Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS
Abstract
1. Introduction
2. Results
2.1. Higher Sequencing Depth Detects a Larger Diversity of the Library
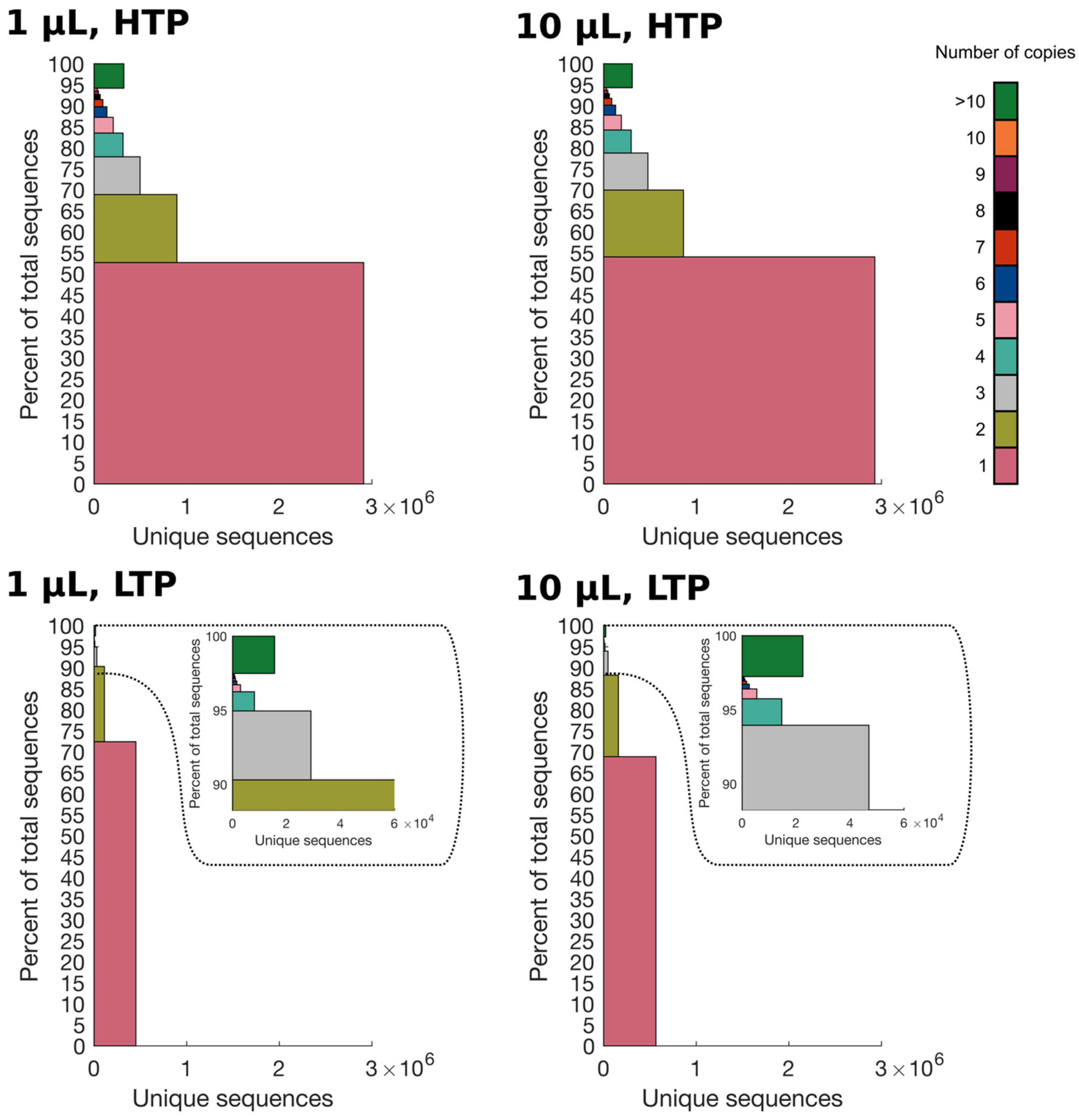
2.2. Higher Sequencing Depth Reveals a Smaller Percentage of Singletons in the Library
2.3. Higher Sequencing Depth Uncovers a More Heterogeneous Distribution of Absolute Frequencies
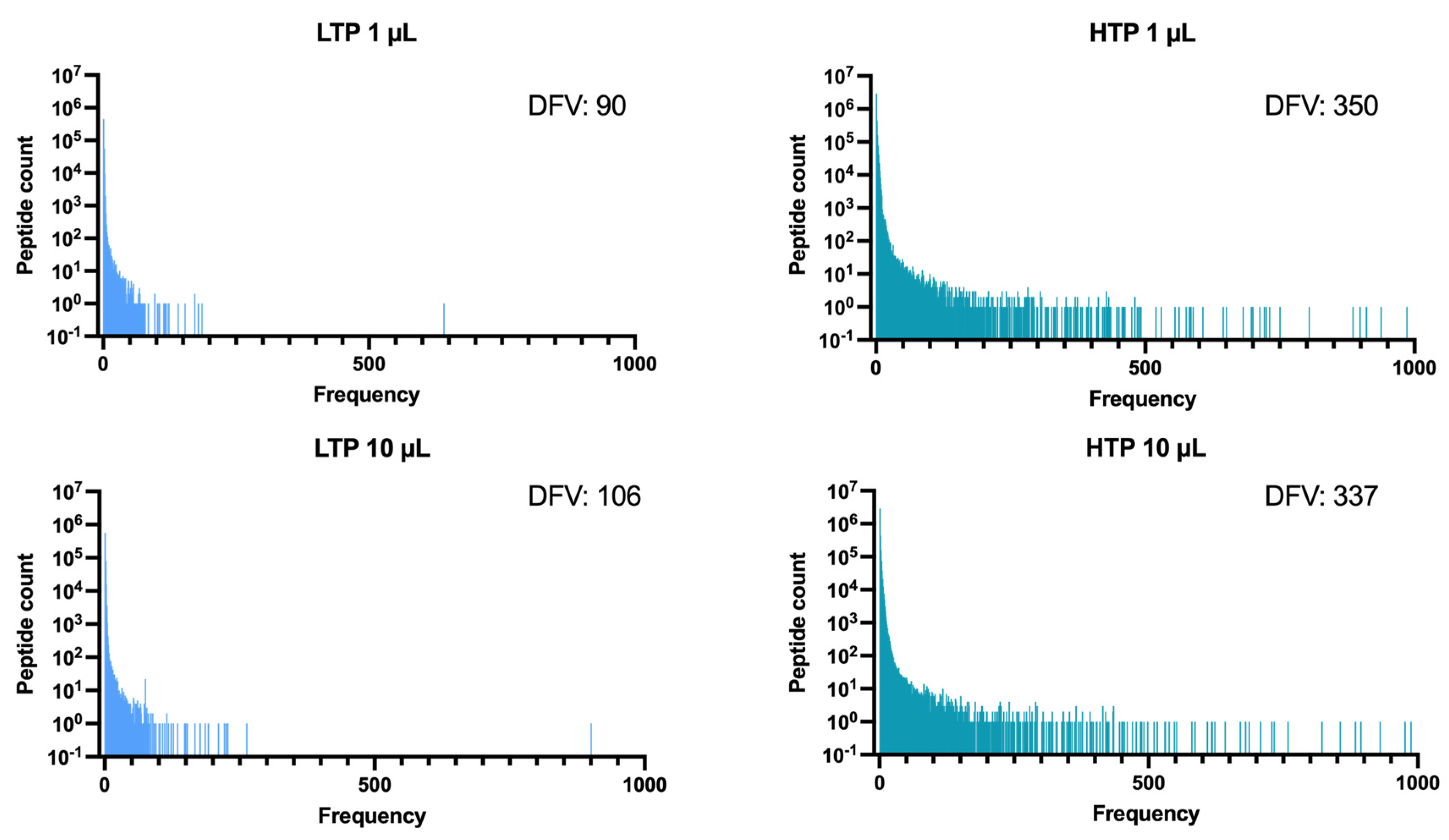
2.4. Higher Sequencing Depth Can Provide a Comparatively Higher Distinguishing Capacity for the Detection of Peptide Frequencies
2.5. The Peptide Composition of Datasets Shows Vast Dissimilarities for Different Sequencing Depths and Volumes
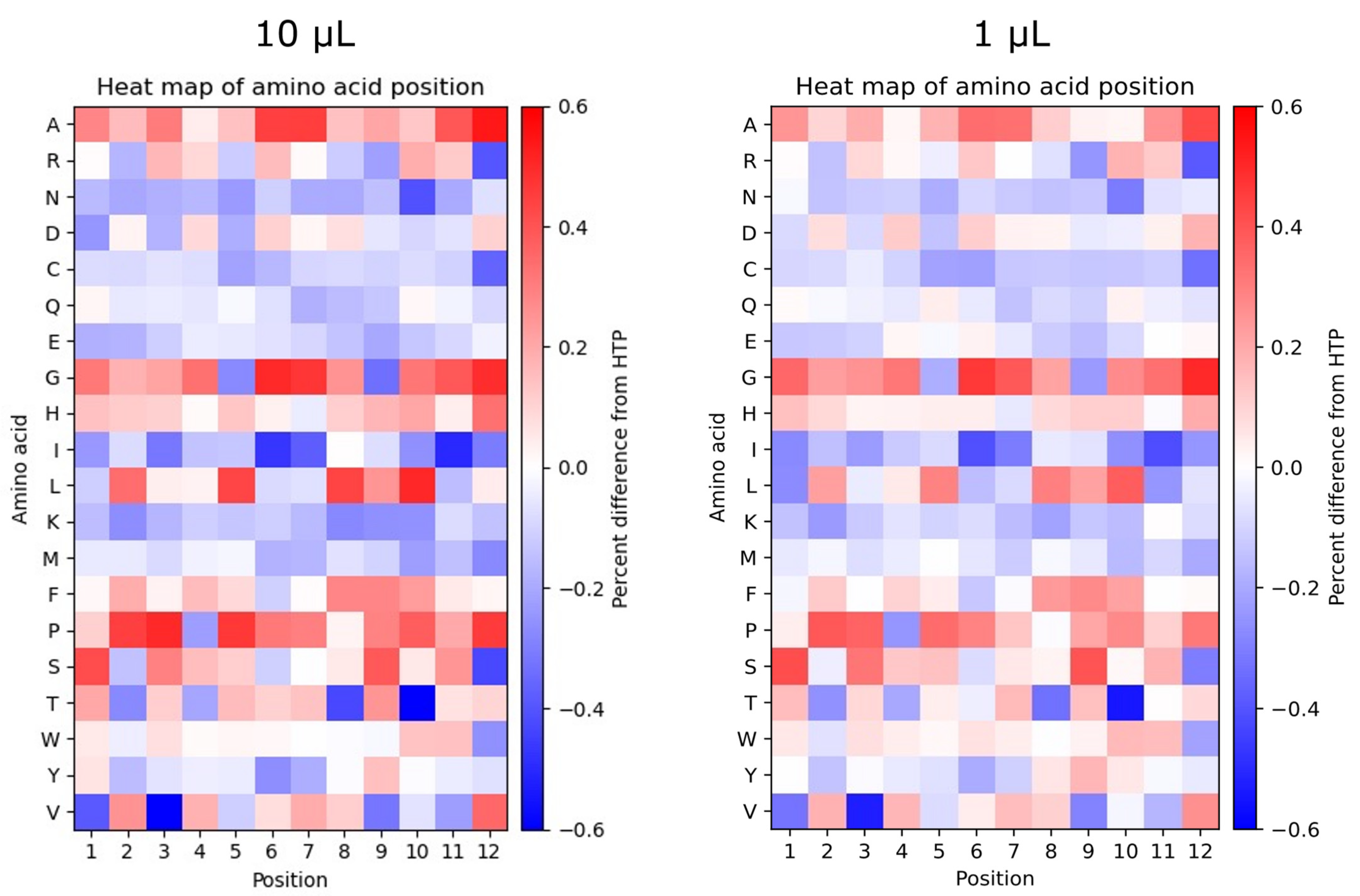
2.6. Different Sequencing Depths Show Discrepancies in the Position-Specific Frequency of Amino Acids
3. Discussion
4. Materials and Methods
4.1. DNA Isolation from Phage Display Peptide Library
4.2. Illumina Sequencing of the Phage Display Peptide Library
4.3. Analysis of Illumina Sequencing Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Mimmi, S.; Maisano, D.; Quinto, I.; Iaccino, E. Phage display: An overview in context to drug discovery. Trends Pharmacol. Sci. 2019, 40, 87–91. [Google Scholar] [CrossRef]
- Newman, M.R.; Benoit, D.S.W. In Vivo Translation of Peptide-Targeted Drug Delivery Systems Discovered by Phage Display. Bioconj. Chem. 2018, 29, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Cochran, F.V.; Cochran, J.R. Phage display and molecular imaging: Expanding fields of vision in living subjects. Biotechnol. Genet. Eng. Rev. 2010, 27, 57–94. [Google Scholar] [CrossRef]
- Deutscher, S.L. Phage Display in Molecular Imaging and Diagnosis of Cancer. Chem. Rev. 2010, 110, 3196–3211. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-B.; Niu, G.; Wang, H.; He, L.; Yang, L.; Ploug, M.; Chen, X. Imaging of urokinase-type plasminogen activator receptor expression using a 64Cu-labeled linear peptide antagonist by microPET. Clin. Cancer Res. 2008, 14, 4758–4766. