Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination
Abstract
1. Introduction
2. The Biochemical Mechanisms Underlying lncRNA Function
3. Approaches for Study of lncRNA–Protein, lncRNA–DNA and lncRNA–RNA Interactions
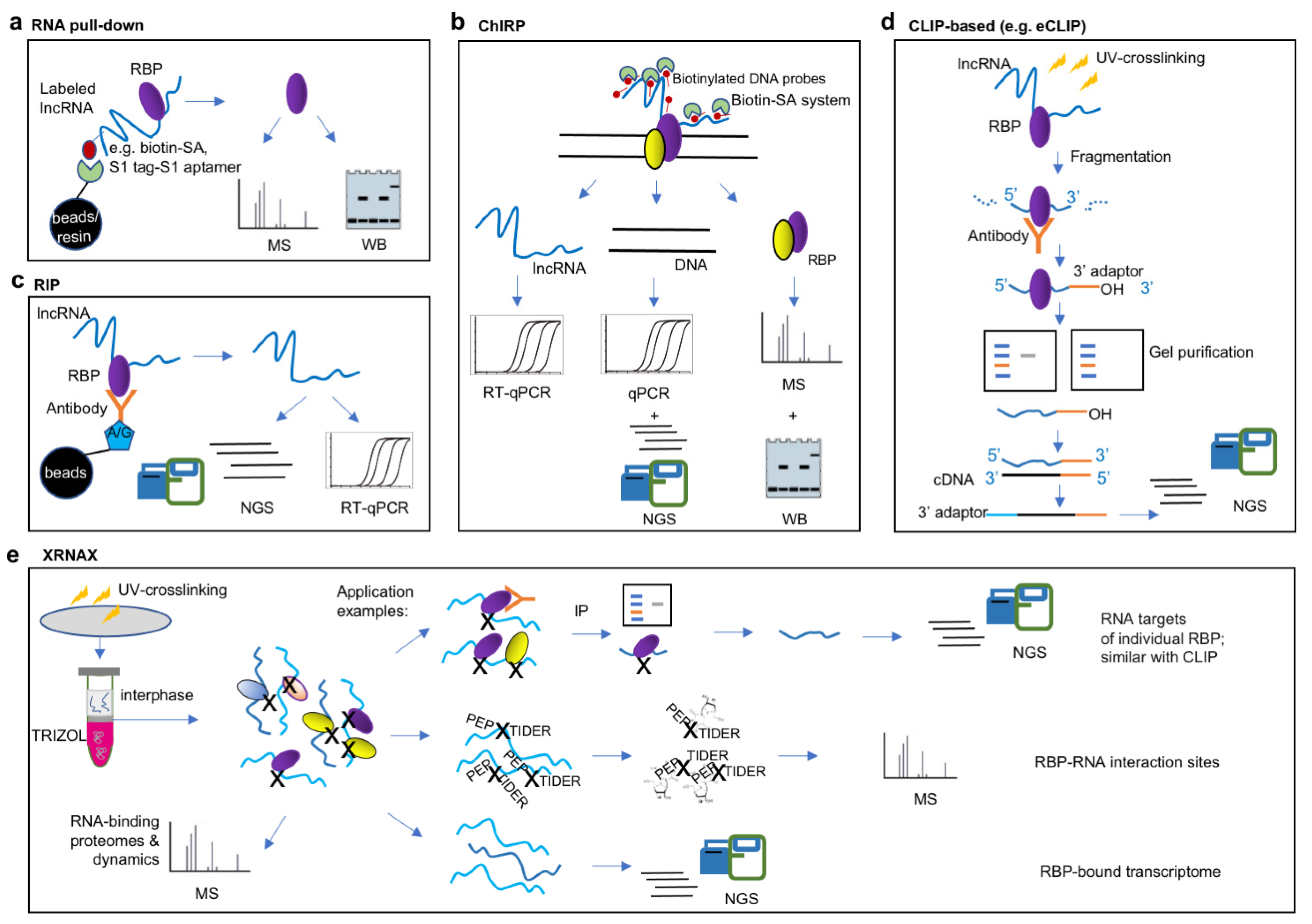
3.1. Approaches for Study of lncRNA–Protein Interactions
3.1.1. lncRNA-Centric Approaches
3.1.2. Protein-Centric Approaches
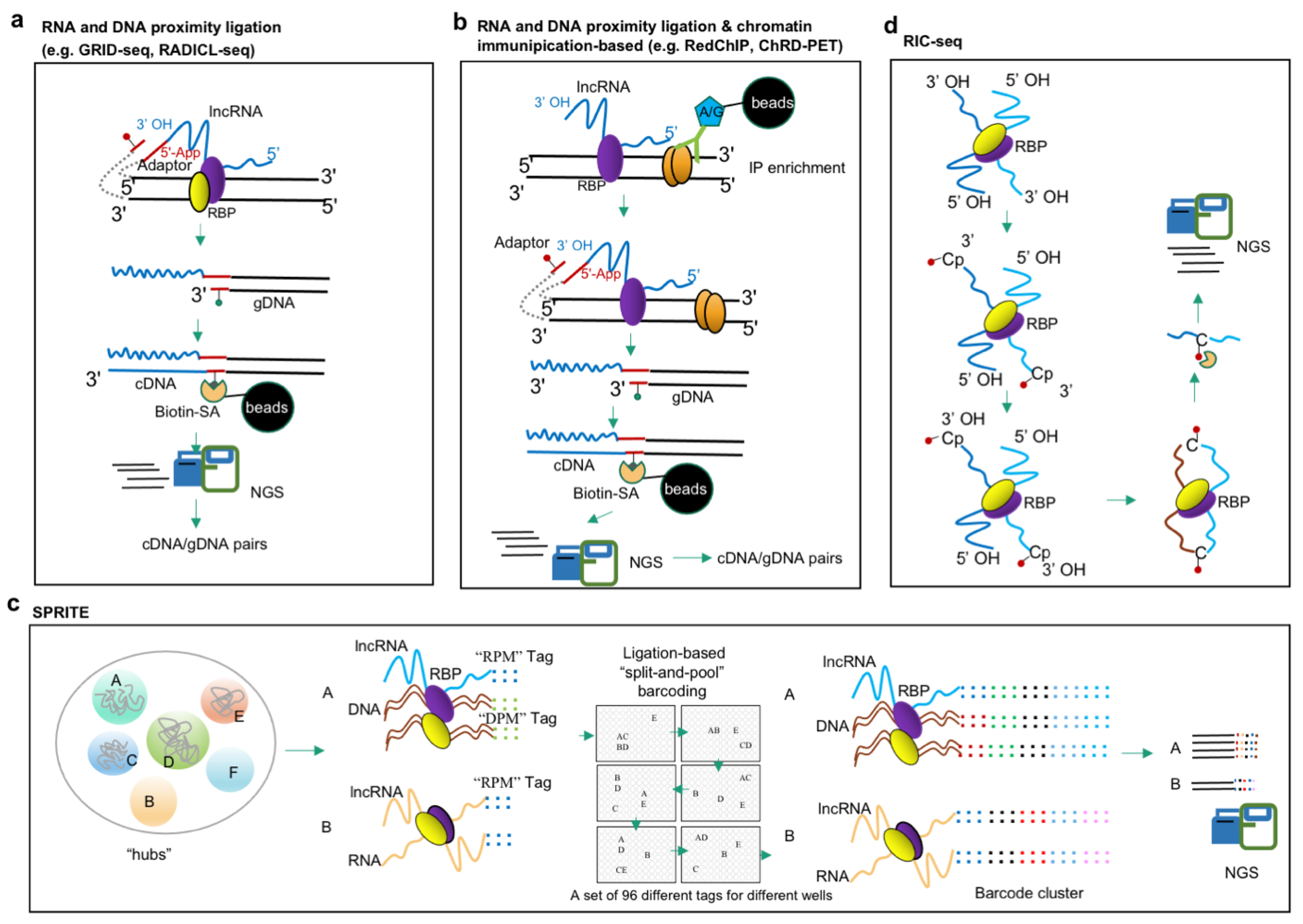
3.2. Approaches for Study of lncRNA–DNA Interactions
3.3. Approaches for Study of lncRNA–RNA Interactions
3.4. Bioinformatic Processes for High-Throughput Approaches in Study of lncRNAs
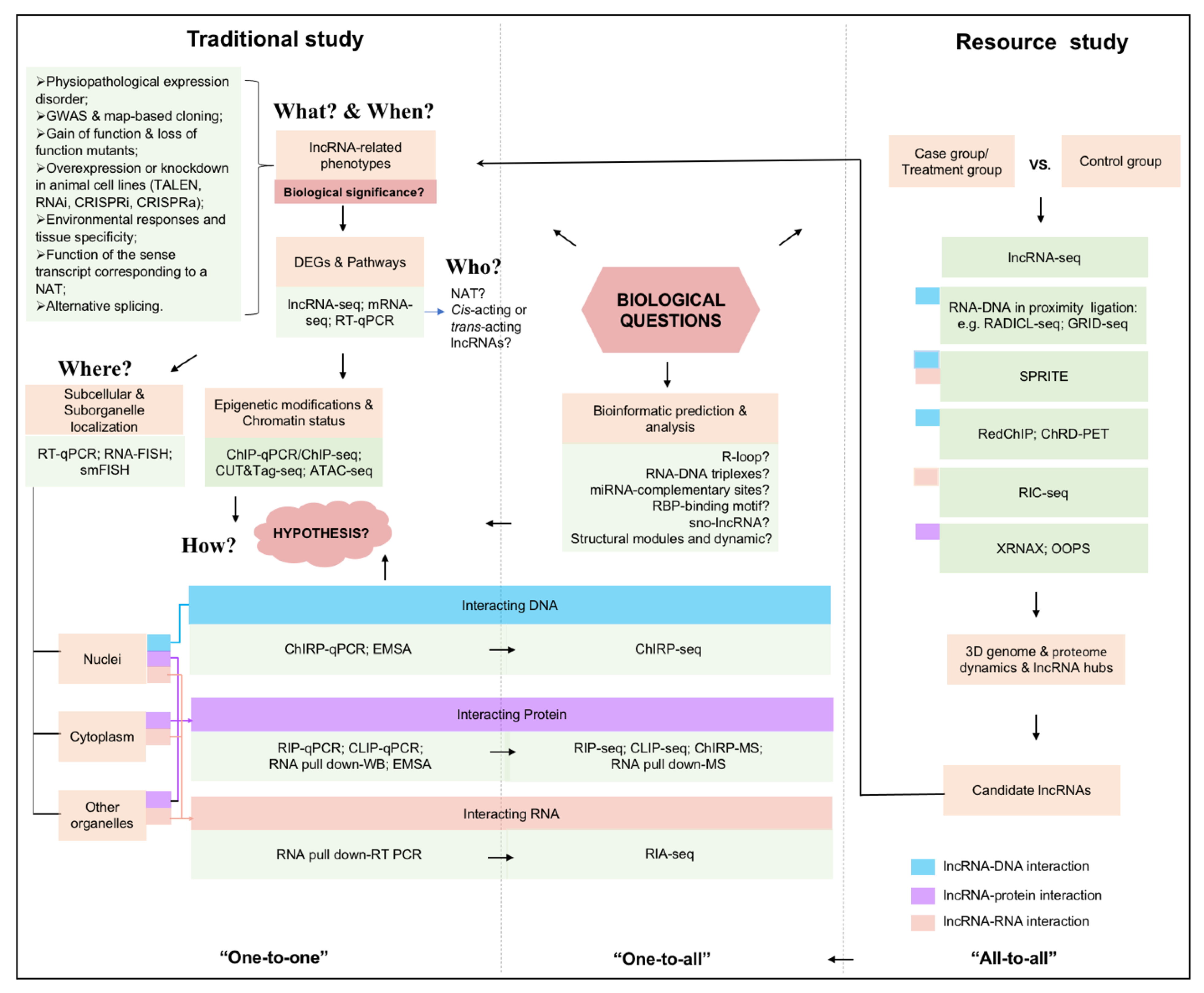
4. A General Mind Map for lncRNA Study
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar]
- Chen, L.; Zhu, Q.-H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Pandyajones, A.; Mcdonel, P.; Shishkin, A.A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X-chromosome*. Science 2013, 341, 1237973. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Wutz, A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011, 12, 542–553. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Fei, J.; Jadaliha, M.; Harmon, T.S.; Li, I.T.S.; Hua, B.; Hao, Q.; Holehouse, A.S.; Reyer, M.; Sun, Q.; Freier, S.M.; et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 2017, 130, 4180–4192. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.B.; Hezroni, H.; Goldrich, M.J.; Ulitsky, I. Regulation of Neuroregeneration by Long Noncoding RNAs. Mol. Cell 2018, 72, 553–567.e5. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef]
- Sánchez, Y.; Segura, V.; Marín-Béjar, O.; Athie, A.; Marchese, F.P.; González, J.; Bujanda, L.; Guo, S.; Matheu, A.; Huarte, M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014, 5, 5812. [Google Scholar] [CrossRef]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 3516. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H.M. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef]
- Kim, D.H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Kim, D.H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312.e4. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.D.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Atianand, M.K.; Caffrey, D.R.; Fitzgerald, K.A. Immunobiology of Long Noncoding RNAs. Annu. Rev. Immunol. 2017, 35, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, L.Z.; Chen, L.L. Understanding lncRNA-protein assemblies with imaging and single-molecule approaches. Curr. Opin. Genet. Dev. 2022, 72, 128–137. [Google Scholar] [CrossRef]
- Chen, L.-L. Towards higher-resolution and in vivo understanding of lncRNA biogenesis and function. Nat. Methods 2022, 19, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e5. [Google Scholar] [CrossRef]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef]
- Zhang, Y.; Long, Y.; Kwoh, C.K. Deep learning based DNA:RNA triplex forming potential prediction. BMC Bioinform. 2020, 21, 522. [Google Scholar] [CrossRef] [PubMed]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915.e4. [Google Scholar] [CrossRef] [PubMed]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Kalwa, M.; Hänzelmann, S.; Otto, S.; Kuo, C.C.; Franzen, J.; Joussen, S.; Fernandez-Rebollo, E.; Rath, B.; Koch, C.; Hofmann, A.; et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016, 44, 10631–10643. [Google Scholar] [CrossRef] [PubMed]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Li, M.; Zhao, T.; Feng, S.; Zhang, H.; Wang, L.; Han, J.; Gao, M.; Lu, K.; Chen, Q.; et al. Neofunctionalization of a polyploidization-activated cotton long intergenic non-coding RNA DAN1 during drought stress regulation. Plant Physiol. 2021, 186, 2152–2168. [Google Scholar] [CrossRef]
