; Doherty, G.P.; Walsh, C.; McInnes, M.; Atri, M. Leiomyomas beyond the Uterus: Unusual Locations, Rare Manifestations. Radiogr. Rev. Publ. Radiol. Soc. N. Am. Inc 2008, 28, 1931–1948. [Google Scholar] [CrossRef]
- Quade, B.J.; McLachlin, C.M.; Soto-Wright, V.; Zuckerman, J.; Mutter, G.L.; Morton, C.C. Disseminated Peritoneal Leiomyomatosis. Clonality Analysis by X Chromosome Inactivation and Cytogenetics of a Clinically Benign Smooth Muscle Proliferation. Am. J. Pathol. 1997, 150, 2153–2166. [Google Scholar]
- Dal Cin, P.; Quade, B.J.; Neskey, D.M.; Kleinman, M.S.; Weremowicz, S.; Morton, C.C. Intravenous Leiomyomatosis Is Characterized by a Der(14)t(12;14)(Q15;Q24). Genes Chromosomes Cancer 2003, 36, 205–206. [Google Scholar] [CrossRef]
- Bowen, J.M.; Cates, J.M.; Kash, S.; Itani, D.; Gonzalez, A.; Huang, D.; Oliveira, A.; Bridge, J.A. Genomic Imbalances in Benign Metastasizing Leiomyoma: Characterization by Conventional Karyotypic, Fluorescence in Situ Hybridization, and Whole Genome SNP Array Analysis. Cancer Genet. 2012, 205, 249–254. [Google Scholar] [CrossRef]
- Wu, R.-C.; Chao, A.-S.; Lee, L.-Y.; Lin, G.; Chen, S.-J.; Lu, Y.-J.; Huang, H.-J.; Yen, C.-F.; Han, C.M.; Lee, Y.-S.; et al. Massively Parallel Sequencing and Genome-Wide Copy Number Analysis Revealed a Clonal Relationship in Benign Metastasizing Leiomyoma. Oncotarget 2017, 8, 47547–47554. [Google Scholar] [CrossRef]
- Jiang, J.; He, M.; Hu, X.; Ni, C.; Yang, L. Deep Sequencing Reveals the Molecular Pathology Characteristics between Primary Uterine Leiomyoma and Pulmonary Benign Metastasizing Leiomyoma. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2018, 20, 1080–1086. [Google Scholar] [CrossRef]
- Ofori, K.; Fernandes, H.; Cummings, M.; Colby, T.; Saqi, A. Benign Metastasizing Leiomyoma Presenting with Miliary Pattern and Fatal Outcome: Case Report with Molecular Analysis & Review of the Literature. Respir. Med. Case Rep. 2019, 27, 100831. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, S.; Liu, Q.; Lu, B. A Clinicopathological and Molecular Analysis in Uterine Leiomyomas and Concurrent/Metachronous Peritoneal Nodules: New Insights into Disseminated Peritoneal Leiomyomatosis. Pathol. Res. Pract. 2020, 216, 152938. [Google Scholar] [CrossRef]
- Nucci, M.R.; Drapkin, R.; Dal Cin, P.; Fletcher, C.D.M.; Fletcher, J.A. Distinctive Cytogenetic Profile in Benign Metastasizing Leiomyoma: Pathogenetic Implications. Am. J. Surg. Pathol. 2007, 31, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Ahvenainen, T.; Khamaiseh, S.; Alkodsi, A.; Mehine, M.; Nevala, R.; Äyräväinen, A.; Bützow, R.; Vahteristo, P. Lung Metastases and Subsequent Malignant Transformation of a Fumarate Hydratase -Deficient Uterine Leiomyoma. Exp. Mol. Pathol. 2022, 126, 104760. [Google Scholar] [CrossRef] [PubMed]
- Ahvenainen, T.V.; Mäkinen, N.M.; von Nandelstadh, P.; Vahteristo, M.E.A.; Pasanen, A.M.; Bützow, R.C.; Vahteristo, P.M. Loss of ATRX/DAXX Expression and Alternative Lengthening of Telomeres in Uterine Leiomyomas. Cancer 2018, 124, 4650–4656. [Google Scholar] [CrossRef] [PubMed]
- Sanada, S.; Ushijima, K.; Yanai, H.; Mikami, Y.; Ohishi, Y.; Kobayashi, H.; Tashiro, H.; Mikami, M.; Miyamoto, S.; Katabuchi, H. A Critical Review of “Uterine Leiomyoma” with Subsequent Recurrence or Metastasis: A Multicenter Study of 62 Cases. J. Obstet. Gynaecol. Res. 2022, 48, 3242–3251. [Google Scholar] [CrossRef]
- Panesar, H.; Dhaliwal, H.S. Iatrogenic Parasitic Leiomyomas: A Late and Uncommon Complication After Laparoscopic Morcellation. Cureus 2022, 14, e24718. [Google Scholar] [CrossRef]
- Holzmann, C.; Kuepker, W.; Rommel, B.; Helmke, B.; Bullerdiek, J. Reasons to Reconsider Risk Associated With Power Morcellation of Uterine Fibroids. Vivo Athens Greece 2020, 34, 1–9. [Google Scholar] [CrossRef]
- Kyozuka, H.; Jin, T.; Sugeno, M.; Kuratsune, K.; Ando, H.; Ito, F.; Odajima, H.; Suzuki, D.; Nomura, Y. A Case of Spontaneous Parasitic Myoma in a Patient without a History of Myomectomy Treated Laparoscopically. Fukushima J. Med. Sci. 2022, 68, 123–127. [Google Scholar] [CrossRef]
- Mittal, K.; Joutovsky, A. Areas with Benign Morphologic and Immunohistochemical Features Are Associated with Some Uterine Leiomyosarcomas. Gynecol. Oncol. 2007, 104, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Mittal, K.R.; Chen, F.; Wei, J.J.; Rijhvani, K.; Kurvathi, R.; Streck, D.; Dermody, J.; Toruner, G.A. Molecular and Immunohistochemical Evidence for the Origin of Uterine Leiomyosarcomas from Associated Leiomyoma and Symplastic Leiomyoma-like Areas. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2009, 22, 1303–1311. [Google Scholar] [CrossRef]
