Proteomic Analysis of Skeletal Muscle and White Adipose Tissue after Aerobic Exercise Training in High Fat Diet Induced Obese Mice
Abstract
1. Introduction
2. Results
2.1. Effect of Aerobic Exercise on Body Weight and the Morphology of Skeletal Muscle and Epididymal Fat Pad
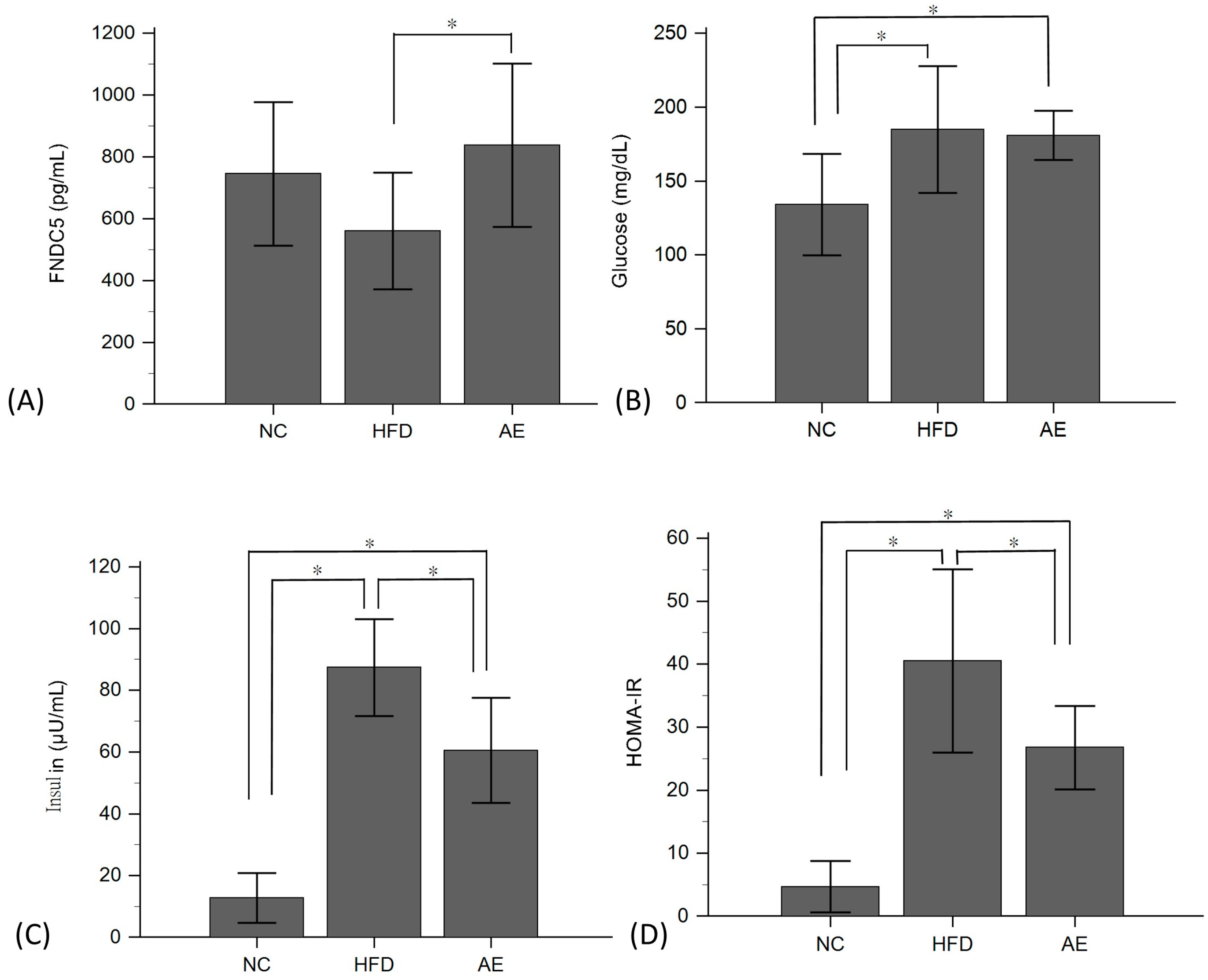
2.2. Effect of Aerobic Exercise on Serum FNDC-5, Glucose, Insulin, and HOMA-IR Levels
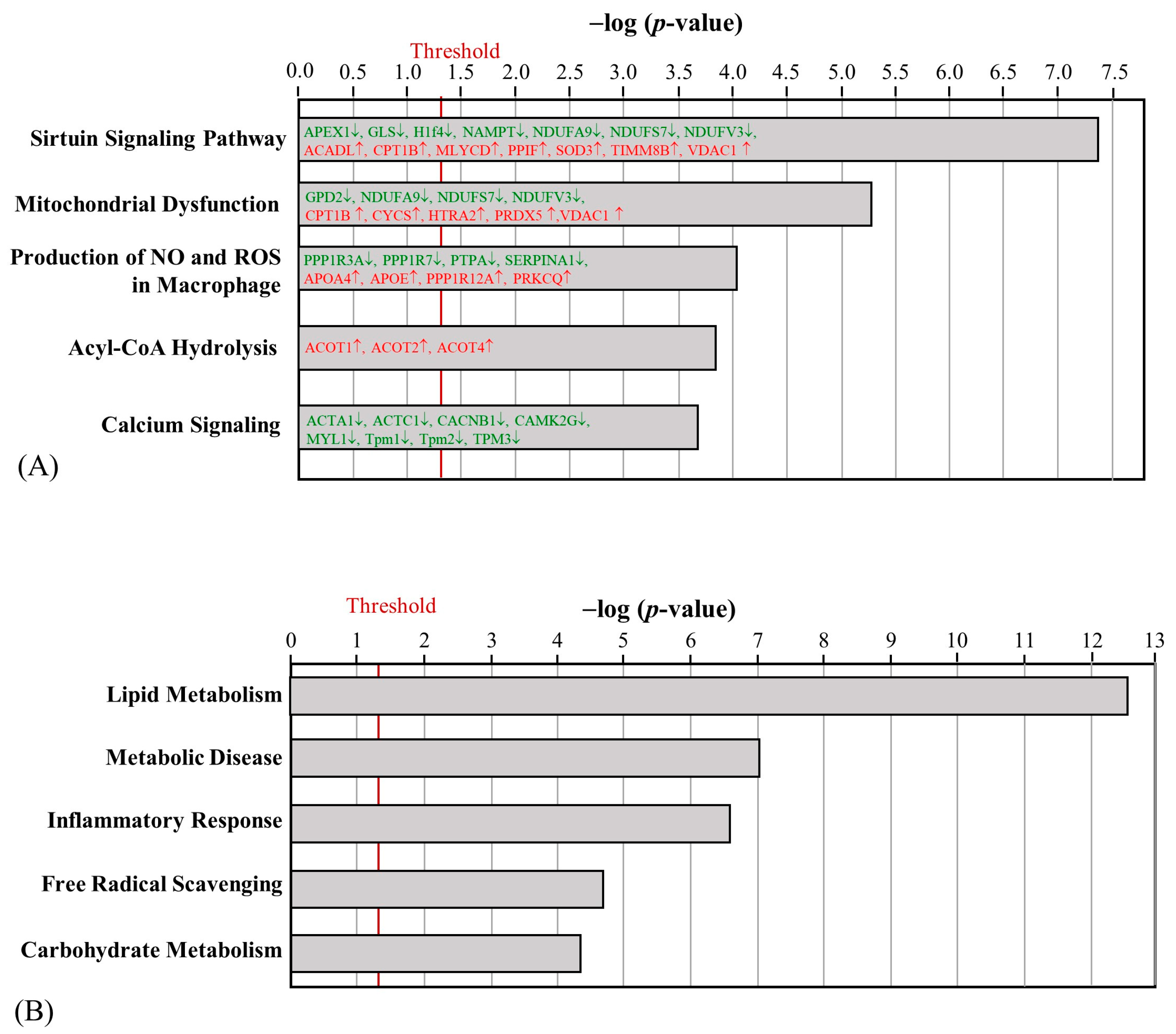
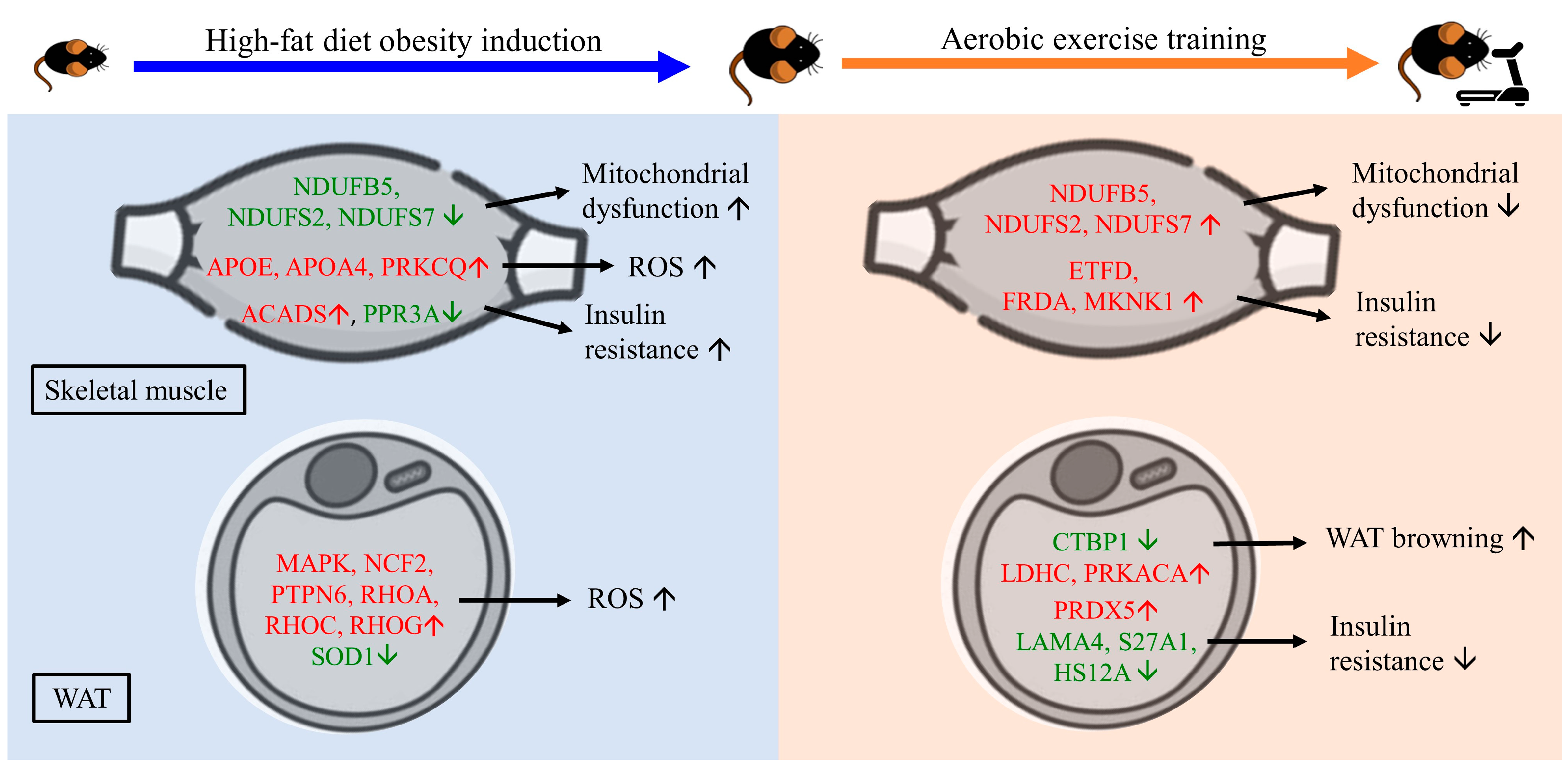
2.3. The Effect of HFD and AE on Skeletal Muscle Proteomic Changes
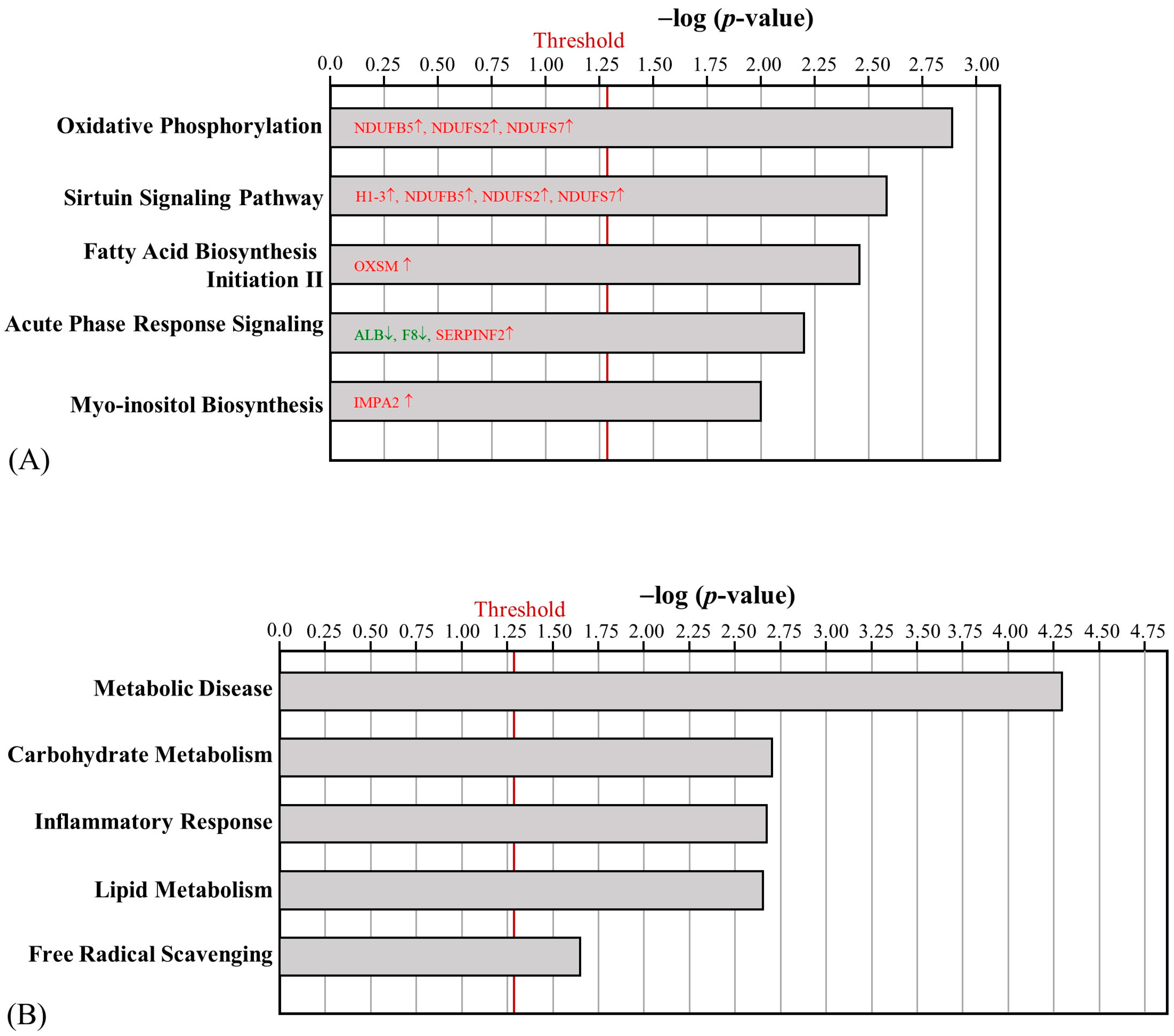
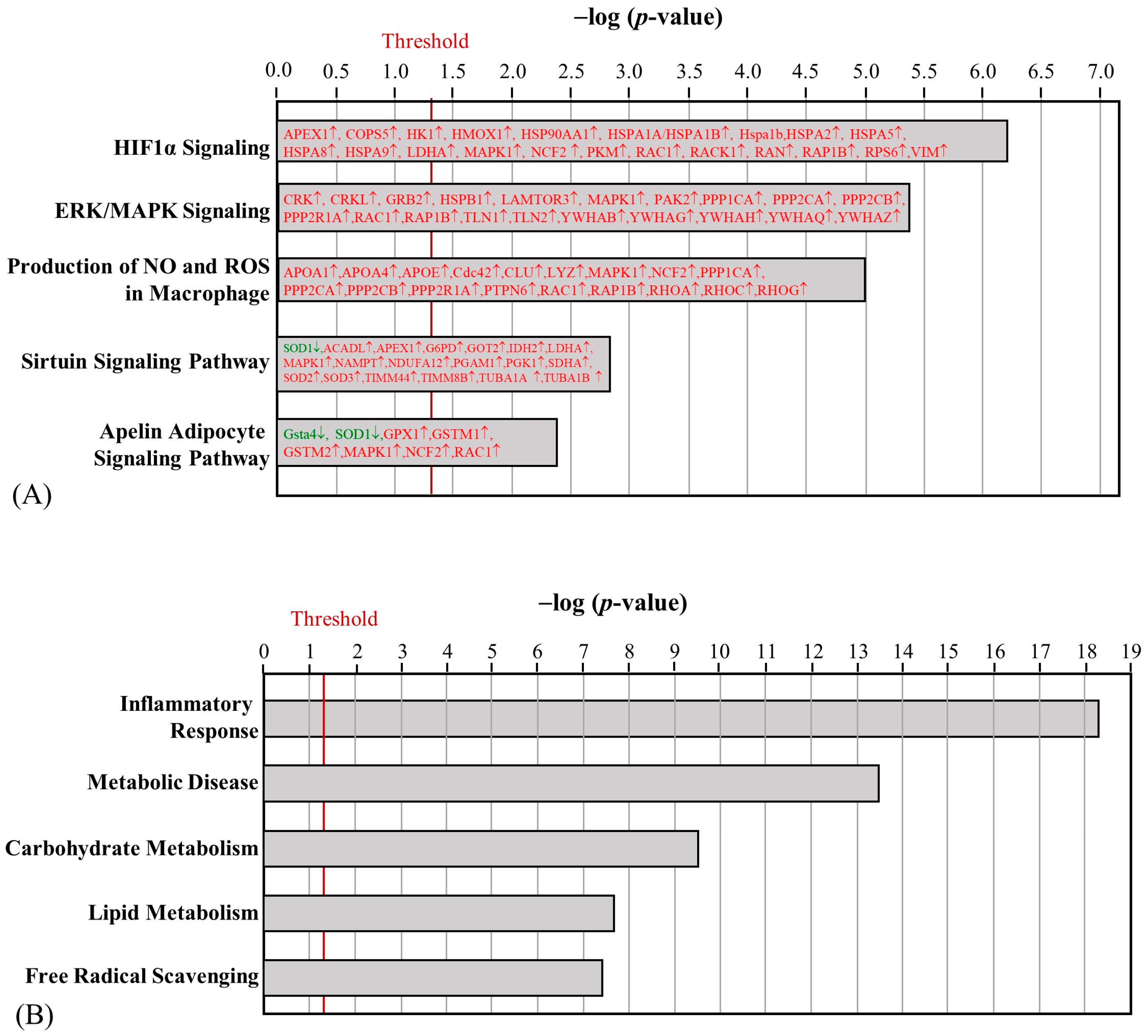
2.4. The Effect of HFD and AE on Epididymal Fat Pad Proteomic Changes
3. Discussion
4. Materials and Methods
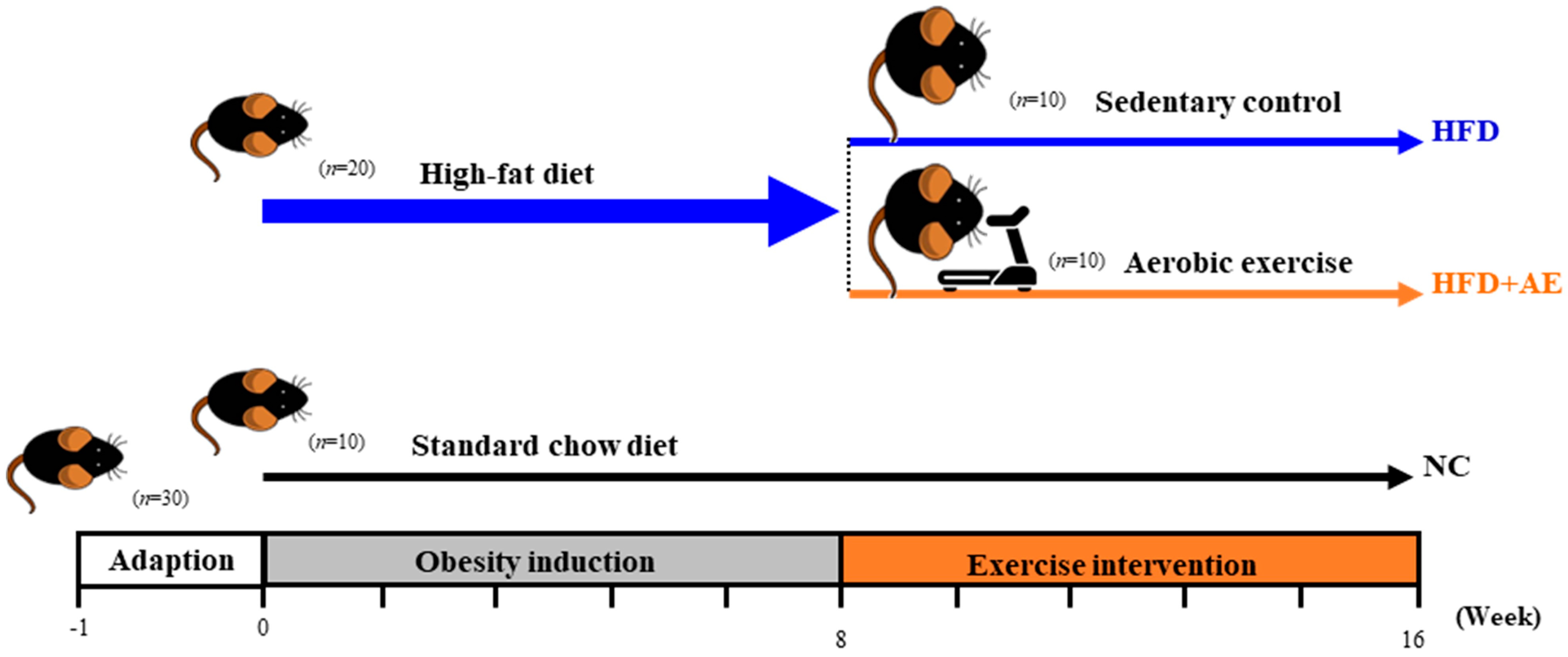
4.1. Animal Experiment
4.2. Exercise Intervention Protocol
4.3. Blood Sample Analysis
4.4. Histology of Skeletal Muscle and Epididymal White Adipose Tissue
4.5. Protein Extraction and Liquid Chromatography–Tandem Mass Spectrometry Analysis
4.6. Mass Spectrometry Data Analysis
4.7. Bioinformatic Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heymsfield, S.B.; Wadden, T.A. Mechanisms, Pathophysiology, and Management of Obesity. N. Engl. J. Med. 2017, 376, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K.; Febbraio, M.A. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef] [PubMed]