- Maldonado, R.; Schwartz, U.; Silberhorn, E.; Langst, G. Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells. Mol. Cell 2019, 73, 1243–1254.e6. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Tan-Wong, S.M.; Dhir, S.; Proudfoot, N.J. R-Loops Promote Antisense Transcription across the Mammalian Genome. Mol. Cell 2019, 76, 600–616.e6. [Google Scholar] [CrossRef]
- Gibbons, H.R.; Aune, T.M. Immunoprecipitation of DNA:RNA Hybrids Using the S9.6 Antibody. Methods Mol. Biol. 2020, 2161, 195–207. [Google Scholar]
- Li, L.; Luo, H.; Lim, D.-H.; Han, L.; Li, Y.; Fu, X.-D.; Qi, Y. Global profiling of RNA–chromatin interactions reveals co-regulatory gene expression networks in Arabidopsis. Nat. Plants 2021, 7, 1364–1378. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schäfer, A.; Grummt, I.; Niehrs, C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Park, Y.J.; Lindroth, A.M.; Schäfer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Liu, C.-X.; Chen, L.-L.; Zhang, Q.C. RNA structure probing uncovers RNA structure-dependent biological functions. Nat. Chem. Biol. 2021, 17, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636.e22. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef]
- Sarma, K.; Cifuentes-Rojas, C.; Ergun, A.; Del Rosario, A.; Jeon, Y.; White, F.; Sadreyev, R.; Lee, J.T. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 2014, 159, 869–883. [Google Scholar] [CrossRef]
- Colognori, D.; Sunwoo, H.; Wang, D.; Wang, C.Y.; Lee, J.T. Xist Repeats A and B Account for Two Distinct Phases of X Inactivation Establishment. Dev. Cell 2020, 54, 21–32.e5. [Google Scholar] [CrossRef]
- Lu, Z.; Guo, J.K.; Wei, Y.; Dou, D.R.; Zarnegar, B.; Ma, Q.; Li, R.; Zhao, Y.; Liu, F.; Choudhry, H.; et al. Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 2020, 11, 6163. [Google Scholar] [CrossRef]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Marquardt, S.; Raitskin, O.; Wu, Z.; Liu, F.; Sun, Q.; Dean, C. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 2014, 54, 156–165. [Google Scholar] [CrossRef]
- Li, P.; Tao, Z.; Dean, C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 2015, 29, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhu, P.; Cheema, J.; Bloomer, R.; Mikulski, P.; Liu, Q.; Zhang, Y.; Dean, C.; Ding, Y. In vivo single-molecule analysis reveals COOLAIR RNA structural diversity. Nature 2022, 609, 394–399. [Google Scholar] [CrossRef]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to study RNA-protein interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- McHugh, C.A.; Guttman, M. RAP-MS: A Method to Identify Proteins that Interact Directly with a Specific RNA Molecule in Cells. Methods Mol. Biol. 2018, 1649, 473–488. [Google Scholar] [PubMed]
- Zeng, F.; Peritz, T.; Kannanayakal, T.J.; Kilk, K.; Eiríksdóttir, E.; Langel, U.; Eberwine, J. A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat. Protoc. 2006, 1, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Tsai, B.P.; Wang, X.; Huang, L.; Waterman, M.L. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteom. 2011, 10, M110.007385. [Google Scholar]
- Matia-González, A.M.; Iadevaia, V.; Gerber, A.P. A versatile tandem RNA isolation procedure to capture in vivo formed mRNA-protein complexes. Methods 2017, 118–119, 93–100. [Google Scholar] [CrossRef]