- Packenham, J.P.; du Manoir, S.; Schrock, E.; Risinger, J.I.; Dixon, D.; Denz, D.N.; Evans, J.A.; Berchuck, A.; Barrett, J.C.; Devereux, T.R.; et al. Analysis of Genetic Alterations in Uterine Leiomyomas and Leiomyosarcomas by Comparative Genomic Hybridization. Mol. Carcinog. 1997, 19, 273–279. [Google Scholar] [CrossRef]
- Ravegnini, G.; Mariño-Enriquez, A.; Slater, J.; Eilers, G.; Wang, Y.; Zhu, M.; Nucci, M.R.; George, S.; Angelini, S.; Raut, C.P.; et al. MED12 Mutations in Leiomyosarcoma and Extrauterine Leiomyoma. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2013, 26, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Bertsch, E.; Qiang, W.; Zhang, Q.; Espona-Fiedler, M.; Druschitz, S.; Liu, Y.; Mittal, K.; Kong, B.; Kurita, T.; Wei, J.-J. MED12 and HMGA2 Mutations: Two Independent Genetic Events in Uterine Leiomyoma and Leiomyosarcoma. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 2014, 27, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.C.; Morton, C.C. Genetic Heterogeneity among Uterine Leiomyomata: Insights into Malignant Progression. Hum. Mol. Genet. 2007, 16 Spec No 1, R7–R13. [Google Scholar] [CrossRef]
- Zhang, Q.; Ubago, J.; Li, L.; Guo, H.; Liu, Y.; Qiang, W.; Kim, J.J.; Kong, B.; Wei, J.-J. Molecular Analyses of 6 Different Types of Uterine Smooth Muscle Tumors: Emphasis in Atypical Leiomyoma. Cancer 2014, 120, 3165–3177. [Google Scholar] [CrossRef] [PubMed]
- Dharajiya, N.G.; Namba, A.; Horiuchi, I.; Miyai, S.; Farkas, D.H.; Almasri, E.; Saldivar, J.-S.; Takagi, K.; Kamei, Y. Uterine Leiomyoma Confounding a Noninvasive Prenatal Test Result. Prenat. Diagn. 2015, 35, 990–993. [Google Scholar] [CrossRef]
- Dharajiya, N.G.; Grosu, D.S.; Farkas, D.H.; McCullough, R.M.; Almasri, E.; Sun, Y.; Kim, S.K.; Jensen, T.J.; Saldivar, J.-S.; Topol, E.J.; et al. Incidental Detection of Maternal Neoplasia in Noninvasive Prenatal Testing. Clin. Chem. 2018, 64, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Ivashchenko, T.E.; Vashukova, E.S.; Kozyulina, P.Y.; Dvoynova, N.M.; Talantova, O.E.; Koroteev, A.L.; Pendina, A.A.; Tikhonov, A.V.; Chiryaeva, O.G.; Petrova, L.I.; et al. Noninvasive Prenatal Testing Using Next Generation Sequencing: Pilot Experience of the D.O. Ott Research Institute of Obstetrics, Gynecology and Reproductology. Russ. J. Genet. 2019, 55, 1208–1213. [Google Scholar] [CrossRef]
- Scott, F.; Menezes, M.; Smet, M.E.; Carey, K.; Hardy, T.; Fullston, T.; Rolnik, D.L.; McLennan, A. Influence of Fibroids on Cell-Free DNA Screening Accuracy. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2022, 59, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Linder, D.; Gartler, S.M. Glucose-6-Phosphate Dehydrogenase Mosaicism: Utilization as a Cell Marker in the Study of Leiomyomas. Science 1965, 150, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Swierczek, S.I.; Piterkova, L.; Jelinek, J.; Agarwal, N.; Hammoud, S.; Wilson, A.; Hickman, K.; Parker, C.J.; Cairns, B.R.; Prchal, J.T. Methylation of AR Locus Does Not Always Reflect X Chromosome Inactivation State. Blood 2012, 119, e100–e109. [Google Scholar] [CrossRef] [PubMed]
- Bullerdiek, J.; Rommel, B. Factors Targeting MED12 to Drive Tumorigenesis? F1000Research 2018, 7, 359. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.S. Molecular Pathways: Transcription Factories and Chromosomal Translocations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 296–300. [Google Scholar] [CrossRef]
- Fritz, A.J.; Sehgal, N.; Pliss, A.; Xu, J.; Berezney, R. Chromosome Territories and the Global Regulation of the Genome. Genes Chromosomes Cancer 2019, 58, 407–426. [Google Scholar] [CrossRef]
- Rogalla, P.; Rohen, C.; Hennig, Y.; Deichert, U.; Bonk, U.; Bullerdiek, J. Telomere Repeat Fragment Sizes Do Not Limit the Growth Potential of Uterine Leiomyomas. Biochem. Biophys. Res. Commun. 1995, 211, 175–182. [Google Scholar] [CrossRef]
- Bonatz, G.; Frahm, S.O.; Andreas, S.; Heidorn, K.; Jonat, W.; Parwaresch, R. Telomere Shortening in Uterine Leiomyomas. Am. J. Obstet. Gynecol. 1998, 179, 591–596. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Hu, J.F.; Vu, T.H.; Oruganti, H.; Giudice, L.C.; Hoffman, A.R. Regulation of Telomerase by Alternate Splicing of Human Telomerase Reverse Transcriptase (HTERT) in Normal and Neoplastic Ovary, Endometrium and Myometrium. Int. J. Cancer 2000, 85, 330–335. [Google Scholar] [CrossRef]
- Oh, B.-K.; Choi, Y.; Choi, J.S. Telomere Shortening and Expression of TRF1 and TRF2 in Uterine Leiomyoma. Mol. Med. Rep. 2021, 24, 606. [Google Scholar] [CrossRef]