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Lo, S.L.; Boulaire, J.; Hong, M.L.W.; Beh, H.M.; Leung, D.S.Y.; Wang, S. A peptide-based carrier for intracellular delivery of proteins into malignant glial cells in vitro. J. Control. Release 2008, 130, 140–145. [Google Scholar] [CrossRef]
- Zang, L.; Shi, L.; Guo, J.; Pan, Q.; Wu, W.; Pan, X.; Wang, J. Screening and identification of a peptide specifically targeted to NCI-H1299 from a phage display peptide library. Cancer Lett. 2009, 281, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.R.; Kelly, K.A.; Mahmood, U.; Weissleder, R.; Deutscher, S.L. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia 2006, 8, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Arap, W.; Pasqualini, R.; Ruoslahti, E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science 1998, 279, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Phage Display: Simple Evolution in a Petri Dish (Nobel Lecture). Angew. Chem. Int. Ed. 2019, 58, 14428–14437. [Google Scholar] [CrossRef] [PubMed]
- Dias-Neto, E.; Nunes, D.N.; Giordano, R.J.; Sun, J.; Botz, G.H.; Yang, K.; Setubal, J.C.; Pasqualini, R.; Arap, W. Next-generation phage display: Integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS ONE 2009, 4, e8338. [Google Scholar] [CrossRef]
- AC’t Hoen, P.; Jirka, S.M.; Bradley, R.; Schultes, E.A.; Aguilera, B.; Pang, K.H.; Heemskerk, H.; Aartsma-Rus, A.; van Ommen, G.J.; den Dunnen, J.T. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012, 421, 622–631. [Google Scholar] [CrossRef]
- Matochko, W.L.; Chu, K.; Jin, B.; Lee, S.W.; Whitesides, G.M.; Derda, R. Deep sequencing analysis of phage libraries using Illumina platform. Methods 2012, 58, 47–55. [Google Scholar] [CrossRef]
- Stellwagen, S.D.; Sarkes, D.A.; Adams, B.L.; Hunt, M.A.; Renberg, R.L.; Hurley, M.M.; Stratis-Cullum, D.N. The next generation of biopanning: Next gen sequencing improves analysis of bacterial display libraries. BMC Biotechnol. 2019, 19, 100. [Google Scholar] [CrossRef]