- van der Kolk, B.W.; Saari, S.; Lovric, A.; Arif, M.; Alvarez, M.; Ko, A.; Miao, Z.; Sahebekhtiari, N.; Muniandy, M.; Heinonen, S.; et al. Molecular pathways behind acquired obesity: Adipose tissue and skeletal muscle multiomics in monozygotic twin pairs discordant for BMI. Cell Rep. Med. 2021, 2, 100226. [Google Scholar] [CrossRef]
- Shulman, G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 2014, 371, 1131–1141. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef]
- Müller, T.D.; Blüher, M.; Tschöp, M.H.; DiMarchi, R.D. Anti-obesity drug discovery: Advances and challenges. Nat. Rev. Drug Discov. 2022, 21, 201–223. [Google Scholar] [CrossRef]
- Ahmed, B.; Sultana, R.; Greene, M.W. Adipose tissue and insulin resistance in obese. Biomed. Pharmacother. 2021, 137, 111315. [Google Scholar] [CrossRef]
- Chow, L.S.; Gerszten, R.E.; Taylor, J.M.; Pedersen, B.K.; van Praag, H.; Trappe, S.; Febbraio, M.A.; Galis, Z.S.; Gao, Y.; Haus, J.M.; et al. Exerkines in health, resilience and disease. Nat. Rev. Endocrinol. 2022, 18, 273–289. [Google Scholar] [CrossRef]
- Contrepois, K.; Wu, S.; Moneghetti, K.J.; Hornburg, D.; Ahadi, S.; Tsai, M.S.; Metwally, A.A.; Wei, E.; Lee-McMullen, B.; Quijada, J.V.; et al. Molecular Choreography of Acute Exercise. Cell 2020, 181, 1112–1130.e16. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, F.; Lachinani, L.; Ghaedi, S.; Nasr-Esfahani, M.H.; Megraw, T.L.; Ghaedi, K. New insights into the cellular activities of Fndc5/Irisin and its signaling pathways. Cell Biosci. 