- Maciejowski, J.; Chatzipli, A.; Dananberg, A.; Chu, K.; Toufektchan, E.; Klimczak, L.J.; Gordenin, D.A.; Campbell, P.J.; de Lange, T. APOBEC3-Dependent Kataegis and TREX1-Driven Chromothripsis during Telomere Crisis. Nat. Genet. 2020, 52, 884–890. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; Yao, X.; Rosiene, J.; Tian, H.; Behr, J.; Bosco, N.; Takai, K.K.; de Lange, T.; Imieliński, M. Structural Variant Evolution after Telomere Crisis. Nat. Commun. 2021, 12, 2093. [Google Scholar] [CrossRef]
- Kuisma, H.; Bramante, S.; Rajamäki, K.; Sipilä, L.J.; Kaasinen, E.; Kaukomaa, J.; Palin, K.; Mäkinen, N.; Sjöberg, J.; Sarvilinna, N.; et al. Parity Associates with Chromosomal Damage in Uterine Leiomyomas. Nat. Commun. 2021, 12, 5448. [Google Scholar] [CrossRef] [PubMed]
- Westhorpe, F.G.; Straight, A.F. The Centromere: Epigenetic Control of Chromosome Segregation during Mitosis. Cold Spring Harb. Perspect. Biol. 2014, 7, a015818. [Google Scholar] [CrossRef]
- Yang, Q.; Mas, A.; Diamond, M.P.; Al-Hendy, A. The Mechanism and Function of Epigenetics in Uterine Leiomyoma Development. Reprod. Sci. Thousand Oaks Calif. 2016, 23, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The Language of Covalent Histone Modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.-H.; Torng, P.-L.; Hsiao, S.-M.; Jeng, Y.-M.; Chen, M.-W.; Chen, C.-A. Histone Deacetylase 6 Regulates Estrogen Receptor Alpha in Uterine Leiomyoma. Reprod. Sci. Thousand Oaks Calif. 2011, 18, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Shahin, S.M.; Sabri, N.A.; Al-Hendy, A.; Yang, Q. Activation of β-Catenin Signaling and Its Crosstalk With Estrogen and Histone Deacetylases in Human Uterine Fibroids. J. Clin. Endocrinol. Metab. 2020, 105, e1517–e1535. [Google Scholar] [CrossRef]
- Sant’Anna, G.D.S.; Brum, I.S.; Branchini, G.; Pizzolato, L.S.; Capp, E.; Corleta, H. von E. Ovarian Steroid Hormones Modulate the Expression of Progesterone Receptors and Histone Acetylation Patterns in Uterine Leiomyoma Cells. Gynecol. Endocrinol. Off. J. Int. Soc. Gynecol. Endocrinol. 2017, 33, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Carbajo-García, M.C.; García-Alcázar, Z.; Corachán, A.; Monleón, J.; Trelis, A.; Faus, A.; Pellicer, A.; Ferrero, H. Histone Deacetylase Inhibition by Suberoylanilide Hydroxamic Acid: A Therapeutic Approach to Treat Human Uterine Leiomyoma. Fertil. Steril. 2022, 117, 433–443. [Google Scholar] [CrossRef]
- Yang, Q.; Nair, S.; Laknaur, A.; Ismail, N.; Diamond, M.P.; Al-Hendy, A. The Polycomb Group Protein EZH2 Impairs DNA Damage Repair Gene Expression in Human Uterine Fibroids. Biol. Reprod. 2016, 94, 69. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Laknaur, A.; Elam, L.; Ismail, N.; Gavrilova-Jordan, L.; Lue, J.; Diamond, M.P.; Al-Hendy, A. Identification of Polycomb Group Protein EZH2-Mediated DNA Mismatch Repair Gene MSH2 in Human Uterine Fibroids. Reprod. Sci. Thousand Oaks Calif. 2016, 23, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Greathouse, K.L.; Bredfeldt, T.; Everitt, J.I.; Lin, K.; Berry, T.; Kannan, K.; Mittelstadt, M.L.; Ho, S.; Walker, C.L. Environmental Estrogens Differentially Engage the Histone Methyltransferase EZH2 to Increase Risk of Uterine Tumorigenesis. Mol. Cancer Res. MCR 2012, 10, 546–557. [Google Scholar] [CrossRef]
- Yu, L.; Ham, K.; Gao, X.; Castro, L.; Yan, Y.; Kissling, G.E.; Tucker, C.J.; Flagler, N.; Dong, R.; Archer, T.K.; et al. Epigenetic Regulation of Transcription Factor Promoter Regions by Low-Dose Genistein through Mitogen-Activated Protein Kinase and Mitogen-and-Stress Activated Kinase 1 Nongenomic Signaling. Cell Commun. Signal. CCS 2016, 14, 18. [Google Scholar] [CrossRef]
- Yu, L.; Liu, J.; Yan, Y.; Burwell, A.; Castro, L.; Shi, M.; Dixon, D. “Metalloestrogenic” Effects of Cadmium Downstream of G Protein-Coupled Estrogen Receptor and Mitogen-Activated Protein Kinase Pathways in Human Uterine Fibroid Cells. Arch. Toxicol. 2021, 95, 1995–2006. [Google Scholar] [CrossRef]
- Berta, D.G.; Kuisma, H.; Välimäki, N.; Räisänen, M.; Jäntti, M.; Pasanen, A.; Karhu, A.; Kaukomaa, J.; Taira, A.; Cajuso, T.; et al. Deficient H2A.Z Deposition Is Associated with Genesis of Uterine Leiomyoma. Nature 2021, 596, 398–403. [Google Scholar] [CrossRef]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional Repression by the Methyl-CpG-Binding Protein MeCP2 Involves a Histone Deacetylase Complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Bochtler, M.; Kolano, A.; Xu, G.-L. DNA Demethylation Pathways: Additional Players and Regulators. BioEssays News Rev. Mol. Cell. Dev. Biol. 2017, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet Proteins in 5mC to 5hmC Conversion, ES-Cell Self-Renewal and Inner Cell Mass Specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.-H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.-T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of α-KG-Dependent Histone and DNA Demethylases by Fumarate and Succinate That Are Accumulated in Mutations of FH and SDH Tumor Suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef]
