Abstract
R2R3-type MYB transcription factors are implicated in drought stress, which is a primary factor limiting the growth and development of woody plants. The identification of R2R3-MYB genes in the Populus trichocarpa genome has been previously reported. Nevertheless, the diversity and complexity of the conserved domain of the MYB gene caused inconsistencies in these identification results. There is still a lack of drought-responsive expression patterns and functional studies of R2R3-MYB transcription factors in Populus species. In this study, we identified a total of 210 R2R3-MYB genes in the P. trichocarpa genome, of which 207 genes were unevenly distributed across all 19 chromosomes. These poplar R2R3-MYB genes were phylogenetically divided into 23 subgroups. Collinear analysis demonstrated that the poplar R2R3-MYB genes underwent rapid expansion and that whole-genome duplication events were a dominant factor in the process of rapid gene expansion. Subcellular localization assays indicated that poplar R2R3-MYB TFs mainly played a transcriptional regulatory role in the nucleus. Ten R2R3-MYB genes were cloned from P. deltoides × P. euramericana cv. Nanlin895, and their expression patterns were tissue-specific. A majority of the genes showed similar drought-responsive expression patterns in two out of three tissues. This study provides a valid cue for further functional characterization of drought-responsive R2R3-MYB genes in poplar and provides support for the development of new poplar genotypes with elevated drought tolerance.
1. Introduction
Over the years, global climate change has led to an increase in the frequency and spatial extent of droughts, severely reducing the stress resistance and resilience of perennial woody plants and even jeopardizing the functioning of forest ecosystems [1,2]. Poplar is a primary tree species that is distributed widely in Northern Hemisphere regions, especially in dry northern and northwestern China [3,4]. Poplar is also a model woody plant for studying abiotic stress response mechanisms, owing to its characteristics, such as a relatively small genome, high genetic diversity, relative ease of vegetative propagation and genetic transformation, short rotation cycle, and fast growth rate [5,6]. However, very little is known about the molecular regulatory mechanism of poplar under soil water deficit.
Transcription factors (e.g., MYB, NAC, and WRKY) are essential components of the transcriptional regulatory network in plant species under abiotic stresses [7]. R2R3-type MYB proteins are the largest MYB subfamily transcription factors related to abiotic stress responses (e.g., drought, dehydration, heat, and salinity) in plants [8,9]. Many studies have documented that plant R2R3-MYB genes might affect drought resistance by thickening leaf cuticular waxes, controlling stomatal aperture, and regulating the expression of ABA signaling pathway genes [10,11]. For example, AtMYB94 and AtMYB96 can influence the expression of wax biosynthesis-related genes (e.g., WSD1 and KCS2) by specifically binding to the MYB binding consensus sequence (MBS) in their promoter region [12,13]. AtMYB44 positively regulated ABA to induce stomatal closure and suppressed the expression of the protein phosphatase 2C (PP2C) genes (e.g., ABI1, ABI2) in Arabidopsis [14,15]. PtrMYB94 (Potri.017G082500) can regulate the expression of ABA and drought stress-related genes (e.g., ABA1, DREB2B and P5CS2) under drought stress [16]. PtoMYB170 plays a role in lignin deposition during poplar wood formation and can also reduce the rate of water evaporation by closing foliar stomata [17]. PtoMYB142 can directly bind to the promoters of wax biosynthesis genes (e.g., CER4 and KCS6) and induce their expression, resulting in increased wax accumulation in poplar leaves to adapt to water scarcity [18].
Accurate identification of R2R3-MYB genes throughout the genome is required to comprehensively analyze their functional roles in various abiotic stress responses. With the increasing number of flowering plant genomes being published, genome-wide identification of the R2R3-MYB gene family has been reported in many dicotyledonous species, such as Arabidopsis thaliana [19], Vitis vinifera [20], Eucalyptus grandis [21], Capsicum annuum [22], Pinus massoniana [23] and Lycium barbarum [24]. Genome-wide identification of the R2R3-MYB genes in the Populus trichocarpa genome (v3.0) has also been reported [25,26,27,28]. These studies identified that the total number of P. trichocarpa R2R3-MYB genes ranged from 191 to 207. The inconsistencies between these resulting R2R3-MYB identifications are partly due to two adjacent conserved MYB repeats at an N-terminal region and a hypervariable regulatory region at a C-terminus [11]. However, a few important R2R3-MYB genes in poplar might be missed owing to the inconsistencies.