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, A.S. Proteomics of Skeletal Muscle: Focus on Insulin Resistance and Exercise Biology. Proteomes 2016, 4, 6. [Google Scholar] [CrossRef]
- Deshmukh, A.S.; Murgia, M.; Nagaraj, N.; Treebak, J.T.; Cox, J.; Mann, M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell Proteom. 2015, 14, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, J.; Zhao, W.G.; Sun, H.; Guo, Z.; Jing, L.; She, Z.; Yuan, T.; Liu, S.N.; Liu, Q.; et al. Comprehensive map and functional annotation of the mouse white adipose tissue proteome. PeerJ 2019, 7, e7352. [Google Scholar] [CrossRef]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Zhang, C.; Yang, S.; Zhang, X.; Liu, Y.; Su, Z. Deficiency in the short-chain acyl-CoA dehydrogenase protects mice against diet-induced obesity and insulin resistance. FASEB J. 2019, 33, 13722–13733. [Google Scholar] [CrossRef]
- Delibegovic, M.; Armstrong, C.G.; Dobbie, L.; Watt, P.W.; Smith, A.J.; Cohen, P.T. Disruption of the striated muscle glycogen targeting subunit PPP1R3A of protein phosphatase 1 leads to increased weight gain, fat deposition, and development of insulin resistance. Diabetes 2003, 52, 596–604. [Google Scholar] [CrossRef]