- Efimova, O.A.; Koltsova, A.S.; Krapivin, M.I.; Tikhonov, A.V.; Pendina, A.A. Environmental Epigenetics and Genome Flexibility: Focus on 5-Hydroxymethylcytosine. Int. J. Mol. Sci. 2020, 21, 3223. [Google Scholar] [CrossRef]
- Nabel, C.S.; Jia, H.; Ye, Y.; Shen, L.; Goldschmidt, H.L.; Stivers, J.T.; Zhang, Y.; Kohli, R.M. AID/APOBEC Deaminases Disfavor Modified Cytosines Implicated in DNA Demethylation. Nat. Chem. Biol. 2012, 8, 751–758. [Google Scholar] [CrossRef]
- Boorstein, R.J.; Cummings, A.; Marenstein, D.R.; Chan, M.K.; Ma, Y.; Neubert, T.A.; Brown, S.M.; Teebor, G.W. Definitive Identification of Mammalian 5-Hydroxymethyluracil DNA N-Glycosylase Activity as SMUG1. J. Biol. Chem. 2001, 276, 41991–41997. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-Mediated Active DNA Demethylation: Mechanism, Function and Beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Ji, D.; Lin, K.; Song, J.; Wang, Y. Effects of Tet-Induced Oxidation Products of 5-Methylcytosine on Dnmt1- and DNMT3a-Mediated Cytosine Methylation. Mol. Biosyst. 2014, 10, 1749–1752. [Google Scholar] [CrossRef]
- Li, S.; Chiang, T.; Richard-Davis, G.; Barrett, J.C.; Mclachlan, J.A. DNA Hypomethylation and Imbalanced Expression of DNA Methyltransferases (DNMT1, 3A, and 3B) in Human Uterine Leiomyoma. Gynecol. Oncol. 2003, 90, 123–130. [Google Scholar] [CrossRef]
- Yamagata, Y.; Maekawa, R.; Asada, H.; Taketani, T.; Tamura, I.; Tamura, H.; Ogane, J.; Hattori, N.; Shiota, K.; Sugino, N. Aberrant DNA Methylation Status in Human Uterine Leiomyoma. Mol. Hum. Reprod. 2009, 15, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Carbajo-García, M.C.; Corachán, A.; Segura-Benitez, M.; Monleón, J.; Escrig, J.; Faus, A.; Pellicer, A.; Cervelló, I.; Ferrero, H. 5-Aza-2′-Deoxycitidine Inhibits Cell Proliferation, Extracellular Matrix Formation and Wnt/β-Catenin Pathway in Human Uterine Leiomyomas. Reprod. Biol. Endocrinol. RBE 2021, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, R.; Yagi, S.; Ohgane, J.; Yamagata, Y.; Asada, H.; Tamura, I.; Sugino, N.; Shiota, K. Disease-Dependent Differently Methylated Regions (D-DMRs) of DNA Are Enriched on the X Chromosome in Uterine Leiomyoma. J. Reprod. Dev. 2011, 57, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Asada, H.; Yamagata, Y.; Taketani, T.; Matsuoka, A.; Tamura, H.; Hattori, N.; Ohgane, J.; Hattori, N.; Shiota, K.; Sugino, N. Potential Link between Estrogen Receptor-Alpha Gene Hypomethylation and Uterine Fibroid Formation. Mol. Hum. Reprod. 2008, 14, 539–545. [Google Scholar] [CrossRef]
- Maekawa, R.; Sato, S.; Yamagata, Y.; Asada, H.; Tamura, I.; Lee, L.; Okada, M.; Tamura, H.; Takaki, E.; Nakai, A.; et al. Genome-Wide DNA Methylation Analysis Reveals a Potential Mechanism for the Pathogenesis and Development of Uterine Leiomyomas. PLoS ONE 2013, 8, e66632. [Google Scholar] [CrossRef]
- Carbajo-García, M.C.; Corachán, A.; Juárez-Barber, E.; Monleón, J.; Payá, V.; Trelis, A.; Quiñonero, A.; Pellicer, A.; Ferrero, H. Integrative Analysis of the DNA Methylome and Transcriptome in Uterine Leiomyoma Shows Altered Regulation of Genes Involved in Metabolism, Proliferation, Extracellular Matrix, and Vesicles. J. Pathol. 2022, 257, 663–673. [Google Scholar] [CrossRef]
- Navarro, A.; Yin, P.; Monsivais, D.; Lin, S.M.; Du, P.; Wei, J.-J.; Bulun, S.E. Genome-Wide DNA Methylation Indicates Silencing of Tumor Suppressor Genes in Uterine Leiomyoma. PLoS ONE 2012, 7, e33284. [Google Scholar] [CrossRef]
- Jiang, W.; Luo, H.; Shen, Z.; Zhang, W.; Chen, A.; Zhu, X. Down-Regulation and Gene Hypermethylation of the 14-3-3 Gamma in Uterine Leiomyoma. Front. Biosci. Landmark Ed. 2016, 21, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Braný, D.; Dvorská, D.; Grendár, M.; Ňachajová, M.; Szépe, P.; Lasabová, Z.; Žúbor, P.; Višňovský, J.; Halášová, E. Different Methylation Levels in the KLF4, ATF3 and DLEC1 Genes in the Myometrium and in Corpus Uteri Mesenchymal Tumours as Assessed by MS-HRM. Pathol. Res. Pract. 2019, 215, 152465. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Maekawa, R.; Tamura, I.; Shirafuta, Y.; Shinagawa, M.; Asada, H.; Taketani, T.; Tamura, H.; Sugino, N. SATB2 and NGR1: Potential Upstream Regulatory Factors in Uterine Leiomyomas. J. Assist. Reprod. Genet. 2019, 36, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Maekawa, R.; Yamagata, Y.; Tamura, I.; Lee, L.; Okada, M.; Jozaki, K.; Asada, H.; Tamura, H.; Sugino, N. Identification of Uterine Leiomyoma-Specific Marker Genes Based on DNA Methylation and Their Clinical Application. Sci. Rep. 2016, 6, 30652. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yin, P.; Xu, J.; Dotts, A.J.; Kujawa, S.A.; Coon V, J.S.; Zhao, H.; Shilatifard, A.; Dai, Y.; Bulun, S.E. Targeting DNA Methylation Depletes Uterine Leiomyoma Stem Cell-Enriched Population by Stimulating Their Differentiation. Endocrinology 2020, 161, bqaa143. [Google Scholar] [CrossRef]
