Pleomorphic Variants of Borreliella (syn. Borrelia) burgdorferi Express Evolutionary Distinct Transcriptomes
Abstract
1. Introduction
2. Results
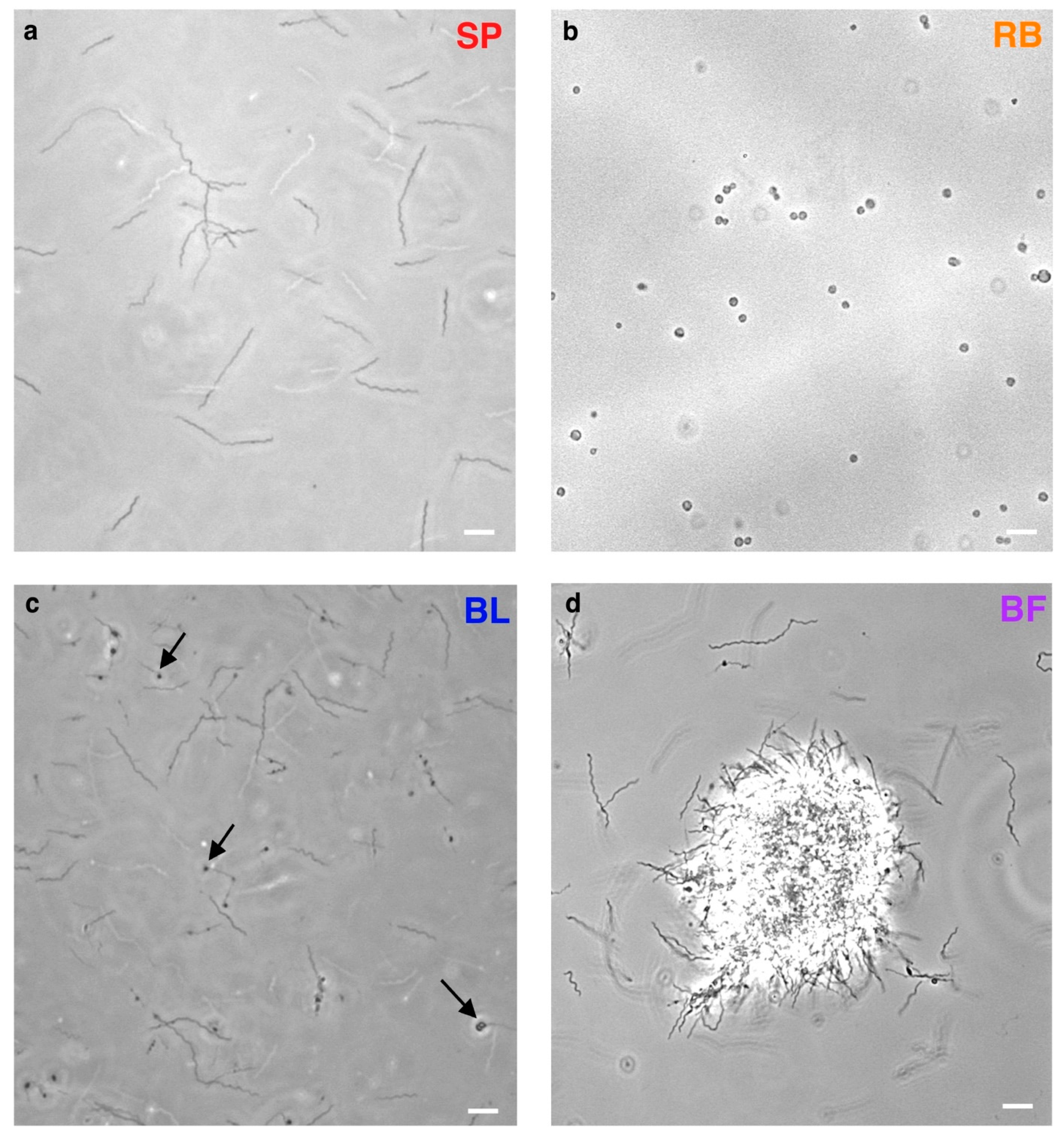
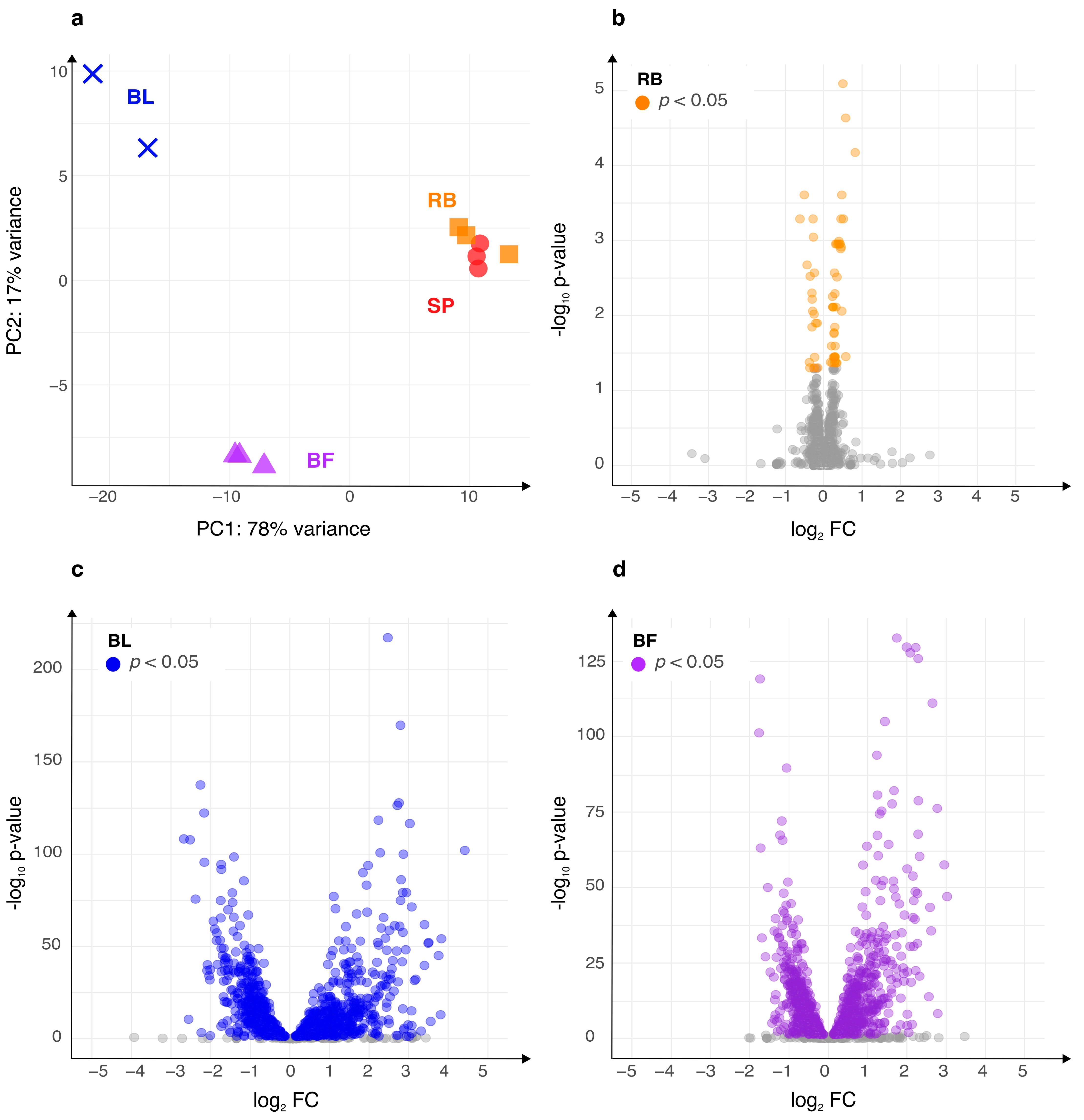
2.1. B. Burgdorferi Morphotypes Show Distinct Transcription Profiles
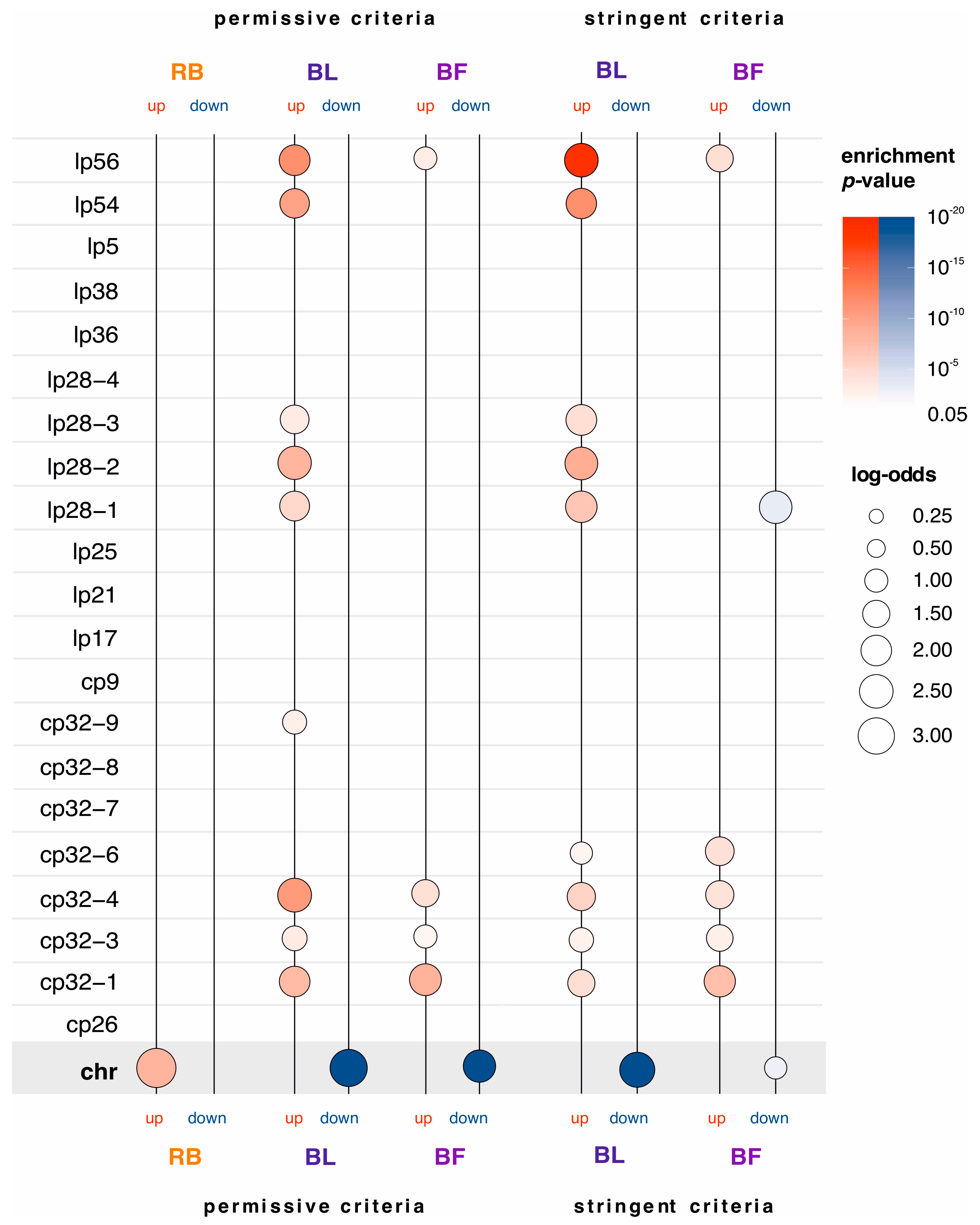
2.2. Morphotype-Specific Functional Enrichments
2.3. Genes Upregulated in Blebs and Biofilms Are Enriched with Plasmid-Encoded Genes
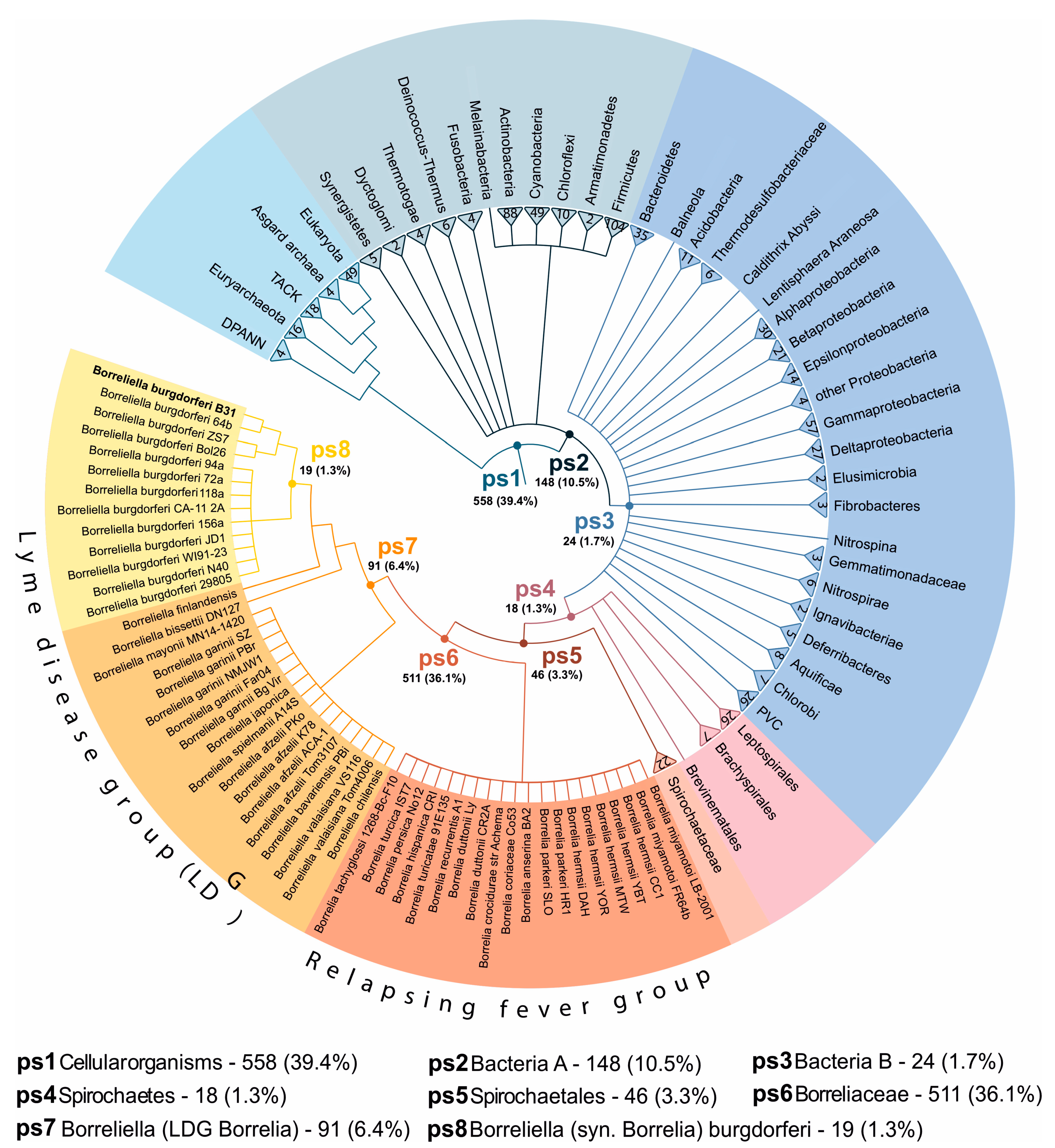
2.4. Biofilms and Blebs Express Evolutionary Younger Genes
2.5. Many B. burgdorferi Virulence Genes Are Differentially Expressed in Blebs and Biofilms
3. Discussion
4. Methods
4.1. Culturing Conditions and Imaging of B. burgdorferi Pleomorphic Forms
4.2. RNA Extraction and Sequencing
4.3. Transcriptome Data Analyses and Functional Annotation
4.4. Phylostratigraphic Analyses
4.5. Enrichment Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caccamo, P.D.; Brun, Y.V. The Molecular Basis of Noncanonical Bacterial Morphology. Trends Microbiol. 2018, 26, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, H.; Hagman, A.; Jules, M.; Sismeiro, O.; Dillies, M.-A.; Gouyette, C.; Kunst, F.; Steinert, M.; Heuner, K.; Coppee, J.-Y.; et al. Virulence Strategies for Infecting Phagocytes Deduced from the in Vivo Transcriptional Program of Legionella Pneumophila. Cell. Microbiol. 2006, 8, 1228–1240. [Google Scholar] [CrossRef]
- Hindré, T.; Brüggemann, H.; Buchrieser, C.; Héchard, Y. Transcriptional Profiling of Legionella Pneumophila Biofilm Cells and the Influence of Iron on Biofilm Formation. Microbiology 2008, 154, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Oliva, G.; Sahr, T.; Buchrieser, C. The Life Cycle of L. Pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 3. [Google Scholar] [CrossRef]
- Gober, J.W.; Marques, M.V. Regulation of Cellular Differentiation in Caulobacter Crescentus. Microbiol. Rev. 1995, 59, 31–47. [Google Scholar] [CrossRef]
- Helaine, S.; Kugelberg, E. Bacterial Persisters: Formation, Eradication, and Experimental Systems. Trends Microbiol. 2014, 22, 417–424. [Google Scholar] [CrossRef]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of Bacterial Persistence during Stress and Antibiotic Exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Michiels, J.E.; Van den Bergh, B.; Verstraeten, N.; Michiels, J. Molecular Mechanisms and Clinical Implications of Bacterial Persistence. Drug Resist. Updates 2016, 29, 76–89. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the Viable but Nonculturable State and Antibiotic Persister Cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Feng, J.; Shi, W.; Zhang, S.; Zhang, Y. Persister Mechanisms in Borrelia Burgdorferi: Implications for Improved Intervention. Emerg. Microbes Infect. 2015, 4, e56. [Google Scholar] [CrossRef] [PubMed]