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; Mack, N.J.; Ahmad, N. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 2018, 28, 643–661. [Google Scholar] [CrossRef]
- Cortés-Rojo, C.; Vargas-Vargas, M.A.; Olmos-Orizaba, B.E.; Rodríguez-Orozco, A.R.; Calderón-Cortés, E. Interplay between NADH oxidation by complex I, glutathione redox state and sirtuin-3, and its role in the development of insulin resistance. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2020, 1866, 165801. [Google Scholar] [CrossRef]
- Li, N.; Shi, H.; Guo, Q.; Gan, Y.; Zhang, Y.; Jia, J.; Zhang, L.; Zhou, Y. Aerobic Exercise Prevents Chronic Inflammation and Insulin Resistance in Skeletal Muscle of High-Fat Diet Mice. Nutrients 2022, 14, 3730. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Niihara, H.; Mizokami, T.; Yada, K.; Suzuki, K. Exercise training attenuates neutrophil infiltration and elastase expression in adipose tissue of high-fat-diet-induced obese mice. Physiol. Rep. 2015, 3, e12534. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Niu, Y.; Liu, X.; Yang, F.; Niu, W.; Fu, L. Proteomic Analysis of Skeletal Muscle in Insulin-Resistant Mice: Response to 6-Week Aerobic Exercise. PLoS ONE 2013, 8, e53887. [Google Scholar] [CrossRef]
- Chan, J.Y.; Bensellam, M.; Lin, R.C.Y.; Liang, C.; Lee, K.; Jonas, J.-C.; Laybutt, D.R. Transcriptome analysis of islets from diabetes-resistant and diabetes-prone obese mice reveals novel gene regulatory networks involved in beta-cell compensation and failure. FASEB J. 2021, 35, e21608. [Google Scholar] [CrossRef]