- Liu, S.; Yin, P.; Xu, J.; Dotts, A.J.; Kujawa, S.A.; Coon V, J.S.; Zhao, H.; Dai, Y.; Bulun, S.E. Progesterone Receptor-DNA Methylation Crosstalk Regulates Depletion of Uterine Leiomyoma Stem Cells: A Potential Therapeutic Target. Stem Cell Rep. 2021, 16, 2099–2106. [Google Scholar] [CrossRef]
- Navarro, A.; Yin, P.; Ono, M.; Monsivais, D.; Moravek, M.B.; Coon, J.S.; Dyson, M.T.; Wei, J.-J.; Bulun, S.E. 5-Hydroxymethylcytosine Promotes Proliferation of Human Uterine Leiomyoma: A Biological Link to a New Epigenetic Modification in Benign Tumors. J. Clin. Endocrinol. Metab. 2014, 99, E2437–E2445. [Google Scholar] [CrossRef]
- Cao, T.; Jiang, Y.; Wang, Z.; Zhang, N.; Al-Hendy, A.; Mamillapalli, R.; Kallen, A.N.; Kodaman, P.; Taylor, H.S.; Li, D.; et al. H19 LncRNA Identified as a Master Regulator of Genes That Drive Uterine Leiomyomas. Oncogene 2019, 38, 5356–5366. [Google Scholar] [CrossRef]
- Zhan, X.; Zhou, H.; Sun, Y.; Shen, B.; Chou, D. Long Non-Coding Ribonucleic Acid H19 and Ten-Eleven Translocation Enzyme 1 Messenger RNA Expression Levels in Uterine Fibroids May Predict Their Postoperative Recurrence. Clin. Sao Paulo Braz. 2021, 76, e2671. [Google Scholar] [CrossRef]
- Efimova, O.A.; Pendina, A.A.; Tikhonov, A.V.; Baranov, V.S. The Evolution of Ideas on the Biological Role of 5-Methylcytosine Oxidative Derivatives in the Mammalian Genome. Russ. J. Genet. Appl. Res. 2018, 8, 11–21. [Google Scholar] [CrossRef]
- Wu, H.; D’Alessio, A.C.; Ito, S.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Genome-Wide Analysis of 5-Hydroxymethylcytosine Distribution Reveals Its Dual Function in Transcriptional Regulation in Mouse Embryonic Stem Cells. Genes Dev. 2011, 25, 679–684. [Google Scholar] [CrossRef]
- Li, W.; Liu, M. Distribution of 5-Hydroxymethylcytosine in Different Human Tissues. J. Nucleic Acids 2011, 2011, 870726. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Ottaviano, R.; Reddington, J.; Sproul, D.; Reinhardt, D.; Dunican, D.; Katz, E.; Dixon, J.M.; Harrison, D.J.; Meehan, R.R. Tissue Type Is a Major Modifier of the 5-Hydroxymethylcytosine Content of Human Genes. Genome Res. 2012, 22, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Efimova, O.A.; Pendina, A.A.; Tikhonov, A.V.; Fedorova, I.D.; Krapivin, M.I.; Chiryaeva, O.G.; Shilnikova, E.M.; Bogdanova, M.A.; Kogan, I.Y.; Kuznetzova, T.V.; et al. Chromosome Hydroxymethylation Patterns in Human Zygotes and Cleavage-Stage Embryos. Reprod. Camb. Engl. 2015, 149, 223–233. [Google Scholar] [CrossRef]
- Efimova, O.A.; Pendina, A.A.; Tikhonov, A.V.; Parfenyev, S.E.; Mekina, I.D.; Komarova, E.M.; Mazilina, M.A.; Daev, E.V.; Chiryaeva, O.G.; Galembo, I.A.; et al. Genome-Wide 5-Hydroxymethylcytosine Patterns in Human Spermatogenesis Are Associated with Semen Quality. Oncotarget 2017, 8, 88294–88307. [Google Scholar] [CrossRef]
- Efimova, O.A.; Pendina, A.A.; Krapivin, M.I.; Kopat, V.V.; Tikhonov, A.V.; Petrovskaia-Kaminskaia, A.V.; Navodnikova, P.M.; Talantova, O.E.; Glotov, O.S.; Baranov, V.S. Inter-Cell and Inter-Chromosome Variability of 5-Hydroxymethylcytosine Patterns in Noncultured Human Embryonic and Extraembryonic Cells. Cytogenet. Genome Res. 2018, 156, 150–157. [Google Scholar] [CrossRef]
- Kudo, Y.; Tateishi, K.; Yamamoto, K.; Yamamoto, S.; Asaoka, Y.; Ijichi, H.; Nagae, G.; Yoshida, H.; Aburatani, H.; Koike, K. Loss of 5-Hydroxymethylcytosine Is Accompanied with Malignant Cellular Transformation. Cancer Sci. 2012, 103, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-C.; Ling, Z.-Q. The Role of TET Family Proteins and 5-Hydroxymethylcytosine in Human Tumors. Histol. Histopathol. 2014, 29, 991–997. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, X.; Guo, L.; Li, Y.; Luo, M.; He, J. Decreased 5-Hydroxymethylcytosine Levels Correlate with Cancer Progression and Poor Survival: A Systematic Review and Meta-Analysis. Oncotarget 2017, 8, 1944–1952. [Google Scholar] [CrossRef]