Here, we identified a total of 210 R2R3-MYB genes in the entire P. trichocarpa genome. We performed a comprehensive bioinformatic analysis of these P. trichocarpa R2R3-MYB genes, including their physicochemical properties, chromosomal distribution, cis-acting element and subcellular location prediction, phylogenetic and collinearity analyses, and RNA-seq profiling of R2R3-MYB genes in three distinct tissues under drought. A total of 10 R2R3-MYB genes were cloned from the poplar genotype NL895 (P. deltoides × P. euramericana cv. Nanlin895), and their tissue-specific and drought-responsive expression patterns were analyzed via quantitative RT-PR (qPCR). In addition, protein subcellular localization analysis indicated that both PdMYB2R032 and PdMYB2R151 proteins were localized in the nucleus of NL895 protoplasts and coincided with their prediction results. These results provide an important clue for screening drought-related R2R3-MYB genes in poplar and for further studying their roles in poplar under drought stress.
2. Results
2.1. Identification and Characterization of Poplar R2R3-MYB Gene Family Members and Their Chromosomal Distribution
According to the hidden Markov model (HMM) file and the structural characteristics of R2R3-MYB, we identified members of the poplar R2R3-MYB gene family. In this research, we identified a total of 210 R2R3-MYB transcription factors (TFs) comprising an adjacent MYB repeat pair from the P. trichocarpa genome (v3.0) (Table S1). These R2R3-MYB TFs were compared with those of four previously published papers (Figure 1) [25,26,27,28]. The union set of all P. trichocarpa R2R3-MYB TFs reported previously in the four papers comprised 211 protein-coding genes, only one (Potri.015G143400.1) of which was not included in the 210 R2R3-MYB genes that we identified. The Potri.015G143400.1 gene was identified as an R2R3-MYB gene only in the results of Chai et al. [25]. By examining the number of MYB repeats on the PFAM, CDD, and SMART web services, the Potri.015G143400.1 protein was found to have only a repeat of the MYB domain and hence belonged to the 1R-MYB gene family. In addition, 189 out of the 210 R2R3-MYB genes had an intersection between our results and four previous genome-wide identification results (Figure 1). The remaining 21 poplar R2R3-MYB proteins were not consistent across any of the five resulting sets and were searched manually for two neighboring MYB repeats at their N-terminal region. Thus, the putative 210 R2R3-MYB genes identified in this study should belong to the R2R3-MYB gene family in P. trichocarpa. In addition, these remaining genes might be arisen by whole-genome duplication (WGD) and dispersed duplication.
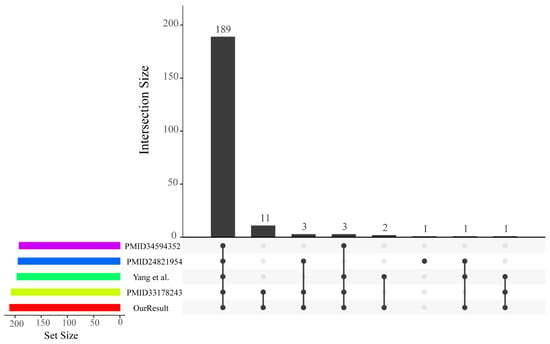
Figure 1.
Visualization of intersecting sets in our and four previous genome-wide identifications of R2R3-MYB genes in P. trichocarpa. Our result for the poplar R2R3-MYB family identification is represented by the red horizontal bar. The other four articles—PMID: 34594352 (https://pubmed.ncbi.nlm.nih.gov/, accessed on 1 May 2022) [28], PMID: 24821954 (https://pubmed.ncbi.nlm.nih.gov/, accessed on 1 May 2022) [25], Yang et al. [27], and PMID: 33178243 (https://pubmed.ncbi.nlm.nih.gov/, accessed on 1 May 2022) [26]—are colored in pink, blue, green, and yellow, respectively. The matrix layout shows all intersections of five R2R3-MYB identification sets. Black solid points in the matrix denote sets that are intersecting.
The location information of these R2R3-MYB genes on poplar chromosomes was obtained from the P. trichocarpa genome annotation file in GFF/GTF format. Only three genes (Potri.t011400.1, Potri.T125000.1, and Potri.T144800.1) among all 210 R2R3-MYB genes were not located on all 19 chromosomes in P. trichocarpa and were renamed PtrMYB2R208, PtrMYB2R209, and PtrMYB2R210, respectively. These three genes are located on scaffolds (Table S1). The other 207 R2R3-MYB genes on the 19 P. trichocarpa chromosomes were separately renamed PtrMYB2R001 to PtrMYB2R207 on the basis of their chromosomal position (Figure 2). As the two with the most R2R3-MYB genes, P. trichocarpa chromosome 1 (Chr01) and chromosome 17 (Chr17) had 23 (PtrMYB2R001-PtrMYB2R023) and 15 (PtrMYB2R173-PtrMYB2R187) PtrMYB2R genes, respectively. Only two genes (PtrMYB2R171 and PtrMYB2R172) were located on P. trichocarpa Chr16, which had the lowest number of R2R3-MYB genes. The R2R3-MYB genes were relatively uniformly distributed over the remaining 16 chromosomes, with nearly 10 R2R3-MYB genes and almost one gene per 2 Mb.