- Simon, M.D. Capture hybridization analysis of RNA targets (CHART). Curr. Protoc. Mol. Biol. 2013, 101, 21.25.1–21.25.16. [Google Scholar]
- Ramanathan, M.; Majzoub, K.; Rao, D.S.; Neela, P.H.; Zarnegar, B.J.; Mondal, S.; Roth, J.G.; Gai, H.; Kovalski, J.R.; Siprashvili, Z.; et al. RNA–protein interaction detection in living cells. Nat. Methods 2018, 15, 207–212. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Peritz, T.; Zeng, F.; Kannanayakal, T.J.; Kilk, K.; Eiríksdóttir, E.; Langel, U.; Eberwine, J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006, 1, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Jensen, K.B.; Ruggiu, M.; Mele, A.; Ule, A.; Darnell, R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science 2003, 302, 1212–1215. [Google Scholar] [CrossRef]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Olivas, W.M. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev. RNA 2011, 2, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Danan, C.; Manickavel, S.; Hafner, M. PAR-CLIP: A Method for Transcriptome-Wide Identification of RNA Binding Protein Interaction Sites. In Post-Transcriptional Gene Regulation; Dassi, E., Ed.; Springer New York: New York, NY, USA, 2016; pp. 153–173. [Google Scholar]
- Huppertz, I.; Attig, J.; D’Ambrogio, A.; Easton, L.E.; Sibley, C.R.; Sugimoto, Y.; Tajnik, M.; König, J.; Ule, J. iCLIP: Protein–RNA interactions at nucleotide resolution. Methods 2014, 65, 274–287. [Google Scholar] [CrossRef]
- Zarnegar, B.J.; Flynn, R.A.; Shen, Y.; Do, B.T.; Chang, H.Y.; Khavari, P.A. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 2016, 13, 489–492. [Google Scholar] [CrossRef]
- Gu, J.; Wang, M.; Yang, Y.; Qiu, D.; Zhang, Y.; Ma, J.; Zhou, Y.; Hannon, G.J.; Yu, Y. GoldCLIP: Gel-omitted Ligation-dependent CLIP. Genom. Proteom. Bioinform. 2018, 16, 136–143. [Google Scholar] [CrossRef]
- Kim, B.; Kim, V.N. fCLIP-seq for transcriptomic footprinting of dsRNA-binding proteins: Lessons from DROSHA. Methods 2019, 152, 3–11. [Google Scholar] [CrossRef]
- Weyn-Vanhentenryck, S.M.; Mele, A.; Yan, Q.; Sun, S.; Farny, N.; Zhang, Z.; Xue, C.; Herre, M.; Silver, P.A.; Zhang, M.Q.; et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014, 6, 1139–1152. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef]
- Trendel, J.; Schwarzl, T.; Horos, R.; Prakash, A.; Bateman, A.; Hentze, M.W.; Krijgsveld, J. The Human RNA-Binding Proteome and Its Dynamics during Translational Arrest. Cell 2019, 176, 391–403.e19. [Google Scholar] [CrossRef] [PubMed]
- Tang, L. Examining global RNA-binding proteomes. Nat. Methods 2019, 16, 144. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, R.M.L.; Smith, T.; Villanueva, E.; Marti-Solano, M.; Monti, M.; Pizzinga, M.; Mirea, D.M.; Ramakrishna, M.; Harvey, R.F.; Dezi, V.; et al. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 2019, 37, 169–178. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Lino Cardenas, C.L.; Kessinger, C.W.; Cheng, Y.; MacDonald, C.; MacGillivray, T.; Ghoshhajra, B.; Huleihel, L.; Nuri, S.; Yeri, A.S.; Jaffer, F.A.; et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun. 2018, 9, 1009. [Google Scholar] [CrossRef]
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr. Biol. 2017, 27, 602–609. [Google Scholar] [CrossRef]
- Bell, J.C.; Jukam, D.; Teran, N.A.; Risca, V.I.; Smith, O.K.; Johnson, W.L.; Skotheim, J.M.; Greenleaf, W.J.; Straight, A.F. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 2018, 7, e27024. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, X.; Luo, D.; Lim, D.H.; Zhou, Y.; Fu, X.D. GRID-seq for comprehensive analysis of global RNA-chromatin interactions. Nat. Protoc. 2019, 14, 2036–2068. [Google Scholar] [CrossRef]