- Tatsch, E.; De Carvalho, J.A.M.; Bollick, Y.S.; Duarte, T.; Duarte, M.; Vaucher, R.A.; Premaor, M.O.; Comim, F.V.; Moresco, R.N. Low frataxin mRNA expression is associated with inflammation and oxidative stress in patients with type 2 diabetes. Diabetes Metab. Res. Rev. 2020, 36, e3208. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Vig, S.; Datta, M.; Jindel, D.; Mathur, A.K.; Mathur, S.K.; Sharma, A. Systems biology approach reveals genome to phenome correlation in type 2 diabetes. PLoS ONE 2013, 8, e53522. [Google Scholar] [CrossRef]
- Henriques, B.J.; Katrine Jentoft Olsen, R.; Gomes, C.M.; Bross, P. Electron transfer flavoprotein and its role in mitochondrial energy metabolism in health and disease. Gene 2021, 776, 145407. [Google Scholar] [CrossRef]
- Chen, J.Q.; Huang, Y.Y.; Gusdon, A.M.; Qu, S. Irisin: A new molecular marker and target in metabolic disorder. Lipids Health Dis. 2015, 14, 2. [Google Scholar] [CrossRef]
- Lo, K.A.; Sun, L. Turning WAT into BAT: A review on regulators controlling the browning of white adipocytes. BioSci. Rep. 2013, 33, e00065. [Google Scholar] [CrossRef]
- Dickson, L.M.; Gandhi, S.; Layden, B.T.; Cohen, R.N.; Wicksteed, B. Protein kinase A induces UCP1 expression in specific adipose depots to increase energy expenditure and improve metabolic health. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2016, 311, R79–R88. [Google Scholar] [CrossRef]