- Meriläinen, L.; Brander, H.; Herranen, A.; Schwarzbach, A.; Gilbert, L. Pleomorphic Forms of Borrelia Burgdorferi Induce Distinct Immune Responses. Microbes Infect. 2016, 18, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Bamm, V.V.; Ko, J.T.; Mainprize, I.L.; Sanderson, V.P.; Wills, M.K.B. Lyme Disease Frontiers: Reconciling Borrelia Biology and Clinical Conundrums. Pathogens 2019, 8, 299. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, R.; Lindsey, N.P.; Fischer, M.; Gregory, C.J.; Hinckley, A.F.; Mead, P.S.; Paz-Bailey, G.; Waterman, S.H.; Drexler, N.A.; Kersh, G.J.; et al. Vital Signs: Trends in Reported Vectorborne Disease Cases—United States and Territories, 2004–2016. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 496–501. [Google Scholar] [CrossRef]
- Radolf, J.D.; Caimano, M.J.; Stevenson, B.; Hu, L.T. Of Ticks, Mice and Men: Understanding the Dual-Host Lifestyle of Lyme Disease Spirochaetes. Nat. Rev. Microbiol. 2012, 10, 87–99. [Google Scholar] [CrossRef]
- Stafford, K.C.; Cartter, M.L.; Magnarelli, L.A.; Ertel, S.-H.; Mshar, P.A. Temporal Correlations between Tick Abundance and Prevalence of Ticks Infected with Borrelia Burgdorferi and Increasing Incidence of Lyme Disease. J. Clin. Microbiol. 1998, 36, 1240–1244. [Google Scholar] [CrossRef]
- Dumic, I.; Severnini, E. “Ticking Bomb”: The Impact of Climate Change on the Incidence of Lyme Disease. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 5719081. [Google Scholar] [CrossRef]
- Wong, K.H.; Shapiro, E.D.; Soffer, G.K. A Review of Post-Treatment Lyme Disease Syndrome and Chronic Lyme Disease for the Practicing Immunologist. Clinic. Rev. Allerg Immunol. 2022, 62, 264–271. [Google Scholar] [CrossRef]
- Rudenko, N.; Golovchenko, M.; Kybicova, K.; Vancova, M. Metamorphoses of Lyme Disease Spirochetes: Phenomenon of Borrelia Persisters. Parasites Vectors 2019, 12, 237. [Google Scholar] [CrossRef]
- Vesey, P.M.; Kuramitsu, H.K. Genetic Analysis of Treponema Denticola ATCC 35405 Biofilm Formation. Microbiology 2004, 150, 2401–2407. [Google Scholar] [CrossRef]
- Ristow, P.; Bourhy, P.; Kerneis, S.; Schmitt, C.; Prevost, M.-C.; Lilenbaum, W.; Picardeau, M. Biofilm Formation by Saprophytic and Pathogenic Leptospires. Microbiology 2008, 154, 1309–1317. [Google Scholar] [CrossRef]
- Umemoto, T.; Namikawa, I.; Yamamoto, M. Colonial Morphology of Treponemes Observed by Electron Microscopy. Microbiol. Immunol. 1984, 28, 11–22. [Google Scholar] [CrossRef]
- Barbour, A.G. Isolation and Cultivation of Lyme Disease Spirochetes. Yale J. Biol. Med. 1984, 57, 521–525. [Google Scholar]
- Vancová, M.; Rudenko, N.; Vaněček, J.; Golovchenko, M.; Strnad, M.; Rego, R.O.M.; Tichá, L.; Grubhoffer, L.; Nebesářová, J. Pleomorphism and Viability of the Lyme Disease Pathogen Borrelia Burgdorferi Exposed to Physiological Stress Conditions: A Correlative Cryo-Fluorescence and Cryo-Scanning Electron Microscopy Study. Front. Microbiol. 2017, 8, 596. [Google Scholar] [CrossRef]