- Rentero Rebollo, I.; Sabisz, M.; Baeriswyl, V.; Heinis, C. Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Nucleic Acids Res. 2014, 42, e169. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, A.M.; Huang, W.; Estes, M.K.; Atmar, R.L.; Palzkill, T. Deep sequencing of phage-displayed peptide libraries reveals sequence motif that detects norovirus. Protein Eng. Des. Sel. 2017, 30, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.W.; Livesay, B.R.; Kacherovsky, N.A.; Cieslewicz, M.; Lutz, E.; Waalkes, A.; Jensen, M.C.; Salipante, S.J.; Pun, S.H. Efficient Identification of Murine M2 Macrophage Peptide Targeting Ligands by Phage Display and Next-Generation Sequencing. Bioconj. Chem. 2015, 26, 1811–1817. [Google Scholar] [CrossRef]
- Matochko, W.L.; Cory Li, S.; Tang, S.K.; Derda, R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Res. 2014, 42, 1784–1798. [Google Scholar] [CrossRef]
- Sloth, A.B.; Bakhshinejad, B.; Jensen, M.; Stavnsbjerg, C.; Liisberg, M.B.; Rossing, M.; Kjaer, A. Analysis of Compositional Bias in a Commercial Phage Display Peptide Library by Next-Generation Sequencing. Viruses 2022, 14, 2402. [Google Scholar] [CrossRef]
- He, B.; Tjhung, K.F.; Bennett, N.J.; Chou, Y.; Rau, A.; Huang, J.; Derda, R. Compositional Bias in Naïve and Chemically-modified Phage-Displayed Libraries uncovered by Paired-end Deep Sequencing. Sci. Rep. 2018, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Alekseyev, Y.O.; Fazeli, R.; Yang, S.; Basran, R.; Maher, T.; Miller, N.S.; Remick, D. A Next-Generation Sequencing Primer—How Does It Work and What Can It Do? Acad. Pathol. 2018, 5, 2374289518766521. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Rodi, D.J.; Soares, A.S.; Makowski, L. Quantitative assessment of peptide sequence diversity in M13 combinatorial peptide phage display libraries. J. Mol. Biol. 2002, 322, 1039–1052. [Google Scholar] [CrossRef]