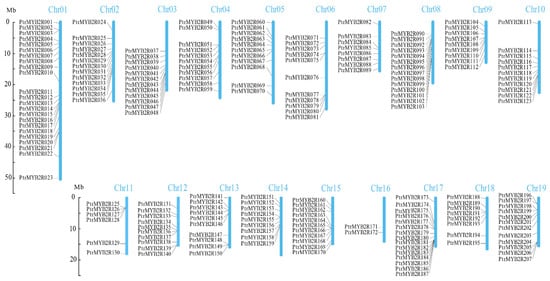
Figure 2.
Distribution of PtrMYB2R genes on the 19 poplar chromosomes. The 19 chromosomes of P. trichocarpa are represented by the blue vertical bars, and their chromosome numbers are at the top of the chromosomal bars. A total of 207 R2R3-MYB genes are distributed over the P. trichocarpa chromosomes. The chromosomal scale bar is 10 Mb.
2.2. Physicochemical Property Analysis of Poplar R2R3-MYB Proteins
To some extent, the physiological and biochemical properties of proteins can provide a reliable theoretical basis for functional research on R2R3-MYB TFs. Therefore, we predicted the physicochemical properties of these poplar R2R3-MYB family members, as shown in Table S2. The protein lengths of PtrMYB2Rs ranged from 162 (PtrMYB2R084) to 1070 (PtrMYB2R142) amino acids, with an average length of 335 amino acids. The molecular weight of the proteins ranged from 18.47 kDa (PtrMYB2R084) to 119.55 kDa (PtrMYB2R142), with a mean of 37.67 kDa. In addition, the isoelectric point (pI) of poplar R2R3-MYB proteins ranged from 4.71 (PtrMYB2R150) to 9.79 (PtrMYB2R179), and the aliphatic index ranged from 50.54 (PtrMYB2R094) to 83.74 (PtrMYB2R049). The aliphatic index determines thermostability of globular proteins, indicating their high thermal stability and flexibility. Among the poplar R2R3-MYB family, only six were stable proteins (PtrMYB2R008, PtrMYB2R018, PtrMYB2R023, PtrMYB2R130, PtrMYB2R182 and PtrMYB2R183), and their instability index was less than 40. The remaining 204 R2R3-MYB proteins were unstable (instability index > 40) and they were randomly distributed in each subgroup. This instability of R2R3-MYBs in poplar is similar in other transcription factor families, such as TIFY [29], ERF [30], and E2F-DP [31]. In addition, the grand average of hydropathicity (GRAVY) of all proteins in this family was less than 0, illustrating that these 210 R2R3-MYB proteins might be all hydrophilic proteins. These results indicated that the great differences in the properties of poplar R2R3-MYB proteins might be implicated in plant biological and abiotic stresses, and these proteins might play important roles in plant growth and development [11].
2.3. Analysis of Cis-Acting Elements in R2R3-MYB Genes
Cis-acting elements are closely related to specific biological functions and may participate in the regulation of gene expression. Hence, we analyzed the cis-acting elements in the promoter upstream of all the PtrMYB2R genes (Figure 3). The promoter region of the PtrMYB2R genes had the highest number of cis-elements involved in abiotic stress, including MYB, MYC, STRE (stress response elements), ABRE (ABA-responsive element), TATA-box, G-box, W-box, and MBS (MYB-binding sites). In addition to STRE, which is involved in osmotic stress and heat shock, other cis-acting elements can bind to transcription factors (e.g., MYB, WRKY, and DREB) to regulate the biological process of the plant response to drought stress [32,33]. We also found that there were many light response elements (Box-4) and oxidation defense elements (e.g., ARE (antioxidant response element), CGTCA motif, and AAGAA motif) [34,35]. In addition, there were some cis-elements related to plant hormone biosynthesis, which were mainly involved in the biosynthesis of MeJA (CAAT-box, CGTCA-motif, and TGACG-motif), ethylene (ERE, ethylene response element), and salicylic acid (TCA element, salicylic acid-responsive elements) [34,36]. Thus, the number of cis-acting elements in the promoter region of the R2R3-MYB genes was large and rich in variety (Table S3).

Figure 3.
Cis-acting elements in the promoter region of all poplar R2R3-MYB genes. The size of a word in the word-cloud image indicates the frequency of the cis-acting elements within the upstream promoter region of all R2R3-MYB genes on the P. trichocarpa genome. The picture is the shape of the P. euramericana leaf.