- Rosa, P.A.; Tilly, K.; Stewart, P.E. The Burgeoning Molecular Genetics of the Lyme Disease Spirochaete. Nat. Rev. Microbiol. 2005, 3, 129–143. [Google Scholar] [CrossRef]
- Kudryashev, M.; Cyrklaff, M.; Baumeister, W.; Simon, M.M.; Wallich, R.; Frischknecht, F. Comparative Cryo-Electron Tomography of Pathogenic Lyme Disease Spirochetes. Mol. Microbiol. 2009, 71, 1415–1434. [Google Scholar] [CrossRef]
- Motaleb, M.A.; Corum, L.; Bono, J.L.; Elias, A.F.; Rosa, P.; Samuels, D.S.; Charon, N.W. Borrelia Burgdorferi Periplasmic Flagella Have Both Skeletal and Motility Functions. Proc. Natl. Acad. Sci. USA 2000, 97, 10899–10904. [Google Scholar] [CrossRef]
- Harman, M.W.; Dunham-Ems, S.M.; Caimano, M.J.; Belperron, A.A.; Bockenstedt, L.K.; Fu, H.C.; Radolf, J.D.; Wolgemuth, C.W. The Heterogeneous Motility of the Lyme Disease Spirochete in Gelatin Mimics Dissemination through Tissue. Proc. Natl. Acad. Sci. USA 2012, 109, 3059–3064. [Google Scholar] [CrossRef]
- Miklossy, J.; Kasas, S.; Zurn, A.D.; McCall, S.; Yu, S.; McGeer, P.L. Persisting Atypical and Cystic Forms of Borrelia Burgdorferi and Local Inflammation in Lyme Neuroborreliosis. J. Neuroinflammation 2008, 5, 40. [Google Scholar] [CrossRef]
- Alban, P.S.; Johnson, P.W.; Nelson, D.R. Serum-Starvation-Induced Changes in Protein Synthesis and Morphology of Borrelia Burgdorferi. Microbiology 2000, 146, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Murgia, R.; Piazzetta, C.; Cinco, M. Cystic Forms of Borrelia Burgdorferi Sensu Lato: Induction, Development, and the Role of RpoS. Wien. Klin. Wochenschr. 2002, 114, 574–579. [Google Scholar]
- Kersten, A.; Poitschek, C.; Rauch, S.; Aberer, E. Effects of Penicillin, Ceftriaxone, and Doxycycline on Morphology of Borrelia Burgdorferi. Antimicrob. Agents. Chemother. 1995, 39, 1127–1133. [Google Scholar] [CrossRef]
- Brorson, Ø.; Brorson, S.H. In Vitro Conversion OfBorrelia Burgdorferi to Cystic Forms in Spinal Fluid, and Transformation to Mobile Spirochetes by Incubation in BSK-H Medium. Infection 1998, 26, 144–150. [Google Scholar] [CrossRef]
- Duray, P.H.; Yin, S.; Ito, Y.; Bezrukov, L.; Cox, C.; Cho, M.; Fitzgerald, W.; Dorward, D.; Zimmerberg, J.; Margolis, L. Invasion of Human Tissue Ex Vivo by Borrelia Burgdorferi. J. Infect. Dis. 2005, 191, 1747–1754. [Google Scholar] [CrossRef]
- Hulínska, D.; Barták, P.; Hercogová, J.; Hančil, J.; Bašta, J.; Schramlová, J. Electron Microscopy of Langerhans Cells and Borrelia Burgdorferi in Lyme Disease Patients. Zentralblatt Für Bakteriologie 1994, 280, 348–359. [Google Scholar] [CrossRef]
- Berndtson, K. Review of Evidence for Immune Evasion and Persistent Infection in Lyme Disease. IJGM 2013, 6, 291–306. [Google Scholar] [CrossRef]