- Fagerlund, A.; Myrset, A.H.; Kulseth, M.A. Construction and characterization of a 9-mer phage display pVIII-library with regulated peptide density. Appl. Microbiol. Biotechnol. 2008, 80, 925–936. [Google Scholar] [CrossRef]
- Glanville, J.; Zhai, W.; Berka, J.; Telman, D.; Huerta, G.; Mehta, G.R.; Ni, I.; Mei, L.; Sundar, P.D.; Day, G.M. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc. Natl. Acad. Sci. USA 2009, 106, 20216–20221. [Google Scholar] [CrossRef] [PubMed]
- Lindner, T.; Kolmar, H.; Haberkorn, U.; Mier, W. DNA libraries for the construction of phage libraries: Statistical and structural requirements and synthetic methods. Molecules 2011, 16, 1625–1641. [Google Scholar] [CrossRef]
- Tsoumpeli, M.T.; Gray, A.; Parsons, A.L.; Spiliotopoulos, A.; Owen, J.P.; Bishop, K.; Maddison, B.C.; Gough, K.C. A Simple Whole-Plasmid PCR Method to Construct High-Diversity Synthetic Phage Display Libraries. Mol. Biotechnol. 2022, 64, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Naqid, I.A.; Owen, J.P.; Maddison, B.C.; Spiliotopoulos, A.; Emes, R.D.; Warry, A.; Tchórzewska, M.A.; Martelli, F.; Gosling, R.J.; Davies, R.H. Mapping polyclonal antibody responses to bacterial infection using next generation phage display. Sci. Rep. 2016, 6, 24232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, S.; Leung, K.K.; O’Donovan, B.; Zou, J.Y.; DeRisi, J.L.; Wells, J.A. Deep profiling of protease substrate specificity enabled by dual random and scanned human proteome substrate phage libraries. Proc. Natl. Acad. Sci. USA 2020, 117, 25464–25475. [Google Scholar] [CrossRef]
- Christiansen, A.; Kringelum, J.V.; Hansen, C.S.; Bøgh, K.L.; Sullivan, E.; Patel, J.; Rigby, N.M.; Eiwegger, T.; Szépfalusi, Z.; de Masi, F.; et al. High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum. Sci. Rep. 2015, 5, 12913. [Google Scholar] [CrossRef] [PubMed]