- Lu, J.-L.; Zhao, L.; Han, S.-C.; Bi, J.-L.; Liu, H.-X.; Yue, C.; Lin, L. MiR-129 Is Involved in the Occurrence of Uterine Fibroid through Inhibiting TET1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4419–4426. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Khorram, O. Regulation of Cell Cycle Regulatory Proteins by MicroRNAs in Uterine Leiomyoma. Reprod. Sci. Thousand Oaks Calif 2019, 26, 250–258. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Rehan, A.; Khorram, O. Tranilast Induces MiR-200c Expression through Blockade of RelA/P65 Activity in Leiomyoma Smooth Muscle Cells. Fertil. Steril. 2020, 113, 1308–1318. [Google Scholar] [CrossRef]
- Lazzarini, R.; Caffarini, M.; Delli Carpini, G.; Ciavattini, A.; Di Primio, R.; Orciani, M. From 2646 to 15: Differentially Regulated MicroRNAs between Progenitors from Normal Myometrium and Leiomyoma. Am. J. Obstet. Gynecol. 2020, 222, 596.e1–596.e9. [Google Scholar] [CrossRef]
- Zota, A.R.; Geller, R.J.; VanNoy, B.N.; Marfori, C.Q.; Tabbara, S.; Hu, L.Y.; Baccarelli, A.A.; Moawad, G.N. Phthalate Exposures and MicroRNA Expression in Uterine Fibroids: The FORGE Study. Epigenetics Insights 2020, 13, 2516865720904057. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kang, D.; Kwon, M.Y.; Lee, H.J.; Kim, M.J. MicroRNAs as Potential Indicators of the Development and Progression of Uterine Leiomyoma. PLoS ONE 2022, 17, e0268793. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-D.; Khorram, O. Cross-Talk between MiR-29c and Transforming Growth Factor-Β3 Is Mediated by an Epigenetic Mechanism in Leiomyoma. Fertil. Steril. 2019, 112, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Xue, H.; Shao, W.; Wang, X.; Liao, H.; Ye, Y. Inhibiting Effect of MiR-29 on Proliferation and Migration of Uterine Leiomyoma via the STAT3 Signaling Pathway. Aging 2022, 14, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Ciebiera, M.; Włodarczyk, M.; Zgliczyński, S.; Łoziński, T.; Walczak, K.; Czekierdowski, A. The Role of MiRNA and Related Pathways in Pathophysiology of Uterine Fibroids-From Bench to Bedside. Int. J. Mol. Sci. 2020, 21, 3016. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhou, L.; Wang, J.; Zhang, X.; Liu, G. Circular RNA Expression Profiling Identifies Novel Biomarkers in Uterine Leiomyoma. Cell. Signal. 2020, 76, 109784. [Google Scholar] [CrossRef] [PubMed]
- Suo, M.; Lin, Z.; Guo, D.; Zhang, A. Hsa_circ_0056686, Derived from Cancer-Associated Fibroblasts, Promotes Cell Proliferation and Suppresses Apoptosis in Uterine Leiomyoma through Inhibiting Endoplasmic Reticulum Stress. PLoS ONE 2022, 17, e0266374. [Google Scholar] [CrossRef]
- Farzaneh, F.; Saravani, M.; Esmailpoor, M.; Mokhtari, M.; Teimoori, B.; Rezaei, M.; Salimi, S. Association of HOTAIR Gene Polymorphisms and Haplotypes with Uterine Leiomyoma Susceptibility in Southeast of Iran. Mol. Biol. Rep. 2019, 46, 4271–4277. [Google Scholar] [CrossRef] [PubMed]
- Falahati, Z.; Mohseni-Dargah, M.; Mirfakhraie, R. Emerging Roles of Long Non-Coding RNAs in Uterine Leiomyoma Pathogenesis: A Review. Reprod. Sci. Thousand Oaks Calif. 2022, 29, 1086–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, G.; Li, B.; Qu, J.; Zhang, Y. LncRNA APTR Promotes Uterine Leiomyoma Cell Proliferation by Targeting ERα to Activate the Wnt/β-Catenin Pathway. Front. Oncol. 2021, 11, 536346. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-D.; Quintanilla, D.; Boos, D.; Khorram, O. Long Noncoding RNA MIAT Modulates the Extracellular Matrix Deposition in Leiomyomas by Sponging MiR-29 Family. Endocrinology 2021, 162, bqab186. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Xue, L.; Li, Z.; Yi, T. Lnc-AL445665.1-4 May Be Involved in the Development of Multiple Uterine Leiomyoma through Interacting with MiR-146b-5p. BMC Cancer 2019, 19, 709. [Google Scholar] [CrossRef]
- Akbari, M.; Yassaee, F.; Aminbeidokhti, M.; Abedin-Do, A.; Mirfakhraie, R. LncRNA SRA1 May Play a Role in the Uterine Leiomyoma Tumor Growth Regarding the MED12 Mutation Pattern. Int. J. Womens Health 2019, 11, 495–500. [Google Scholar] [CrossRef]
- Zota, A.R.; Geller, R.J.; Calafat, A.M.; Marfori, C.Q.; Baccarelli, A.A.; Moawad, G.N. Phthalates Exposure and Uterine Fibroid Burden among Women Undergoing Surgical Treatment for Fibroids: A Preliminary Study. Fertil. Steril. 2019, 111, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Kim, S.; Bastiaensen, M.; Malarvannan, G.; Poma, G.; Caballero Casero, N.; Gys, C.; Covaci, A.; Lee, S.; Lim, J.-E.; et al. Exposure to Organophosphate Esters, Phthalates, and Alternative Plasticizers in Association with Uterine Fibroids. Environ. Res. 2020, 189, 109874. [Google Scholar] [CrossRef]
- Lee, J.; Jeong, Y.; Mok, S.; Choi, K.; Park, J.; Moon, H.-B.; Choi, G.; Kim, H.-J.; Kim, S.Y.; Choi, S.R.; et al. Associations of Exposure to Phthalates and Environmental Phenols with Gynecological Disorders. Reprod. Toxicol. Elmsford N. Y. 2020, 95, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, L.; Castro, L.; Yan, Y.; Clayton, N.P.; Bushel, P.; Flagler, N.D.; Scappini, E.; Dixon, D. Short-Term Tetrabromobisphenol A Exposure Promotes Fibrosis of Human Uterine Fibroid Cells in a 3D Culture System through TGF-Beta Signaling. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22101. [Google Scholar] [CrossRef]