- Malge, A.; Ghai, V.; Reddy, P.J.; Baxter, D.; Kim, T.-K.; Moritz, R.L.; Wang, K. MRNA Transcript Distribution Bias between Borrelia Burgdorferi Bacteria and Their Outer Membrane Vesicles. FEMS Microbiol. Lett. 2018, 365, fny135. [Google Scholar] [CrossRef]
- Karvonen, K.; Tammisto, H.; Nykky, J.; Gilbert, L. Borrelia Burgdorferi Outer Membrane Vesicles Contain Antigenic Proteins, but Do Not Induce Cell Death in Human Cells. Microorganisms 2022, 10, 212. [Google Scholar] [CrossRef]
- Whitmire, W.M.; Garon, C.F. Specific and Nonspecific Responses of Murine B Cells to Membrane Blebs of Borrelia Burgdorferi. Infect. Immun. 1993, 61, 1460–1467. [Google Scholar] [CrossRef]
- Sapi, E.; Bastian, S.L.; Mpoy, C.M.; Scott, S.; Rattelle, A.; Pabbati, N.; Poruri, A.; Burugu, D.; Theophilus, P.A.S.; Pham, T.V.; et al. Characterization of Biofilm Formation by Borrelia Burgdorferi In Vitro. PLoS ONE 2012, 7, e48277. [Google Scholar] [CrossRef] [PubMed]
- Futo, M.; Opašić, L.; Koska, S.; Čorak, N.; Široki, T.; Ravikumar, V.; Thorsell, A.; Lenuzzi, M.; Kifer, D.; Domazet-Lošo, M.; et al. Embryo-Like Features in Developing Bacillus Subtilis Biofilms. Mol. Biol. Evol. 2021, 38, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Futo, M.; Široki, T.; Koska, S.; Čorak, N.; Tušar, A.; Domazet-Lošo, M.; Domazet-Lošo, T. A Novel Time-Lapse Imaging Method for Studying Developing Bacterial Biofilms. Sci. Rep. 2022, 12, 21120. [Google Scholar] [CrossRef] [PubMed]
- Sapi, E.; Kasliwala, R.S.; Ismail, H.; Torres, J.P.; Oldakowski, M.; Markland, S.; Gaur, G.; Melillo, A.; Eisendle, K.; Liegner, K.B.; et al. The Long-Term Persistence of Borrelia Burgdorferi Antigens and DNA in the Tissues of a Patient with Lyme Disease. Antibiotics 2019, 8, 183. [Google Scholar] [CrossRef]
- Sapi, E.; Theophilus, P.A.S.; Pham, T.V.; Burugu, D.; Luecke, D.F. Effect of RpoN, RpoS and LuxS Pathways on the Biofilm Formation and AntibiotIc Sensitivity of Borrelia Burgdorferi. Eur. J. Microbiol. Immunol. 2016, 6, 272–286. [Google Scholar] [CrossRef]
- Lantos, P.M.; Auwaerter, P.G.; Wormser, G.P. A Systematic Review of Borrelia Burgdorferi Morphologic Variants Does Not Support a Role in Chronic Lyme Disease. Clin. Infect. Dis. 2014, 58, 663–671. [Google Scholar] [CrossRef]
- Al-Robaiy, S.; Dihazi, H.; Kacza, J.; Seeger, J.; Schiller, J.; Huster, D.; Knauer, J.; Straubinger, R.K. Metamorphosis of Borrelia Burgdorferi Organisms—RNA, Lipid and Protein Composition in Context with the Spirochetes’ Shape. J. Basic Microbiol. 2010, 50, S5–S17. [Google Scholar] [CrossRef]
- Drecktrah, D.; Lybecker, M.; Popitsch, N.; Rescheneder, P.; Hall, L.S.; Samuels, D.S. The Borrelia Burgdorferi RelA/SpoT Homolog and Stringent Response Regulate Survival in the Tick Vector and Global Gene Expression during Starvation. PLoS Pathog 2015, 11, e1005160. [Google Scholar] [CrossRef]
- Domazet-Lošo, T.; Brajković, J.; Tautz, D. A Phylostratigraphy Approach to Uncover the Genomic History of Major Adaptations in Metazoan Lineages. Trends Genet. 2007, 23, 533–539. [Google Scholar] [CrossRef]