- Bonetti, A.; Agostini, F.; Suzuki, A.M.; Hashimoto, K.; Pascarella, G.; Gimenez, J.; Roos, L.; Nash, A.J.; Ghilotti, M.; Cameron, C.J.F.; et al. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat. Commun. 2020, 11, 1018. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Sultanov, R.I.; Magnitov, M.D.; Galitsyna, A.A.; Dashinimaev, E.B.; Lieberman Aiden, E.; Razin, S.V. RedChIP identifies noncoding RNAs associated with genomic sites occupied by Polycomb and CTCF proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2116222119. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Huang, X.; Zhang, Y.; Xu, W.; Yang, Y.; Zhang, Q.; Hu, Z.; Xing, F.; Sun, Q.; Li, G.; et al. The landscape of promoter-centred RNA-DNA interactions in rice. Nat. Plants 2022, 8, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790.e30. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Bhat, P.; Chovanec, P.; Jachowicz, J.W.; Ollikainen, N.; Detmar, E.; Soehalim, E.; Guttman, M. SPRITE: A genome-wide method for mapping higher-order 3D interactions in the nucleus using combinatorial split-and-pool barcoding. Nat. Protoc. 2022, 17, 36–75. [Google Scholar] [CrossRef]
- Gavrilov, A.A.; Zharikova, A.A.; Galitsyna, A.A.; Luzhin, A.V.; Rubanova, N.M.; Golov, A.K.; Petrova, N.V.; Logacheva, M.D.; Kantidze, O.L.; Ulianov, S.V.; et al. Studying RNA-DNA interactome by Red-C identifies noncoding RNAs associated with various chromatin types and reveals transcription dynamics. Nucleic Acids Res. 2020, 48, 6699–6714. [Google Scholar] [CrossRef]
- Shishkin, A.A.; Giannoukos, G.; Kucukural, A.; Ciulla, D.; Busby, M.; Surka, C.; Chen, J.; Bhattacharyya, R.P.; Rudy, R.F.; Patel, M.M.; et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat. Methods 2015, 12, 323–325. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef]
- Cao, C.; Cai, Z.; Ye, R.; Su, R.; Hu, N.; Zhao, H.; Xue, Y. Global in situ profiling of RNA-RNA spatial interactions with RIC-seq. Nat. Protoc. 2021, 16, 2916–2946. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Levin, M.; Butter, F.; Scheibe, M. Quantitative Proteomics to Identify Nuclear RNA-Binding Proteins of Malat1. Int. J. Mol. Sci. 2020, 21, 1166. [Google Scholar] [CrossRef]
- Kim, J.; Piao, H.-L.; Kim, B.-J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef]
- Tao, X.; Feng, S.; Zhao, T.; Guan, X. Efficient chromatin profiling of H3K4me3 modification in cotton using CUT&Tag. Plant Methods 2020, 16, 120. [Google Scholar]
- Yan, F.; Powell, D.R.; Curtis, D.J.; Wong, N.C. From reads to insight: A hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 2020, 21, 22. [Google Scholar] [CrossRef]
- Nakato, R.; Sakata, T. Methods for ChIP-seq analysis: A practical workflow and advanced applications. Methods 2021, 187, 44–53. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, D.Y.; Greenblatt, J.F.; Zhang, Z. RIPSeeker: A statistical package for identifying protein-associated transcripts from RIP-seq experiments. Nucleic Acids Res. 2013, 41, e94. [Google Scholar] [CrossRef] [PubMed]
- Kucukural, A.; Özadam, H.; Singh, G.; Moore, M.J.; Cenik, C. ASPeak: An abundance sensitive peak detection algorithm for RIP-Seq. Bioinformatics 2013, 29, 2485–2486. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yan, Z.; Nguyen, T.C.; Bouman Chen, Z.; Chien, S.; Zhong, S. Mapping RNA–chromatin interactions by sequencing with iMARGI. Nat. Protoc. 2019, 14, 3243–3272. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.; Forcato, M.; Ferrari, F. Hi-C analysis: From data generation to integration. Biophys. Rev. 2019, 11, 67–78. [Google Scholar] [CrossRef]