2.4. Phylogenetic Analysis of the Poplar R2R3-MYB Gene Family
To better understand the evolutionary relationship between poplar R2R3-MYB genes, we constructed an unrooted ML (maximum likelihood) tree using all the poplar R2R3-MYB proteins. More than 90% of branches in the ML tree had a bootstrapping value of more than 80%, indicating the reliability of subgroup classification (Figure 4). All 210 R2R3-MYB proteins in the entire P. trichocarpa genome were distinctly divided into 23 subgroups, the S16 subgroup of which had only one gene member. However, unlike in the study of Zhao et al. [26], there was no S12 subgroup in our gene family classification result. Based on the bidirectional best hits (BBH) between 210 PtrMYB2R genes and the AthR2R3-MYB genes, no PtrMYB2R genes were found to be homologous to the AthR2R3-MYB genes belonging to the S12 subgroup. In addition, a total of 21 R2R3-MYB genes were clustered together in a single cluster, and they shared high sequence similarity with AtR2R3-MYB genes in nine subgroups. However, they were not classified into any of the known subgroups in A. thaliana and were thus assigned to the subgroup “Other”.
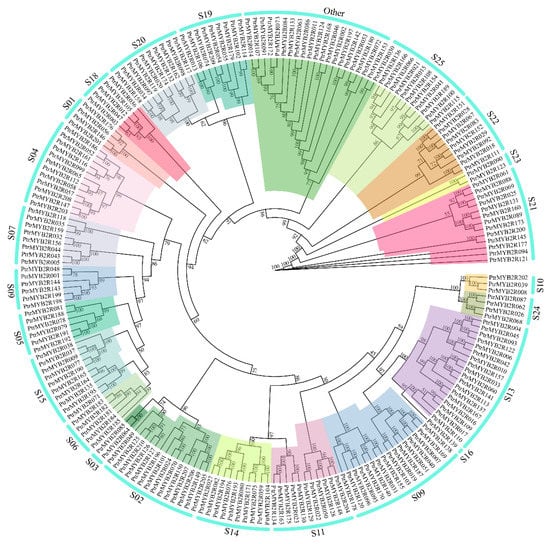
Figure 4.
Phylogenetic tree of poplar R2R3-MYB gene family. The poplar R2R3-MYB family is divided into 23 subfamilies which are marked in different colors. The outer circle shows the numbers of these 23 subgroups. The bootstrap values are shown at branch nodes of the phylogenetic tree.
2.5. Collinearity Analysis of R2R3-MYB Genes
Gene duplication was one of the primary driving forces in plant genome evolution. Recent studies have shown that tandem and segmental duplication events are likely to be key factors in the diversification and functional evolution of R2R3-MYB genes [37]. Hence, we performed collinearity analysis and discovered that the poplar R2R3-MYB gene family had two major gene expansion patterns: WGD/segmental duplication events and tandem duplication (Table S4). Twelve pairs of tandem duplication events were detected, and these clusters of tandem duplication genes were mainly located on the Chr01, Chr03, Chr11, Chr13, Chr17, and Chr19 chromosomes, involving 19 genes in the S02, S04, S06, and S09 subgroups (Figure 5). A total of 171 PtrMYB2R genes (81.4%) were implicated in the WGD/segmental duplication event, which was mainly caused by whole-genome replication. These results suggested that the evolutionary development of the poplar R2R3-MYB gene family was influenced by both tandem duplication events and WGD/segmental duplication events, in which WGD might play a crucial role. WGD events certainly contributed to the expansion and functional diversification of R2R3-MYB genes in the P. trichocarpa genome, providing adequate preparation for angiosperms in response to drastic environmental changes [10]. A total of 156 duplicated gene pairs in the R2R3-MYB family were identified (Figure 5 and Table S5), and nonsynonymous (Ka)/synonymous (Ks) analyses of these gene pairs were subsequently performed. The results showed that the Ka/Ks ratios of all duplicate gene pairs were less than 1 (Table S5), indicating that the poplar R2R3-MYB gene family might be subject to purifying selection during genome evolution [38]. In addition, the Ks value is proportional to the occurrence time of the gene duplication events, which can be used to estimate the divergence time of duplicated gene pairs [39]. Therefore, we analyzed their Ks divergence time and found that 48% of the homologous gene pairs were produced between 8 and 31 million years ago (Mya), which was after the separation of Populus and Salix (45 Mya) [40]. Approximately 24% of the duplicated gene pairs were generated after the divergence of Populus and Arabidopsis lineages (100–120 Mya) [5]. This indicated that a large number of WGD and tandem duplication events occurred after the differentiation of Populus with Salix and Arabidopsis, which led to the rapid expansion of the R2R3-MYB gene family in Populus species.