- Domazet-Lošo, T.; Tautz, D. A Phylogenetically Based Transcriptome Age Index Mirrors Ontogenetic Divergence Patterns. Nature 2010, 468, 815–818. [Google Scholar] [CrossRef]
- Domazet-Lošo, T.; Tautz, D. Phylostratigraphic Tracking of Cancer Genes Suggests a Link to the Emergence of Multicellularity in Metazoa. BMC Biol. 2010, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Domazet-Lošo, T.; Carvunis, A.-R.; Mar Albà, M.; Sebastijan Šestak, M.; Bakarić, R.; Neme, R.; Tautz, D. No Evidence for Phylostratigraphic Bias Impacting Inferences on Patterns of Gene Emergence and Evolution. Mol. Biol. Evol. 2017, 4, msw284. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.K.; Savage, C.R.; Brissette, C.A.; Seshu, J.; Livny, J.; Stevenson, B. RNA-Seq of Borrelia Burgdorferi in Multiple Phases of Growth Reveals Insights into the Dynamics of Gene Expression, Transcriptome Architecture, and Noncoding RNAs. PLoS ONE 2016, 11, e0164165. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Guan, G.; Liu, Z.; Li, Y.; Luo, J.; Yin, H. RNA-Seq-Based Analysis of Changes in Borrelia Burgdorferi Gene Expression Linked to Pathogenicity. Parasites Vectors 2015, 8, 155. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on MRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Byrgazov, K.; Vesper, O.; Moll, I. Ribosome Heterogeneity: Another Level of Complexity in Bacterial Translation Regulation. Curr. Opin. Microbiol. 2013, 16, 133–139. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Molecular. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Fraser, C.M.; Casjens, S.; Huang, W.M.; Sutton, G.G.; Clayton, R.; Lathigra, R.; White, O.; Ketchum, K.A.; Dodson, R.; Hickey, E.K.; et al. Genomic Sequence of a Lyme Disease Spirochaete, Borrelia Burgdorferi. Nature 1997, 390, 580–586. [Google Scholar] [CrossRef]
- Casjens, S.; Palmer, N.; Van Vugt, R.; Mun Huang, W.; Stevenson, B.; Rosa, P.; Lathigra, R.; Sutton, G.; Peterson, J.; Dodson, R.J.; et al. A Bacterial Genome in Flux: The Twelve Linear and Nine Circular Extrachromosomal DNAs in an Infectious Isolate of the Lyme Disease Spirochete Borrelia Burgdorferi: Borrelia Plasmids. Mol. Microbiol. 2000, 35, 490–516. [Google Scholar] [CrossRef]
- Kurokawa, C.; Lynn, G.E.; Pedra, J.H.F.; Pal, U.; Narasimhan, S.; Fikrig, E. Interactions between Borrelia Burgdorferi and Ticks. Nat. Rev. Microbiol. 2020, 18, 587–600. [Google Scholar] [CrossRef]