- Kim, J.H. Analysis of the in Vitro Effects of Di-(2-Ethylhexyl) Phthalate Exposure on Human Uterine Leiomyoma Cells. Exp. Ther. Med. 2018, 15, 4972–4978. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Das, P.; Vall, A.J.; Yan, Y.; Gao, X.; Sifre, M.I.; Bortner, C.D.; Castro, L.; Kissling, G.E.; Moore, A.B.; et al. Bisphenol A Induces Human Uterine Leiomyoma Cell Proliferation through Membrane-Associated ERα36 via Nongenomic Signaling Pathways. Mol. Cell. Endocrinol. 2019, 484, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, Q.; Ding, B.; Xu, J.; Shen, Y. Bisphenol A Promotes the Proliferation of Leiomyoma Cells by GPR30-EGFR Signaling Pathway. J. Obstet. Gynaecol. Res. 2019, 45, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Mahalingaiah, S.; Hart, J.E.; Wise, L.A.; Terry, K.L.; Boynton-Jarrett, R.; Missmer, S.A. Prenatal Diethylstilbestrol Exposure and Risk of Uterine Leiomyomata in the Nurses’ Health Study II. Am. J. Epidemiol. 2014, 179, 186–191. [Google Scholar] [CrossRef]
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Toppari, J.; Zoeller, R.T. EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr. Rev. 2015, 36, E1–E150. [Google Scholar] [CrossRef]
- Katz, T.A.; Yang, Q.; Treviño, L.S.; Walker, C.L.; Al-Hendy, A. Endocrine-Disrupting Chemicals and Uterine Fibroids. Fertil. Steril. 2016, 106, 967–977. [Google Scholar] [CrossRef]
- Prusinski Fernung, L.E.; Yang, Q.; Sakamuro, D.; Kumari, A.; Mas, A.; Al-Hendy, A. Endocrine Disruptor Exposure during Development Increases Incidence of Uterine Fibroids by Altering DNA Repair in Myometrial Stem Cells. Biol. Reprod. 2018, 99, 735–748. [Google Scholar] [CrossRef]
- Chen, M.; Guo, J.; Ruan, J.; Yang, Z.; He, C.; Zuo, Z. Neonatal Exposure to Environment-Relevant Levels of Tributyltin Leads to Uterine Dysplasia in Rats. Sci. Total Environ. 2020, 720, 137615. [Google Scholar] [CrossRef]
- Elkafas, H.; Ali, M.; Elmorsy, E.; Kamel, R.; Thompson, W.E.; Badary, O.; Al-Hendy, A.; Yang, Q. Vitamin D3 Ameliorates DNA Damage Caused by Developmental Exposure to Endocrine Disruptors in the Uterine Myometrial Stem Cells of Eker Rats. Cells 2020, 9, 1459. [Google Scholar] [CrossRef]
- Bulun, S.E.; Simpson, E.R.; Word, R.A. Expression of the CYP19 Gene and Its Product Aromatase Cytochrome P450 in Human Uterine Leiomyoma Tissues and Cells in Culture. J. Clin. Endocrinol. Metab. 1994, 78, 736–743. [Google Scholar] [CrossRef]
- Bulun, S.E.; Noble, L.S.; Takayama, K.; Michael, M.D.; Agarwal, V.; Fisher, C.; Zhao, Y.; Hinshelwood, M.M.; Ito, Y.; Simpson, E.R. Endocrine Disorders Associated with Inappropriately High Aromatase Expression. J. Steroid Biochem. Mol. Biol. 1997, 61, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, H.; Kitawaki, J.; Kado, N.; Koshiba, H.; Fushiki, S.; Honjo, H. Gonadotropin-Releasing Hormone Agonist and Danazol Normalize Aromatase Cytochrome P450 Expression in Eutopic Endometrium from Women with Endometriosis, Adenomyosis, or Leiomyomas. Fertil. Steril. 2003, 79 (Suppl. 1), 735–742. [Google Scholar] [CrossRef]
- Calzada-Mendoza, C.C.; Sánchez, E.C.; Campos, R.R.; Becerril, A.M.; Madrigal, E.B.; Sierra, A.R.; Mendez, E.B.; Ocharán, E.H.; Herrera, N.G.; Ceballos-Reyes, G. Differential Aromatase (CYP19) Expression in Human Arteries from Normal and Neoplasic Uterus: An Immunohistochemical and in Situ Hybridization Study. Front. Biosci. J. Virtual Libr. 2006, 11, 389–393. [Google Scholar] [CrossRef]
- Bulun, S.E.; Imir, G.; Utsunomiya, H.; Thung, S.; Gurates, B.; Tamura, M.; Lin, Z. Aromatase in Endometriosis and Uterine Leiomyomata. J. Steroid Biochem. Mol. Biol. 2005, 95, 57–62. [Google Scholar] [CrossRef]
- Hatok, J.; Zubor, P.; Galo, S.; Kirschnerova, R.; Dobrota, D.; Danko, J.; Racay, P. Endometrial Aromatase MRNA as a Possible Screening Tool for Advanced Endometriosis and Adenomyosis. Gynecol. Endocrinol. Off. J. Int. Soc. Gynecol. Endocrinol. 2011, 27, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Noble, L.S.; Simpson, E.R.; Johns, A.; Bulun, S.E. Aromatase Expression in Endometriosis. J. Clin. Endocrinol. Metab. 1996, 81, 174–179. [Google Scholar] [CrossRef]
- Noble, L.S.; Takayama, K.; Zeitoun, K.M.; Putman, J.M.; Johns, D.A.; Hinshelwood, M.M.; Agarwal, V.R.; Zhao, Y.; Carr, B.R.; Bulun, S.E. Prostaglandin E2 Stimulates Aromatase Expression in Endometriosis-Derived Stromal Cells. J. Clin. Endocrinol. Metab. 1997, 82, 600–606. [Google Scholar] [CrossRef]