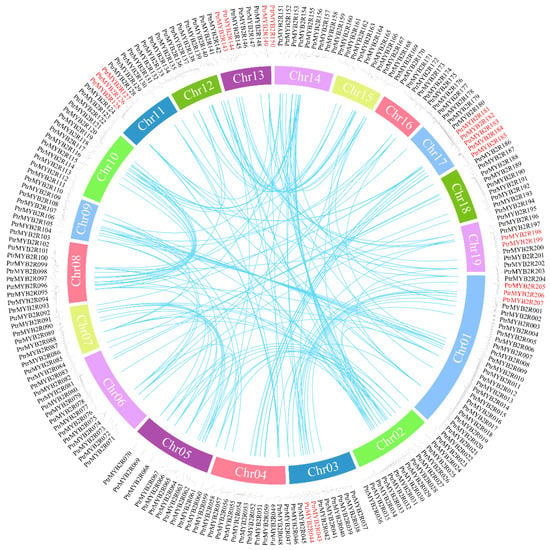
Figure 5.
Collinearity analysis of poplar R2R3-MYB genes. The outer circle shows the chromosomal distribution of these R2R3-MYB genes. The tandem duplicated genes on the outer circle are marked in red. The duplicated gene pairs are indicated by blue lines in the inner ring.
To understand the phylogenetic relationship of R2R3-MYB genes between poplar and the other four species, we performed an interspecific collinearity analysis (Figure 6). The four species consisted of two Salix species (S. purpurea and S. suchowensis), A. thaliana, and O. sativa. The number of orthologous R2R3-MYB pairs of P. trichocarpa with S. purpurea, S. suchowensis, A. thaliana, and O. sativa were 276, 266, 89, and 53, respectively. It can be seen that there was stronger collinearity between P. trichocarpa and the two Salix species (S. purpurea and S. suchowensis), followed by A. thaliana and finally O. sativa. This phenomenon could be because the Salix and Populus genera were a sister group in the family Salicaceae. The species belonging to the Salix and Populus genera shared a relatively close evolutionary relationship and a high level of sequence similarity within their R2R3-MYB genes. It seemed obvious that a large proportion of the orthologous R2R3-MYB pairs between P. trichocarpa and the two Salix species were located on the collinear blocks between the genomes of P. trichocarpa and the two Salix species. This finding also implied that some of the R2R3-MYB genes in the P. trichocarpa genome suffered from WGD/segmental duplication events during their evolution.
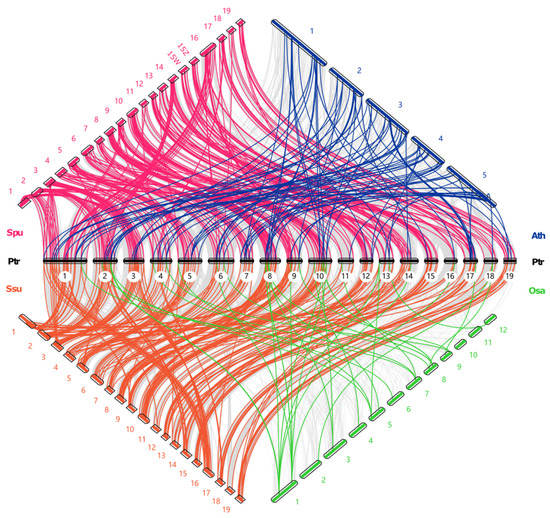
Figure 6.
The interspecific collinearity analysis of R2R3-MYB genes in poplar and four different species. The chromosomes of the P. trichocarpa, S. purpurea, S. suchowensis, A. thaliana and O. sativa genomes are marked in black, pink, orange, blue, and green, respectively. The collinearity blocks between P. trichocarpa and other four plant species are marked by gray lines.
2.6. Poplar R2R3-MYB Gene RNA-Seq Analysis
To select putative drought-responsive R2R3-MYB genes in P. trichocarpa, we reanalyzed RNA-seq profiling from the NCBI SRA database of PtrMYB2Rs in three different tissues under drought treatment. And then we selected 10 R2R3-MYB genes for further expression pattern analysis. As shown in Figure 7A, PtrMYB2R028 and PtrMYB2R151 were highly expressed in all tissues (log2(FPKM + 1) ≥ 7), and PtrMYB2R060 was moderately expressed in all tissues (7 > log2(FPKM + 1) ≥ 5). Some genes showed tissue-specific expression. For example, PtrMYB2R176 was only highly expressed in leaves, PtrMYB2R089 was preferentially expressed in stems, and PtrMYB2R111 and PtrMYB2R119 were specifically expressed in roots.