- Schwartz, I.; Margos, G.; Casjens, S.R.; Qiu, W.-G.; Eggers, C.H. Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and Prophages. Curr. Issues Mol. Biol. 2022, 42, 409–454. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-Negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Adeolu, M.; Gupta, R.S. A Phylogenomic and Molecular Marker Based Proposal for the Division of the Genus Borrelia into Two Genera: The Emended Genus Borrelia Containing Only the Members of the Relapsing Fever Borrelia, and the Genus Borreliella Gen. Nov. Containing the Members of the Lyme Disease Borrelia (Borrelia Burgdorferi Sensu Lato Complex). Antonie van Leeuwenhoek 2014, 105, 1049–1072. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S. Distinction between Borrelia and Borreliella Is More Robustly Supported by Molecular and Phenotypic Characteristics than All Other Neighbouring Prokaryotic Genera: Response to Margos’ et al. “The Genus Borrelia Reloaded” (PLoS ONE 13(12): E0208432). PLoS ONE 2019, 14, e0221397. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.G.; Gupta, R.S. The Family Borreliaceae (Spirochaetales), a Diverse Group in Two Genera of Tick-Borne Spirochetes of Mammals, Birds, and Reptiles. J. Med. Entomol. 2021, 58, 1513–1524. [Google Scholar] [CrossRef]
- Margos, G.; Gofton, A.; Wibberg, D.; Dangel, A.; Marosevic, D.; Loh, S.-M.; Oskam, C.; Fingerle, V. The Genus Borrelia Reloaded. PLoS ONE 2018, 13, e0208432. [Google Scholar] [CrossRef]
- Margos, G.; Fingerle, V.; Cutler, S.; Gofton, A.; Stevenson, B.; Estrada-Peña, A. Controversies in Bacterial Taxonomy: The Example of the Genus Borrelia. Ticks Tick-Borne Dis. 2020, 11, 101335. [Google Scholar] [CrossRef]
- Arahal, D.R.; Bull, C.T.; Busse, H.-J.; Christensen, H.; Chuvochina, M.; Dedysh, S.N.; Fournier, P.-E.; Konstantinidis, K.T.; Parker, C.T.; Rossello-Mora, R.; et al. Judicial Opinions 123–127. Int. J. Syst. Evol. Microbiol. 2022, 72, 005708. [Google Scholar] [CrossRef]
- Gupta, R.S.; Mahmood, S.; Adeolu, M. A Phylogenomic and Molecular Signature Based Approach for Characterization of the Phylum Spirochaetes and Its Major Clades: Proposal for a Taxonomic Revision of the Phylum. Front. Microbiol. 2013, 4, 217. [Google Scholar] [CrossRef]
- Zhang, J.-R.; Hardham, J.M.; Barbour, A.G.; Norris, S.J. Antigenic Variation in Lyme Disease Borreliae by Promiscuous Recombination of VMP-like Sequence Cassettes. Cell 1997, 89, 275–285. [Google Scholar] [CrossRef]
- Anderson, C.; Brissette, C.A. The Brilliance of Borrelia: Mechanisms of Host Immune Evasion by Lyme Disease-Causing Spirochetes. Pathogens 2021, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Caine, J.A.; Coburn, J. Multifunctional and Redundant Roles of Borrelia Burgdorferi Outer Surface Proteins in Tissue Adhesion, Colonization, and Complement Evasion. Front. Immunol. 2016, 7, 442. [Google Scholar] [CrossRef] [PubMed]
- Shoberg, R.J.; Thomas, D.D. Specific Adherence of Borrelia Burgdorferi Extracellular Vesicles to Human Endothelial Cells in Culture. Infect. Immun. 1993, 61, 3892–3900. [Google Scholar] [CrossRef]
- Coburn, J.; Garcia, B.; Hu, L.T.; Jewett, M.W.; Kraiczy, P.; Norris, S.J.; Skare, J. Lyme Disease Pathogenesis. Curr. Issues Mol. Biol. 2022, 42, 473–518. [Google Scholar] [CrossRef]