- Guilford, B.L.; Parson, J.C.; Grote, C.W.; Vick, S.N.; Ryals, J.M.; Wright, D.E. Increased FNDC5 is associated with insulin resistance in high fat-fed mice. Physiol. Rep. 2017, 5, e13319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.M.; Xu, M.J.; Cai, Y.; Zhao, G.; Guan, Y.; Kong, W.; Tang, C.; Wang, X. Mitochondrial reactive oxygen species promote p65 nuclear translocation mediating high-phosphate-induced vascular calcification in vitro and in vivo. Kidney Int. 2011, 79, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.S.; Irwin, N.; O’Harte, F.P. Potential Therapeutic Role for Apelin and Related Peptides in Diabetes: An Update. Clin. Med. Insights Endocrinol. Diabetes 2022, 15, 11795514221074679. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Park, S.J.; Kim, J.H.; Seong, J.B.; Kim, K.M.; Woo, H.A.; Lee, D.S. Peroxiredoxin 5 regulates adipogenesis-attenuating oxidative stress in obese mouse models induced by a high-fat diet. Free Radic. Biol. Med. 2018, 123, 27–38. [Google Scholar] [CrossRef]
- Vaicik, M.K.; Blagajcevic, A.; Ye, H.; Morse, M.C.; Yang, F.; Goddi, A.; Brey, E.M.; Cohen, R.N. The Absence of Laminin α4 in Male Mice Results in Enhanced Energy Expenditure and Increased Beige Subcutaneous Adipose Tissue. Endocrinology 2018, 159, 356–367. [Google Scholar] [CrossRef]