- Uhl, M.; Houwaart, T.; Corrado, G.; Wright, P.R.; Backofen, R. Computational analysis of CLIP-seq data. Methods 2017, 118–119, 60–72. [Google Scholar] [CrossRef]
- Holgersen, E.M.; Gillespie, A.; Leavy, O.C.; Baxter, J.S.; Zvereva, A.; Muirhead, G.; Johnson, N.; Sipos, O.; Dryden, N.H.; Broome, L.R.; et al. Identifying high-confidence capture Hi-C interactions using CHiCANE. Nat. Protoc. 2021, 16, 2257–2285. [Google Scholar] [CrossRef]
- Ramani, V.; Deng, X.; Qiu, R.; Gunderson, K.L.; Steemers, F.J.; Disteche, C.M.; Noble, W.S.; Duan, Z.; Shendure, J. Massively multiplex single-cell Hi-C. Nat. Methods 2017, 14, 263–266. [Google Scholar] [CrossRef]
- de Goede, O.M.; Nachun, D.C.; Ferraro, N.M.; Gloudemans, M.J.; Rao, A.S.; Smail, C.; Eulalio, T.Y.; Aguet, F.; Ng, B.; Xu, J.; et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 2021, 184, 2633–2648.e19. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Lagarde, J.; Uszczynska-Ratajczak, B.; Carbonell, S.; Perez-Lluch, S.; Abad, A.; Davis, C.; Gingeras, T.R.; Frankish, A.; Harrow, J.; Guigo, R.; et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017, 49, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Hagemann-Jensen, M.; Ziegenhain, C.; Chen, P.; Ramskold, D.; Hendriks, G.J.; Larsson, A.J.M.; Faridani, O.R.; Sandberg, R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020, 38, 708–714. [Google Scholar] [CrossRef]
- Gupta, I.; Collier, P.G.; Haase, B.; Mahfouz, A.; Joglekar, A.; Floyd, T.; Koopmans, F.; Barres, B.; Smit, A.B.; Sloan, S.A.; et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 2018, 36, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Philpott, M.; Watson, J.; Thakurta, A.; Brown, T.; Brown, T.; Oppermann, U.; Cribbs, A.P. Nanopore sequencing of single-cell transcriptomes with scCOLOR-seq. Nat. Biotechnol. 2021, 39, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.; Olsson, T.S.G.; Hartley, M.; Dean, C.; Rosa, S. Single Molecule RNA FISH in Arabidopsis Root Cells. Bio-Protocol 2017, 7, e2240. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, C.; Zhang, S.; Yan, H.; Zhang, L.; Jiang, A.; Liu, Y.; Feng, Y.; Li, D.; Guo, Y.; et al. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 2021, 56, 702–715.e8. [Google Scholar] [CrossRef]
- Rosa, S.; Duncan, S.; Dean, C. Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 2016, 7, 13031. [Google Scholar] [CrossRef]
- Marin-Bejar, O.; Mas, A.M.; Gonzalez, J.; Martinez, D.; Athie, A.; Morales, X.; Galduroz, M.; Raimondi, I.; Grossi, E.; Guo, S.; et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017, 18, 202. [Google Scholar] [CrossRef]
- Fanucchi, S.; Fok, E.T.; Dalla, E.; Shibayama, Y.; Borner, K.; Chang, E.Y.; Stoychev, S.; Imakaev, M.; Grimm, D.; Wang, K.C.; et al. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 2019, 51, 138–150. [Google Scholar] [CrossRef]
- Patel, H.P.; Brouwer, I.; Lenstra, T.L. Optimized protocol for single-molecule RNA FISH to visualize gene expression in S. cerevisiae. STAR Protoc. 2021, 2, 100647. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.A.; Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019, 14, 1734–1755. [Google Scholar] [CrossRef]