- Croucher, N.J.; Thomson, N.R. Studying Bacterial Transcriptomes Using RNA-Seq. Curr. Opin. Microbiol. 2010, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Hör, J.; Gorski, S.A.; Vogel, J. Bacterial RNA Biology on a Genome Scale. Molecular. Cell. 2018, 70, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Couzin-Frankel, J. Long-Underfunded Lyme Disease Research Gets an Injection of Money—And Ideas. Science 2019. [Google Scholar] [CrossRef]
- Domazet-Lošo, M.; Široki, T.; Domazet-Lošo, T. Macroevolutionary Dynamics of Gene Family Gain and Loss along Multicellular Eukaryotic Lineages; Evolutionary Biology. bioRxiv 2022. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a Bacterial World, a New Imperative for the Life Sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef]
- Domazet-Loso, T. An Evolutionary Analysis of Orphan Genes in Drosophila. Genome Res. 2003, 13, 2213–2219. [Google Scholar] [CrossRef]
- Khalturin, K.; Hemmrich, G.; Fraune, S.; Augustin, R.; Bosch, T.C.G. More than Just Orphans: Are Taxonomically-Restricted Genes Important in Evolution? Trends Genet. 2009, 25, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Domazet-Lošo, T. The Evolutionary Origin of Orphan Genes. Nat. Rev. Genet. 2011, 12, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Derouiche, A.; Pandit, S.; Rahimi, S.; Kalantari, A.; Futo, M.; Ravikumar, V.; Jers, C.; Mokkapati, V.R.S.S.; Vlahoviček, K.; et al. Evolutionary Analysis of the Bacillus Subtilis Genome Reveals New Genes Involved in Sporulation. Mol. Biol. Evol. 2020, 37, msaa035. [Google Scholar] [CrossRef]
- Drecktrah, D.; Samuels, D.S. Genetic Manipulation of Borrelia Spp. In Spirochete Biology: The Post Genomic Era; Adler, B., Ed.; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, Switzerland, 2017; Volume 415, pp. 113–140. ISBN 978-3-319-89637-3. [Google Scholar]
- Rosa, P.A.; Jewett, M.W. Genetic Manipulation of Borrelia. Curr. Issues Mol. Biol. 2022, 42, 307–332. [Google Scholar] [CrossRef] [PubMed]
- Zückert, W.R. Laboratory Maintenance of Borrelia Burgdorferi. CP Microbiol. 2007, 4, 12C.1.1-10. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by EggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Seshadri, R.; Varghese, N.J.; Eloe-Fadrosh, E.A.; Meier-Kolthoff, J.P.; Göker, M.; Coates, R.C.; Hadjithomas, M.; Pavlopoulos, G.A.; Paez-Espino, D.; et al. 1,003 Reference Genomes of Bacterial and Archaeal Isolates Expand Coverage of the Tree of Life. Nat. Biotechnol. 2017, 35, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A New View of the Tree of Life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Paster, B.J.; Dewhirst, F.E. Phylogenetic Foundation of Spirochetes. J. Mol. Microbiol. Biotechnol. 2000, 2, 341–344. [Google Scholar] [PubMed]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The Two-Domain Tree of Life Is Linked to a New Root for the Archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef] [PubMed]