- Wu, Q.; Ortegon, A.M.; Tsang, B.; Doege, H.; Feingold, K.R.; Stahl, A. FATP1 is an insulin-sensitive fatty acid transporter involved in diet-induced obesity. Mol. Cell Biol. 2006, 26, 3455–3467. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Qi, T.; Kong, Q.; Cheng, H.; Cao, X.; Li, Y.; Li, C.; Liu, L.; Ding, Z. HSPA12A is required for adipocyte differentiation and diet-induced obesity through a positive feedback regulation with PPAR? Cell Death Differ. 2019, 26, 2253–2267. [Google Scholar] [CrossRef]
- Schork, K.; Podwojski, K.; Turewicz, M.; Stephan, C.; Eisenacher, M. Important Issues in Planning a Proteomics Experiment: Statistical Considerations of Quantitative Proteomic Data. Methods Mol. Biol. 2021, 2228, 1–20. [Google Scholar] [CrossRef]
- Lehnig, A.C.; Stanford, K.I. Exercise-induced adaptations to white and brown adipose tissue. J. Exp. Biol. 2018, 221 (Suppl. S1), jeb161570. [Google Scholar] [CrossRef]
- Xu, X.; Ying, Z.; Cai, M.; Xu, Z.; Li, Y.; Jiang, S.Y.; Tzan, K.; Wang, A.; Parthasarathy, S.; He, G.; et al. Exercise ameliorates high-fat diet-induced metabolic and vascular dysfunction, and increases adipocyte progenitor cell population in brown adipose tissue. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2011, 300, R1115–R1125. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.J.; Lu, C.W.; Liao, C.C.; Chiang, C.H.; Huang, C.C.; Huang, K.C. Ovariectomy Interferes with Proteomes of Brown Adipose Tissue in Rats. Int. J. Med. Sci. 2022, 19, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, T.-J.; Lu, C.-W.; Lin, L.-Y.; Hsu, Y.-J.; Huang, C.-C.; Huang, K.-C. Proteomic Analysis of Skeletal Muscle and White Adipose Tissue after Aerobic Exercise Training in High Fat Diet Induced Obese Mice. Int. J. Mol. Sci. 2023, 24, 5743. https://doi.org/10.3390/ijms24065743
Chou T-J, Lu C-W, Lin L-Y, Hsu Y-J, Huang C-C, Huang K-C. Proteomic Analysis of Skeletal Muscle and White Adipose Tissue after Aerobic Exercise Training in High Fat Diet Induced Obese Mice. International Journal of Molecular Sciences. 2023; 24(6):5743. https://doi.org/10.3390/ijms24065743
Chicago/Turabian StyleChou, Tzu-Jung, Chia-Wen Lu, Li-Yu Lin, Yi-Ju Hsu, Chi-Chang Huang, and Kuo-Chin Huang. 2023. "Proteomic Analysis of Skeletal Muscle and White Adipose Tissue after Aerobic Exercise Training in High Fat Diet Induced Obese Mice" International Journal of Molecular Sciences 24, no. 6: 5743. https://doi.org/10.3390/ijms24065743
APA StyleChou, T.-J., Lu, C.-W., Lin, L.-Y., Hsu, Y.-J., Huang, C.-C., & Huang, K.-C. (2023). Proteomic Analysis of Skeletal Muscle and White Adipose Tissue after Aerobic Exercise Training in High Fat Diet Induced Obese Mice. International Journal of Molecular Sciences, 24(6), 5743. https://doi.org/10.3390/ijms24065743