- Torres, M.; Becquet, D.; Guillen, S.; Boyer, B.; Moreno, M.; Blanchard, M.P.; Franc, J.L.; François-Bellan, A.M. RNA Pull-down Procedure to Identify RNA Targets of a Long Non-coding RNA. J. Vis. Exp. 2018, 57379. [Google Scholar]
- Chu, C.; Quinn, J.; Chang, H.Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012, 61, e3912. [Google Scholar]
- Hagège, H.; Klous, P.; Braem, C.; Splinter, E.; Dekker, J.; Cathala, G.; de Laat, W.; Forné, T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007, 2, 1722–1733. [Google Scholar] [CrossRef]
- Li, X.; Luo, O.J.; Wang, P.; Zheng, M.; Wang, D.; Piecuch, E.; Zhu, J.J.; Tian, S.Z.; Tang, Z.; Li, G.; et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat. Protoc. 2017, 12, 899–915. [Google Scholar] [CrossRef]
- Lafontaine, D.L.; Yang, L.; Dekker, J.; Gibcus, J.H. Hi-C 3.0: Improved Protocol for Genome-Wide Chromosome Conformation Capture. Curr. Protoc. 2021, 1, e198. [Google Scholar] [CrossRef] [PubMed]
- Jäger, R.; Migliorini, G.; Henrion, M.; Kandaswamy, R.; Speedy, H.E.; Heindl, A.; Whiffin, N.; Carnicer, M.J.; Broome, L.; Dryden, N.; et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat. Commun. 2015, 6, 6178. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yao, T.; Yang, W.; Wu, P.; Liu, Y.; Yang, B. Protocol to detect nucleotide-protein interaction in vitro using a non-radioactive competitive electrophoretic mobility shift assay. STAR Protoc. 2022, 3, 101730. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jin, S.; Kim, S.Y.; Kim, W.; Ahn, J.H. A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods 2017, 13, 42. [Google Scholar] [CrossRef]
- Zhao, L.; Xie, L.; Zhang, Q.; Ouyang, W.; Deng, L.; Guan, P.; Ma, M.; Li, Y.; Zhang, Y.; Xiao, Q.; et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 2020, 11, 2658. [Google Scholar] [CrossRef]
- Sullivan, A.E.; Santos, S.D.M. An Optimized Protocol for ChIP-Seq from Human Embryonic Stem Cell Cultures. STAR Protoc. 2020, 1, 100062. [Google Scholar] [CrossRef]
- Kaya-Okur, H.S.; Janssens, D.H.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 2020, 15, 3264–3283. [Google Scholar]
- Zhang, B.; Srivastava, A.; Mimitou, E.; Stuart, T.; Raimondi, I.; Hao, Y.; Smibert, P.; Satija, R. Characterizing cellular heterogeneity in chromatin state with scCUT&Tag-pro. Nat. Biotechnol. 2022, 40, 1220–1230. [Google Scholar]
- Jegu, T.; Blum, R.; Cochrane, J.C.; Yang, L.; Wang, C.Y.; Gilles, M.E.; Colognori, D.; Szanto, A.; Marr, S.K.; Kingston, R.E.; et al. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 2019, 26, 96–109. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.D.; Meyer, M.J.; Abramzon, V.; Ad, O.; Adcock, P.; Ahmad, F.R.; Alppay, G.; Ball, J.A.; Beach, J.; Belhachemi, D.; et al. Real-time dynamic single-molecule protein sequencing on an integrated semiconductor device. Science 2022, 378, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Özcan, A.; Krajeski, R.; Ioannidi, E.; Lee, B.; Gardner, A.; Makarova, K.S.; Koonin, E.V.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 2021, 597, 720–725. [Google Scholar] [CrossRef]
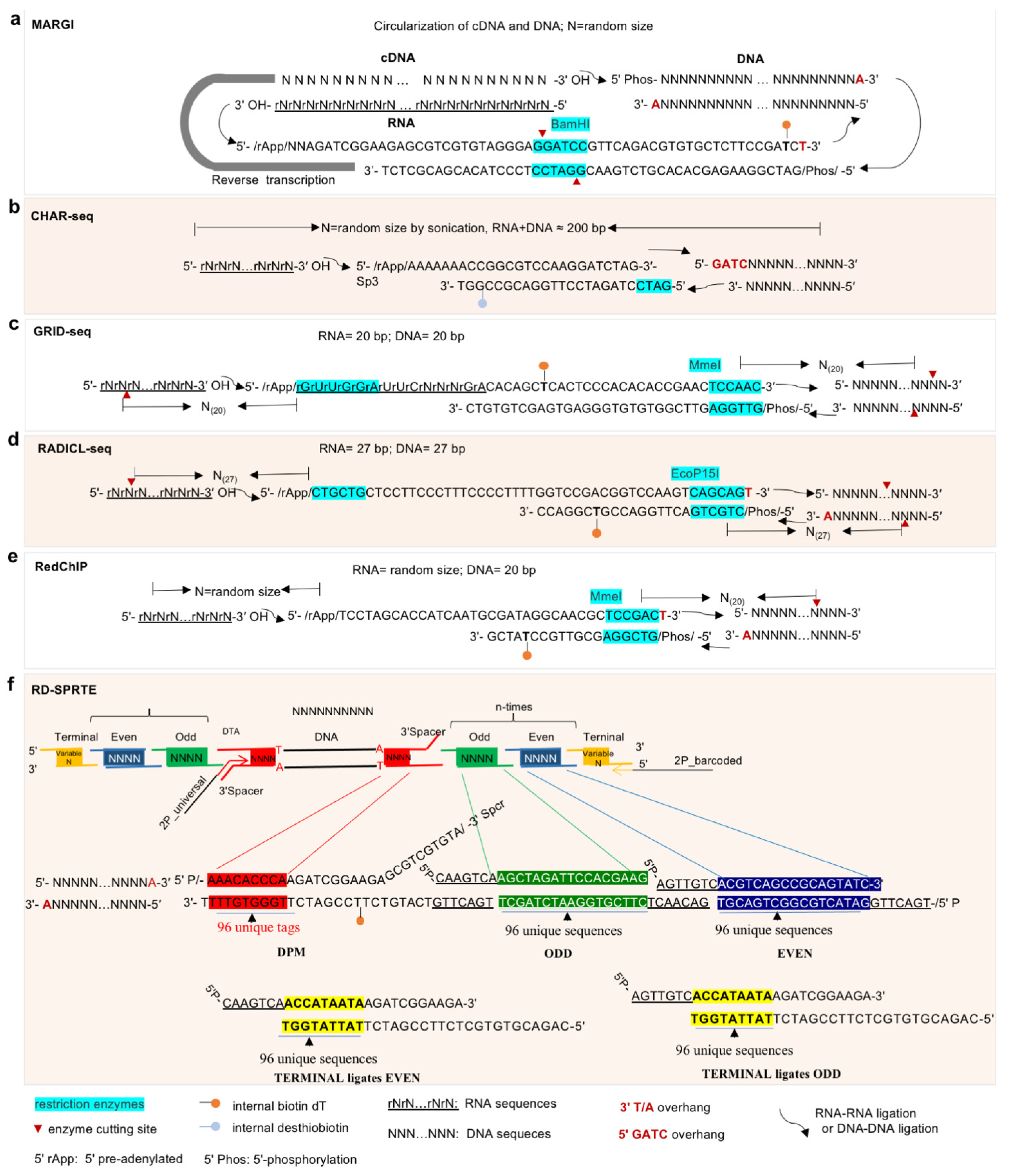
| MARGI | CHAR-Seq | GRID-Seq | RADICL-Seq | RedChIP | ChRD-PET | SPRITE | |
|---|---|---|---|---|---|---|---|
| Purpose | RNA–DNA interaction | RNA–DNA–protein interaction | RNA–DNA and DNA–DNA interactions | ||||
| Mechanism | RNA and DNA proximity ligation | Combination of RNA and DNA proximity ligation and ChIP | Split-and-pool barcoding ligation | ||||
| Applied organism/cells | Mammalian cells | Drosophila cells | Mammalian cells | Mammalian cells | Mammalian cells | Leaves of rice seedlings | Mammalian cells |