- Di, L.; Pagan, P.E.; Packer, D.; Martin, C.L.; Akther, S.; Ramrattan, G.; Mongodin, E.F.; Fraser, C.M.; Schutzer, S.E.; Luft, B.J.; et al. BorreliaBase: A Phylogeny-Centered Browser of Borrelia Genomes. BMC Bioinform. 2014, 15, 233. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the Phylogeny and Coding Potential of Microbial Dark Matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A Phylogeny-Driven Genomic Encyclopaedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef]
- Richter, D.; Postic, D.; Sertour, N.; Livey, I.; Matuschka, F.-R.; Baranton, G. Delineation of Borrelia Burgdorferi Sensu Lato Species by Multilocus Sequence Analysis and Confirmation of the Delineation of Borrelia Spielmanii Sp. Nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 873–881. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]



| Permissive Criteria | Stringent Criteria | |||||
|---|---|---|---|---|---|---|
| DE Cutoff | p < 0.05 | p < 0.05 and Fold-Change > 2 | ||||
| N (%) | Up | Down | Total | Up | Down | Total |
| RB | 44 (2.85) | 23 (1.49) | 67 (4.34) | 0 (0) | 0 (0) | 0 (0) |
| BL | 529 (34.26) | 522 (33.81) | 1051 (68.07) | 274 (17.75) | 142 (9.20) | 416 (26.94) |
| BF | 467 (30.25) | 464 (30.01) | 931 (60.30) | 156 (10.10) | 60 (3.89) | 216 (13.99) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čorak, N.; Anniko, S.; Daschkin-Steinborn, C.; Krey, V.; Koska, S.; Futo, M.; Široki, T.; Woichansky, I.; Opašić, L.; Kifer, D.; et al. Pleomorphic Variants of Borreliella (syn. Borrelia) burgdorferi Express Evolutionary Distinct Transcriptomes. Int. J. Mol. Sci. 2023, 24, 5594. https://doi.org/10.3390/ijms24065594
Čorak N, Anniko S, Daschkin-Steinborn C, Krey V, Koska S, Futo M, Široki T, Woichansky I, Opašić L, Kifer D, et al. Pleomorphic Variants of Borreliella (syn. Borrelia) burgdorferi Express Evolutionary Distinct Transcriptomes. International Journal of Molecular Sciences. 2023; 24(6):5594. https://doi.org/10.3390/ijms24065594
Chicago/Turabian StyleČorak, Nina, Sirli Anniko, Christina Daschkin-Steinborn, Viktoria Krey, Sara Koska, Momir Futo, Tin Široki, Innokenty Woichansky, Luka Opašić, Domagoj Kifer, and et al. 2023. "Pleomorphic Variants of Borreliella (syn. Borrelia) burgdorferi Express Evolutionary Distinct Transcriptomes" International Journal of Molecular Sciences 24, no. 6: 5594. https://doi.org/10.3390/ijms24065594
APA StyleČorak, N., Anniko, S., Daschkin-Steinborn, C., Krey, V., Koska, S., Futo, M., Široki, T., Woichansky, I., Opašić, L., Kifer, D., Tušar, A., Maxeiner, H.-G., Domazet-Lošo, M., Nicolaus, C., & Domazet-Lošo, T. (2023). Pleomorphic Variants of Borreliella (syn. Borrelia) burgdorferi Express Evolutionary Distinct Transcriptomes. International Journal of Molecular Sciences, 24(6), 5594. https://doi.org/10.3390/ijms24065594