- Shozu, M.; Sumitani, H.; Segawa, T.; Yang, H.-J.; Murakami, K.; Kasai, T.; Inoue, M. Overexpression of Aromatase P450 in Leiomyoma Tissue Is Driven Primarily through Promoter I.4 of the Aromatase P450 Gene (CYP19). J. Clin. Endocrinol. Metab. 2002, 87, 2540–2548. [Google Scholar] [CrossRef]
- Imir, A.G.; Lin, Z.; Yin, P.; Deb, S.; Yilmaz, B.; Cetin, M.; Cetin, A.; Bulun, S.E. Aromatase Expression in Uterine Leiomyomata Is Regulated Primarily by Proximal Promoters I.3/II. J. Clin. Endocrinol. Metab. 2007, 92, 1979–1982. [Google Scholar] [CrossRef]
- Ishikawa, H.; Reierstad, S.; Demura, M.; Rademaker, A.W.; Kasai, T.; Inoue, M.; Usui, H.; Shozu, M.; Bulun, S.E. High Aromatase Expression in Uterine Leiomyoma Tissues of African-American Women. J. Clin. Endocrinol. Metab. 2009, 94, 1752–1756. [Google Scholar] [CrossRef]
- Thompson, P.A.; Khatami, M.; Baglole, C.J.; Sun, J.; Harris, S.A.; Moon, E.-Y.; Al-Mulla, F.; Al-Temaimi, R.; Brown, D.G.; Colacci, A.; et al. Environmental Immune Disruptors, Inflammation and Cancer Risk. Carcinogenesis 2015, 36 (Suppl. 1), S232–S253. [Google Scholar] [CrossRef]
- Sacco, K.; Portelli, M.; Pollacco, J.; Schembri-Wismayer, P.; Calleja-Agius, J. The Role of Prostaglandin E2 in Endometriosis. Gynecol. Endocrinol. Off. J. Int. Soc. Gynecol. Endocrinol. 2012, 28, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Jung, G.Y.; Park, S.B.; Cho, Y.J.; Han, M. Assessment of the Effects of Prostaglandins on Myometrial and Leiomyoma Cells in Vitro through MicroRNA Profiling. Mol. Med. Rep. 2018, 18, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Barcikowska, Z.; Rajkowska-Labon, E.; Grzybowska, M.E.; Hansdorfer-Korzon, R.; Zorena, K. Inflammatory Markers in Dysmenorrhea and Therapeutic Options. Int. J. Environ. Res. Public Health 2020, 17, 1191. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, X.; Jeschke, U.; von Schönfeldt, V. COX-2-PGE2-EPs in Gynecological Cancers. Arch. Gynecol. Obstet. 2020, 301, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Baranov, V.S.; Ivaschenko, T.E.; Yarmolinskaya, M.I. Comparative Systems Genetics View of Endometriosis and Uterine Leiomyoma: Two Sides of the Same Coin? Syst. Biol. Reprod. Med. 2016, 62, 93–105. [Google Scholar] [CrossRef]
- Roy, D.; Liehr, J.G. Estrogen, DNA Damage and Mutations. Mutat. Res. 1999, 424, 107–115. [Google Scholar] [CrossRef]
- Yager, J.D. Mechanisms of Estrogen Carcinogenesis: The Role of E2/E1-Quinone Metabolites Suggests New Approaches to Preventive Intervention--A Review. Steroids 2015, 99, 56–60. [Google Scholar] [CrossRef]
- Tian, H.; Gao, Z.; Wang, G.; Li, H.; Zheng, J. Estrogen Potentiates Reactive Oxygen Species (ROS) Tolerance to Initiate Carcinogenesis and Promote Cancer Malignant Transformation. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 141–150. [Google Scholar] [CrossRef]
- Bulun, S.E.; Moravek, M.B.; Yin, P.; Ono, M.; Coon, J.S.; Dyson, M.T.; Navarro, A.; Marsh, E.E.; Zhao, H.; Maruyama, T.; et al. Uterine Leiomyoma Stem Cells: Linking Progesterone to Growth. Semin. Reprod. Med. 2015, 33, 357–365. [Google Scholar] [CrossRef]
- Kastner, P.; Krust, A.; Turcotte, B.; Stropp, U.; Tora, L.; Gronemeyer, H.; Chambon, P. Two Distinct Estrogen-Regulated Promoters Generate Transcripts Encoding the Two Functionally Different Human Progesterone Receptor Forms A and B. EMBO J. 1990, 9, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Omar, M.; Laknaur, A.; Al-Hendy, A.; Yang, Q. Myometrial Progesterone Hyper-Responsiveness Associated with Increased Risk of Human Uterine Fibroids. BMC Womens Health 2019, 19, 92. [Google Scholar] [CrossRef] [PubMed]
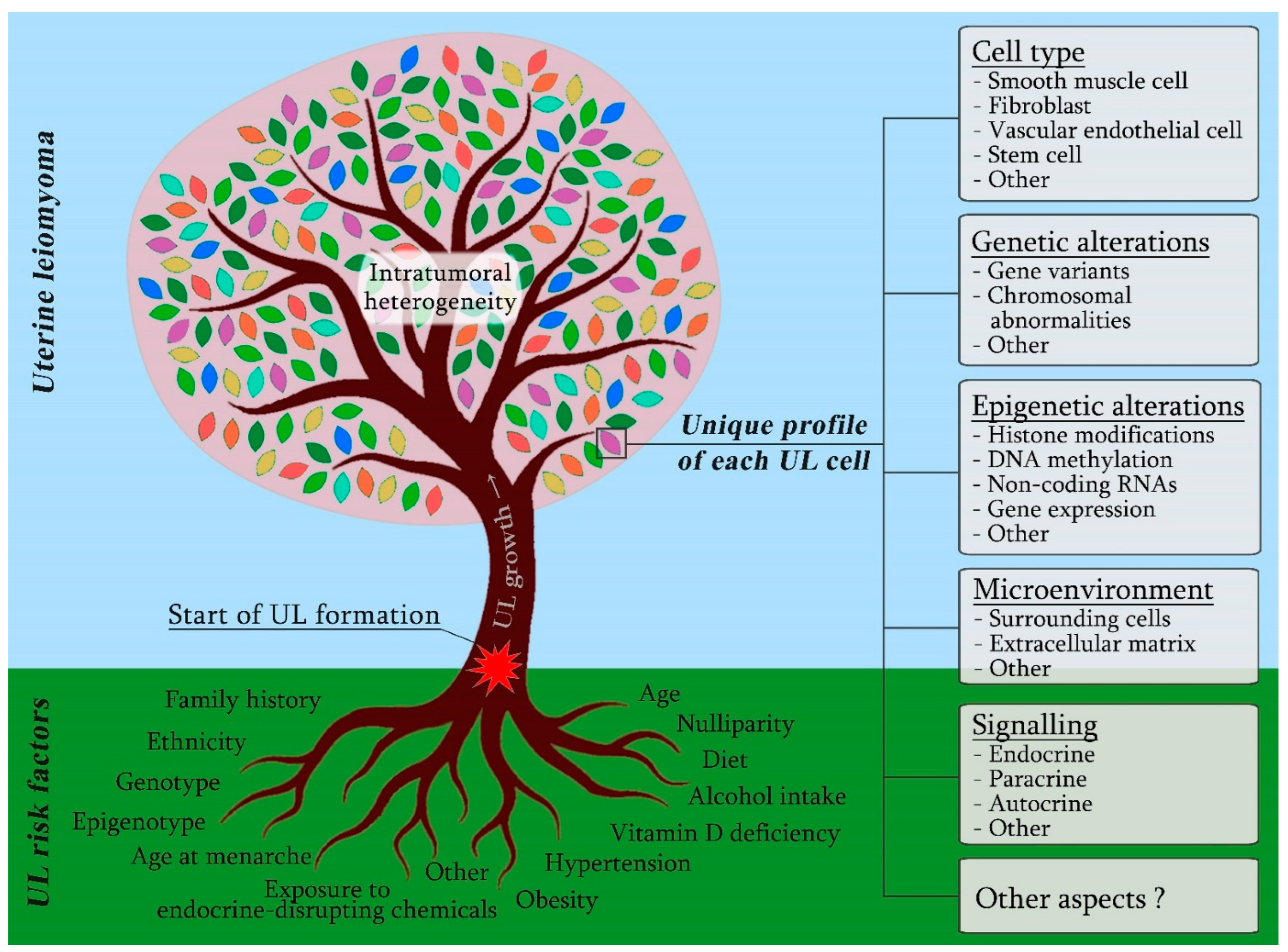
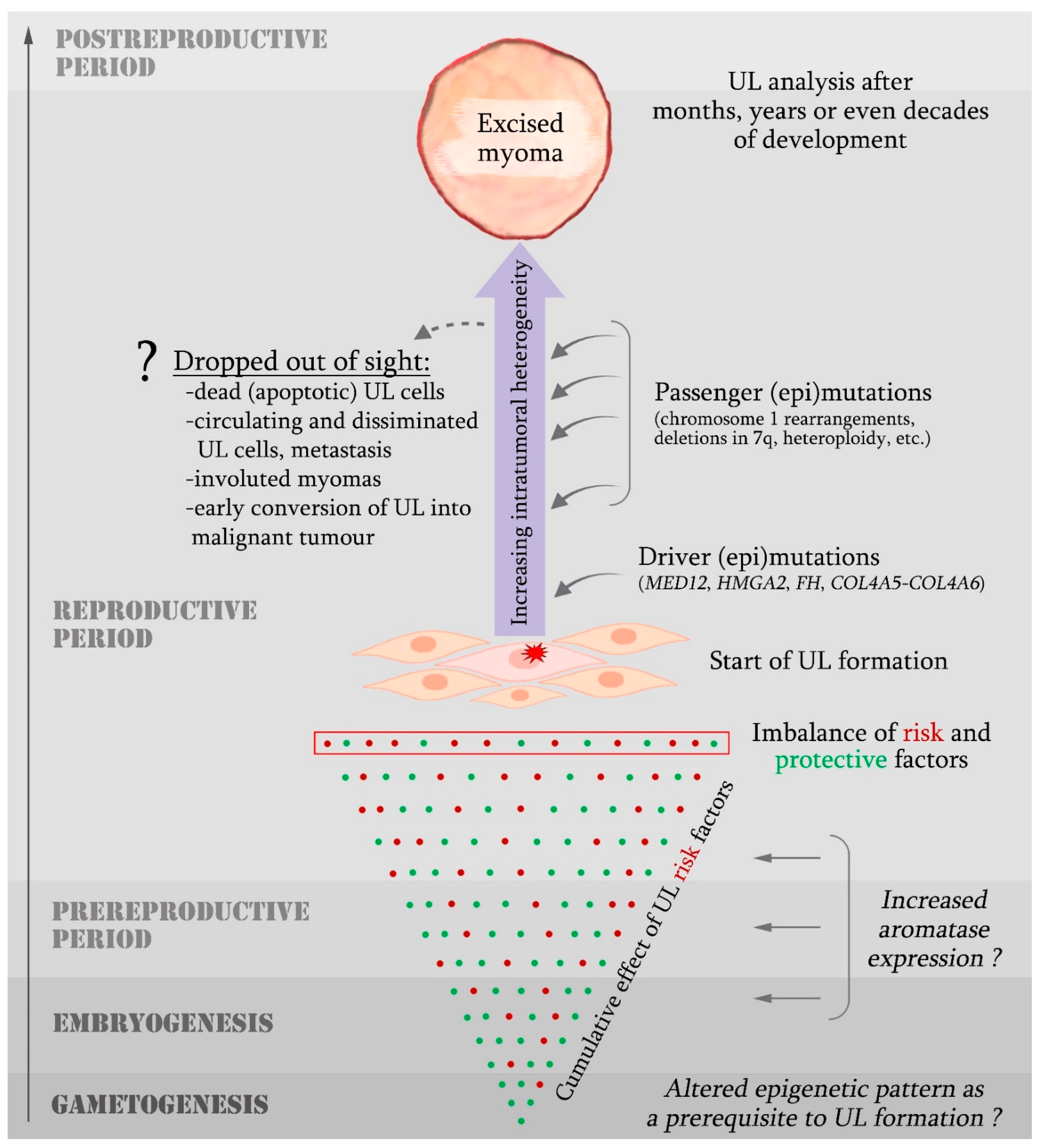
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koltsova, A.S.; Efimova, O.A.; Pendina, A.A. A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity. Int. J. Mol. Sci. 2023, 24, 5752. https://doi.org/10.3390/ijms24065752
Koltsova AS, Efimova OA, Pendina AA. A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity. International Journal of Molecular Sciences. 2023; 24(6):5752. https://doi.org/10.3390/ijms24065752
Chicago/Turabian StyleKoltsova, Alla S., Olga A. Efimova, and Anna A. Pendina. 2023. "A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity" International Journal of Molecular Sciences 24, no. 6: 5752. https://doi.org/10.3390/ijms24065752
APA StyleKoltsova, A. S., Efimova, O. A., & Pendina, A. A. (2023). A View on Uterine Leiomyoma Genesis through the Prism of Genetic, Epigenetic and Cellular Heterogeneity. International Journal of Molecular Sciences, 24(6), 5752. https://doi.org/10.3390/ijms24065752