- Makowski, L.; Soares, A. Estimating the diversity of peptide populations from limited sequence data. Bioinformatics 2003, 19, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Galán, A.; Comor, L.; Horvatić, A.; Kuleš, J.; Guillemin, N.; Mrljak, V.; Bhide, M. Library-based display technologies: Where do we stand? Mol. BioSyst. 2016, 12, 2342–2358. [Google Scholar] [CrossRef]
- Kuzmicheva, G.; Jayanna, P.; Sorokulova, I.; Petrenko, V. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009, 22, 9–18. [Google Scholar] [CrossRef]
- Sclavons, C.; Burtea, C.; Boutry, S.; Laurent, S.; Vander Elst, L.; Muller, R.N. Phage display screening for tumor necrosis factor-α-binding peptides: Detection of inflammation in a mouse model of hepatitis. Int. J. Pept. 2013, 2013, 348409. [Google Scholar] [CrossRef] [PubMed]
- Brinton, L.T.; Bauknight, D.K.; Dasa, S.S.K.; Kelly, K.A. PHASTpep: Analysis software for discovery of cell-selective peptides via phage display and next-generation sequencing. PLoS ONE 2016, 11, e0155244. [Google Scholar] [CrossRef]
- Kamstrup Sell, D.; Sloth, A.B.; Bakhshinejad, B.; Kjaer, A. A White Plaque, Associated with Genomic Deletion, Derived from M13KE-Based Peptide Library Is Enriched in a Target-Unrelated Manner during Phage Display Biopanning Due to Propagation Advantage. Int. J. Mol. Sci. 2022, 23, 3308. [Google Scholar] [CrossRef] [PubMed]

| Sample | Unique Cleaned Reads | Unique Removed Reads (Number and Percentage) | Total Unique Reads | Absolute Cleaned Reads | Absolute Removed Reads (Number and Percentage) | Total Absolute Reads |
|---|---|---|---|---|---|---|
| HTP 1 μL | 3.70 × 106 | 7.03 × 105 (15.95 %) | 4.41 × 106 | 5.52 × 106 | 1.49 × 106 (21.21 %) | 7.01 × 106 |
| HTP 10 μL | 3.69 × 106 | 7.13 × 105 (16.19 %) | 4.40 × 106 | 5.42 × 106 | 1.52 × 106 (21.93 %) | 6.94 × 106 |
| LTP 1 μL | 5.21 × 105 | 7.89 × 104 (13.17 %) | 5.10 × 105 | 6.23 × 105 | 1.60 × 105 (20.47 %) | 7.84 × 105 |
| LTP 10 μL | 6.66 × 105 | 1.06 × 105 (13.70 %) | 7.72 × 105 | 8.20 × 105 | 2.15 × 105 (20.80 %) | 1.04 × 106 |
| Peptide Sequence | 10 μL LTP | 10 μL HTP | ||||
|---|---|---|---|---|---|---|