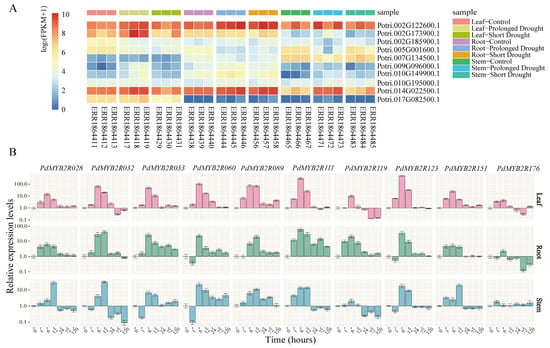
Figure 7.
Expression patterns of 10 R2R3-MYB genes in poplar. (A). RNA-seq expression profiles of 10 PtrMYB2R genes in the P. trichocarpa under drought stress. The gene ID for the 10 PtrMYB2R genes are as follows: PtrMYB2R028, PtrMYB2R032, PtrMYB2R033, PtrMYB2R060, PtrMYB2R089, PtrMYB2R111, PtrMYB2R119, PtrMYB2R123, PtrMYB2R151, and PtrMYB2R176. The heatmap is plotted using the log2-transformed (FPKM+1) values. There are three samples per treatment for each tissue, and the sample information is shown on the right side, such as leaf control (SRA accession no. ERR1864411, ERR1864412, and ERR1864413), leaf prolonged drought (ERR1864417, ERR1864418, and ERR1864419), and so on. (B). qRT-PCR profiles of 10 PdMYB2Rs in NL895 with PEG6000 treatment. The relative expression values of these PdMYB2R genes are log2-transformed. The leaves, stems, and roots are harvested at 0, 1, 6, 12, 24, 72, and 120 h after 10% PEG6000 treatment.
We also compared the transcript abundance of these PtrMYB2R genes under normal conditions and drought stress (short-term and long-term drought stress). Under long-term drought stress, the expression levels of most PtrMYB2R genes were significantly changed (p < 0.05) and most of them were upregulated, with the exception of PtrMYB2R033 and PtrMYB2R060. In contrast, the expression levels of most genes did not change significantly after short-term drought treatment (p > 0.05). The results indicated that PtrMYB2R genes might play an important regulatory role in response to drought stress, especially when plants are faced with prolonged drought.
2.7. PdMYB2R Gene Cloning and the Expression Patterns of PdMYB2R Genes in Different Tissues
We cloned and sequenced these 10 R2R3-MYB genes from the poplar genotype NL895 (P. deltoides × P. euramericana cv. Nanlin895). They were named according to their corresponding PtrMYB2R gene names (PdMYB2R028, PdMYB2R032, PdMYB2R033, PdMYB2R060, PdMYB2R089, PdMYB2R111, PdMYB2R119, PdMYB2R123, PdMYB2R151, and PdMYB2R176). The ORFs (open reading frames) of these genes ranged from roughly 700 to 1400 bp in length (Figure S1). The sequences of these cloned PdMYB2R genes had an average similarity of more than 99% with their corresponding PtrMYB2R genes.
We then analyzed the expression levels of these PdMYB2R genes among three tissues (roots, stems and leaves) of NL895 using qRT-PR. As shown in Figure S2, these genes showed multiple expression patterns among tissues and had certain tissue specificity, which was similar to the results of the RNA-seq data analysis. For example, PdMYB2R032/089/123/151 genes had similar expression patterns, and their expression levels were significantly higher in roots than in stems and leaves. Among them, the expression level of PdMYB2R089 in roots was particularly significant compared to that in leaves and stems, and its expression level was 290 times higher than that in leaves. The expression levels of PdMYB2R111/119 in leaves were approximately 1.5–3 times those of the other tissues. PdMYB2R176 had a similar expression pattern, but it is worth noting that the expression level of PdMYB2R176 in leaves was approximately 25.5 times that in roots and approximately 2.7 times that in stems. However, PdMYB2R033 had a significantly higher expression level in stems than in the other two tissues. The remaining two genes, PdMYB2R028 and PdMYB2R060, were not significantly different between the three tissues.
2.8. Analysis of the Expression Pattern of Poplar PdMYB2R Genes under Drought Stress
Drought is one of the important environmental factors restricting the growth and development of woody plants, and even severe drought may result in whole-plant mortality [41]. Hence, we further explored the drought-induced expression patterns of PdMYB2R genes in three different tissues using qRT-PCR. The change trends of different genes at different drought treatment time points were shown in Figure 7B. Nine genes excluding PdMYB2R119 showed similar patterns in leaves, all exhibiting a trend of first increasing and then decreasing. The relative expression levels of most genes peaked at 6 h or 12 h after PEG6000 treatment and then decreased to the initial expression levels. The expression patterns of PdMYB2R genes in roots and stems fluctuated at individual time points. For example, PdMYB2R032, PdMYB2R033, PdMYB2R060, PdMYB2R123, and PdMYB2R176 in the roots and/or stems showed a trend of slight decrease at 1 h, rapid increase to peak value at 6 h, and then rapid decline. In addition, at the late stage of drought treatment (3 d or 5 d), individual genes were slightly upregulated in some tissues, such as PdMYB2R033, PdMYB2R060 (roots and stems), and PdMYB2R111 (roots), but the changes were not significant and did not affect the overall trend.