| Crosslinking | 1% FA; or FA + DSG | 1% FA | DSG + 3% FA | 1% or 2% FA | 1% FA | 1% FA | DSG +3% FA |
| Nuclei isolation condition | NP-40 and SDS | Igepal, SDS, and Triton X-100 | SDS | NP-40 | NP40, SDS, and Triton X-100 | Triton X-100 and SDS | Triton X-100 and NP-40 |
| Chromatin fragmentation | Sonication or HaeIII | DpnII | AluI | DNase I | NlaIII | Sonication | Sonication and DNase |
| Bridge linker | See Figure 3 | ||||||
| Reduction in nascent transcription | N/A | N/A | N/A | RNase H | N/A | RNase H | N/A |
| Carrier between enzymatic steps | N/A | N/A | N/A | AMPure XP magnetic beads | N/A | Protein G magnetic beads | NHS-activated magnetic beads |
| Enzymes in ligation | T4 RNA Ligase 2, truncated KQ and T4 DNA ligase | T4 RNA Ligase 1 and T4 DNA ligase | |||||
| Length of RNA and DNA pairs | Random size (~150 bp) | Random size (RNA + DNA ≈ 200 bp) | RNA = 20 bp; DNA = 20 bp | RNA = 27 bp; DNA = 27 bp | RNA: random size; DNA = 20 bp | Random size (~150 bp) | Random size (280 bp–1.3 kb) |
| Depth and sensitivity | 105 million unique mapped read pairs corresponding to 2864 non-coding pxRNAs (proximal interaction) and 747 non-coding diRNAs (direct interaction) | 22.2 million unique mapped read pairs corresponding to ~16,800 RNA transcripts | ~40 million unique mapped read pairs corresponding to 868 mRNAs and 72 ncRNAs | ~8.4 million unique mapped read pairs corresponding to 288,065 RNA–DNA interacting loci and 14,001 transcripts | 18 ncRNAs specificly for CTCF and EZH2 proteins | ~12.5 million unique mapped read pairs corresponding to 68,758 RNA–DNA interaction clusters | 8 billion reads corresponding to 720 billion SPRITE clusters and ~650 lncRNAs |
| References | [90] | [91] | [92,93] | [94] | [95] | [96] | [97,98,99] |
| Technology | Software/Code Used | References |
|---|---|---|
| CLIP or CLIP-based | Trimmomatic, TopHat, Piranha, PARalyzer, CLIPper, or Block-based peak calling | [117] |
| RNA immunoprecipitation (RIP); RIP-seq | RIPSeeker; ASPeak | [113,114] |
| Chromosome confromation capture assay (3C)-based techniques (3C-qPCR; ChIA-PET; Hi-C; Capture Hi-C) | Myriad tools including compartments caller, TAD callers, and interaction callers, along with visualization tools as summarized by Pal et al., 2019; CHiCANE toolkit | Capture Hi-C [118]; single cell Hi-C [116,119] |
| RIC-seq | FastQC, Trimmomatic, cutadapt, STAR; in-house script: remove_PCR_duplicates.pl, collect_pair_tags.pl, collect_pair_tags.pl, separate_intra_inter.pl, category_intra_reads.pl, cluster_intra_reads.pl, MonteCarlo_simulation.pl (GitHub: https://github.com/caochch/RICpipe; accessed on 21 May 2021) | [106] |
| ChIRP-seq | Bowtie, macs2 | [34] |
| ChIP-seq; CUT&Tag; ATAC-seq | Fastp, hisat2, picard, macs2, deepTools, ChIPseeker | [110,111,112] |
| MARGI; iMARGI | iMARGI-Docker | [115] |
| CHAR-seq | FlyPipe (https://github.com/straightlab/flypipe; accessed on 20 February 2019), Super Deduper, Trimmomatic, bowtie2, SAMtools, BEDtools, MACS2, Circos, R, GraphPad Prism v7.0, deepTools2, HOMER, Geneious | [91] |
| GRID-seq | Cutadapt, bwa, samtools, GridTools.py, bgzip, tabix, Cytoscape | [93] |
| RADICL-seq | TagDust2, TagDust, RNAdust, FastUniq, BWA, samtools, bedtools, CAGEr package, ScoreMatrixBin package | [94] |
| RedChIP | RedClib (GitHub: https://github.com/agalitsyna/RedClib; accessed on 9 July 2020) | [100] |