| Absolute Frequency | Relative Frequency (%) | 95% CI | Absolute Frequency | Relative Frequency (%) | 95% CI | |
| TLHFKPPNVTML | 20 | 2.44 × 10−3 | [5;35] | 87 | 1.61 × 10−3 | [56;118] |
| ERSMYWEDITPM | 90 | 1.66 × 10−3 | [58;122] | |||
| ASNGTDHTRTPF | 189 | 3.49 × 10−3 | [143;235] | |||
| AHTMQQISPNHC | 85 | 1.57 × 10−3 | [54;116] | |||
| SDSLFWNMMTDV | 170 | 3.14 × 10−3 | [127;213] | |||
| AWPPTGILPMLN | 161 | 2.97 × 10−3 | [119;203] | |||
| AMDRQPTWSVAN | 25 | 3.05 × 10−3 | [8;42] | 144 | 2.66 × 10−3 | [104;184] |
| HTSPRHYSSMSA | 155 | 2.86 × 10−3 | [114;196] | |||
| NTVYAQPTGVLS | 120 | 2.21 × 10−3 | [84;156] | |||
| TAPRPQSILNGL | 128 | 2.36 × 10−3 | [90;166] | |||
| LLPTGNVLENFP | 120 | 2.21 × 10−3 | [84;156] | |||
| TPPQISTPTQPV | 30 | 3.66 × 10−3 | [12;48] | 133 | 2.45 × 10−3 | [95;171] |
| NLNDSYGLSSDR | 256 | 4.72 × 10−3 | [203;309] | |||
| NLVSTGLAYQSL | 35 | 4.27 × 10−3 | [15;55] | 163 | 3.01 × 10−3 | [121;205] |
| GNMGYMRPGHNN | 160 | 2.95 × 10−3 | [118;202] | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sloth, A.B.; Bakhshinejad, B.; Stavnsbjerg, C.; Rossing, M.; Kjaer, A. Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS. Int. J. Mol. Sci. 2023, 24, 5396. https://doi.org/10.3390/ijms24065396
Sloth AB, Bakhshinejad B, Stavnsbjerg C, Rossing M, Kjaer A. Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS. International Journal of Molecular Sciences. 2023; 24(6):5396. https://doi.org/10.3390/ijms24065396
Chicago/Turabian StyleSloth, Ane Beth, Babak Bakhshinejad, Camilla Stavnsbjerg, Maria Rossing, and Andreas Kjaer. 2023. "Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS" International Journal of Molecular Sciences 24, no. 6: 5396. https://doi.org/10.3390/ijms24065396
APA StyleSloth, A. B., Bakhshinejad, B., Stavnsbjerg, C., Rossing, M., & Kjaer, A. (2023). Depth of Sequencing Plays a Determining Role in the Characterization of Phage Display Peptide Libraries by NGS. International Journal of Molecular Sciences, 24(6), 5396. https://doi.org/10.3390/ijms24065396