Under drought conditions, 10 genes were induced to varying degrees in roots, stems, and leaves. Most genes (except PdMYB2R119) had higher expression levels in leaves than in roots and stems. Especially when poplar was subjected to drought stress for 6 h, the expression levels of PdMYB2R060, PdMYB2R111, and PdMYB2R123 in leaves were significantly upregulated and were respectively approximately 100, 310, and 500 times higher than those under normal conditions. It is worth noting that PdMYB2R119 was preferentially upregulated in response to instant drought treatment in roots but significantly downregulated in stems and leaves under 2–5 days of drought treatment. Similarly, PdMYB2R032 was also significantly downregulated in leaves and stems under 2–5 days of drought stress, suggesting that PdMYB2R032 and PdMYB2R119 might be tissue-specific in different stress periods. In addition, we discovered that all PdMYB2R genes had similar change trends in the two tissues, and there were no genes with completely identical change trends among the three tissues, which further validated that PdMYB2R has certain tissue specificity.
Our results demonstrated that the relative expression levels of most genes changed significantly under short-term drought stress (within 1 day), but the changes were not obvious under 1- to 5-day PEG6000 treatment, and the expression levels of most PdMYB2R genes tended to be stable at 1 day after exposure to the drought treatment. There was a slight difference from the P. trichocarpa RNA-seq data, which might be attributed to the different methods of drought treatment and plant materials. In this study, the longest stress time was 5 days, so there was a certain uncertainty about the longer stress time point. In conclusion, the transcriptome analysis of P. trichocarpa and the qPCR results of PdMYB2R showed that the expression levels of these 10 R2R3-MYB genes all changed significantly under the drought environment. These results indicated that these R2R3-MYB genes might play an important role in Populus species under drought stress. However, their specific regulatory patterns and mechanisms need to be further investigated.
2.9. Subcellular Localization and Transcriptional Activation Activity
The subcellular location of R2R3-MYB TFs might be involved in their regulatory role in the transcriptional regulatory network. Thus, we used three online software tools to predict the subcellular localization of poplar R2R3-MYB protein. The results showed that only PtrMYB2R041 (Potri.003G123800.1) and PtrMYB2R124 (Potri.010G240800.1) of all 210 PtrMYB2R TFs were predicted to be located in the extracellular and cytoplasmic regions. The remaining 208 R2R3-MYB proteins were predicted to have a nuclear localization signal (Table S6). Then, we randomly selected two putative nuclear-located R2R3-MYB genes for validating subcellular localization. The subcellular localization assay showed that the encoded proteins of PdMYB2R032 and PdMYB2R151 only exhibited fluorescent signal in the nucleus but not in the cytoplasm and cell membrane (Figure 8A). These findings indicated that the two R2R3-MYB TFs might function in the nucleus of Populus species.
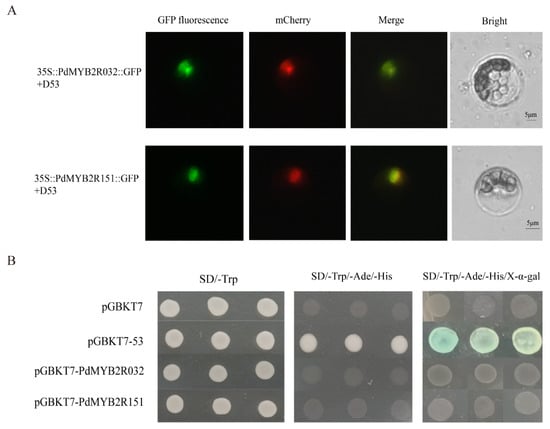
Figure 8.
Subcellular localization and autoactivation activity of PdMYB2R032 and PdMYB2R151 proteins. (A) Subcellular localization. mCherry-D53, a red and nuclear-localized marker. Scale bar: 5 μm. (B) Analysis of autoactivation activity. pGBKT7 vector was the negative control, and pGBKT7-53 vector was the positive control. Yeast cells were grown on SD medium at 29 °C for 48 h.