| ChRD-PET | FastQC, Cutadapt, flash, BWA-MEM, BWA-ALN, HIAST2, BOWTIE2, BEDTools, HTSeq, MACS2, deepTools, plotBedpe function in the Sushi package in R, Seqtk, ggplot2 package, ggtern package | [96] |
| SPRITE | Conda, Snakemake, astq2json.py, config.yaml, Trim galore!, Cutadapt, Bowtie2, Bedtools, Multiqc, Samtools, Pigz, Fastqc, Python packages (Pysam, Numpy, R packages, Ggplot2, Gplots, Readr, Optparse); SPRITE pipeline (https://github.com/GuttmanLab/sprite-pipeline/wiki; accessed on 10 January 2022) | [99] |
| Technology | Aim | Example lncRNAs (Refs) | Recommended Protocols |
|---|---|---|---|
| smRNA FISH | Visualization and localization of lncRNA | COOLAIR [130]; TINCR [13]; ANRIL [83]; LINC-PINT [131]; UMLILO [132] | Arabidopsis [128]; Yeast [133] |
| CLIP or CLIP-based | Protein-centered method to identify specific RBP-associated lncRNAs | ANRIL [83]; LINC-PINT [131] | eCLIP [35] |
| RNA immunoprecipitation (RIP); RIP-seq | Protein-centered method to identify specific RBP-associated lncRNA | DINO [134]; TINCR [13]; ANRIL [83]; UMLILO [132]; COLDAIR [26]; COLDWRAP [27]; LINC-PINT [131]; MEG3 [33] | [135] |
| DNA-RNA duplex immunopurification; DRIPc-seq | Immunopurification detection of R-loop | APOLO [136] | [137] |
| RNA pull-down | LncRNA-centered method to identify specific lncRNA-associated RNAs (pull down-PCR/pull down-seq) or proteins (pull down-WB/ pull down-MS) | lncRNA-PNUTS [105]; TINCR [13]; ANRIL [83]; COLDWRAP [27]; LINC-PINT [131] | [138] |
| ChIRP | Analysis of lncRNA chromatin targets (ChIRP-seq or ChIRP–qPCR) or protein interactors (ChIRP-MS) | roX2 [34]; TERC [34]; HOTAIR [34]; COOLAIR [60]; APOLO [136]; MEG3 [33]; Xist [7]; DINO [134]; UMLILO [132]; | protocol with video [139] |
| Chromosome conformation capture assay (3C)-based techniques | 3D genome architecture | APOLO [136]; UMLILO [132] | 3C-qPCR [140]; ChIA-PET [141]; Hi-C [142]; Capture Hi-C [143]; Single cell Hi-C [144] |
| Electrophoretic mobility shift assay (EMSA) | In vitro detection of lncRNA–DNA triplex structures and lncRNA–protein binding | MEG3 [33]; ANRIL [83] | [145] |
| ChIP | Immunopurification detection of DNA targets of lncRNA protein interactors (e.g., TFs and polycomb group proteins) and profiling of histone modifications | COOLAIR [60]; APOLO [136]; MEG3 [33]; ANRIL [83]; UMLILO [132]; LINC-PINT [131] | Plant cells [146]; eChIP-Seq [147]; Animal cells [148] |
| CUT&Tag | Enzyme-tethered method to analyze DNA targets of proteins and histone modifications with low input | MALAT1 [129] | Animal cells [149]; Plant cells [110]; scCUT&Tag [150] |
| ATAC-seq | Chromatin accessibility analysis | DINO [134]; Xist [151] | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, X.; Li, S.; Chen, G.; Wang, J.; Xu, S. Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination. Int. J. Mol. Sci. 2023, 24, 5562. https://doi.org/10.3390/ijms24065562
Tao X, Li S, Chen G, Wang J, Xu S. Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination. International Journal of Molecular Sciences. 2023; 24(6):5562. https://doi.org/10.3390/ijms24065562
Chicago/Turabian StyleTao, Xiaoyuan, Sujuan Li, Guang Chen, Jian Wang, and Shengchun Xu. 2023. "Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination" International Journal of Molecular Sciences 24, no. 6: 5562. https://doi.org/10.3390/ijms24065562
APA StyleTao, X., Li, S., Chen, G., Wang, J., & Xu, S. (2023). Approaches for Modes of Action Study of Long Non-Coding RNAs: From Single Verification to Genome-Wide Determination. International Journal of Molecular Sciences, 24(6), 5562. https://doi.org/10.3390/ijms24065562