Then, we investigated the autoactivation activities of the two TFs by transforming yeast. The two fusion plasmids, including pGBKT7-PdMYB2R032 and pGBKT7-PdMYB2R151, were transformed into yeast cells AH109. The results of transcriptional activation activity showed that both PdMYB2R032 and PdMYB2R151 proteins could grow on SD/Trp medium as well as negative and positive controls, indicating that they were nontoxic. As shown in Figure 8B, only the transformants containing pGBKT7-35 can grow on the triple deficient medium (SD/-Trp/-Ade/-His), and showed blue after adding X-α-Gal. However, pGBKT7, pGBKT7-PdMYB2R032 and pGBKT7-PdMYB2R151 could not grow on the triple deficient medium, indicating that the full-length sequences of PdMYB2R032 and PdMYB2R151 did not possess transcriptional activation activity for activating the expression of downstream reporter genes.
3. Discussion
3.1. Identification and Evolution of the Poplar R2R3-MYB Gene Family
To the best of our knowledge, the identification of R2R3-MYB TFs in the entire P. trichocarpa genome (v3.0) has been reported in four papers. However, some differences persisted in the identification results of the previously published papers. These differences were mainly due to the diversity and complexity of the conserved domain of MYB genes, resulting in unclear or incorrect gene classification of MYB subfamilies [11]. In the present study, we identified a total of 210 PtrMYB2R gene members in P. trichocarpa. The poplar R2R3-MYB gene family had a larger number of gene members than other identified dicotyledonous families, such as A. thaliana (126) [42], Eucalyptus grandis (141) [21], Malus domestica (186) [43], Cinnamomum camphora (96) [44], and Camellia sinensis (112) [45]. The R2R3-MYB gene family may have undergone functional diversification during genome evolution in plants, thus forming different subfamilies [11]. In this study, the poplar R2R3-MYB gene family was phylogenetically clustered into 23 subgroups. Although the functions of most PtrMYB2R genes have not been characterized, 90% of the PtrMYB2R genes are clustered in the functional groups of Arabidopsis. Many PtrMYB2R genes were grouped into subgroups S13, S15, and S22, the members of which are involved in abiotic stress responses in Arabidopsis. Consequently, phylogenetic analysis will aid in the identification of PtrMYB2R genes related to the drought stress response [46].
Many studies have shown that whole-genome duplication (WGD) and fragment duplication events are important mechanisms by which the R2R3-MYB gene family has rapidly expanded during poplar genome evolution [28]. Chromosomal mapping analysis showed that 207 R2R3-MYB genes were unevenly distributed over 19 P. trichocarpa chromosomes, and most R2R3-MYB genes were concentrated on certain chromosomal regions. This finding was similar to the uneven chromosomal distribution of poplar MYB-related genes, which might be caused by multiple DNA duplications, such as WGD, tandem duplication and dispersed duplication [47]. Then, we found a large number of WGD events and small-scale tandem duplication events among the poplar R2R3-MYB gene family, and most paralogous gene pairs diverged after the separation of Populus and Salix (45 Mya). The results showed that WGD events played a leading role in the rapid expansion of the poplar R2R3-MYB gene family, and small tandem repeat events also played a role in promoting the expansion [11]. The poplar R2R3-MYB family was also likely to suffer rapid expansion after its divergence with monocotyledons and Salix species. In addition, we discovered that the R2R3-MYB duplicated gene pairs of poplar might have been mainly affected by purifying selection (Ka/Ks < 1) during evolution, which was consistent with the results obtained for other plants, such as Liriodendron chinense and Solanum tuberosum [48,49]. These duplicated R2R3-MYB genes might be associated with the gain of new functions that increase ecological resilience to a variety of adverse environmental stresses [48].
3.2. Analysis of Functional Sites of Poplar R2R3-MYB Proteins
Understanding the spatial information of the proteins encoded by the R2R3-MYB genes in cells can provide a reference for judging its protein function. The subcellular localization prediction results showed that 205 of the 210 (97.6%) PtrR2R3-MYB proteins might be localized in the nucleus, which is consistent with the R2R3-MYB protein research of other species [45,50]. The subcellular localization experiment results confirmed our prediction, indicating that they were nuclear-located proteins, which was consistent with the theory that TFs normally functioned in the nucleus [51,52]. TFs can regulate target genes by binding specifically to the cis-element of their promoters in the nucleus [53]. R2R3-MYB TFs could bind specifically to the cis-acting elements of drought-responsive genes, resulting in control of their expression levels [7].
This study showed that PdMYB2R032 and PdMYB2R151 transcription factors have no non-autoactivation activity and can be directly used for subsequent protein interaction experiments with drought-related proteins [54]. These results suggest that these R2R3-MYB proteins may interact with other proteins to form transcription complexes to regulate downstream gene expression [55,56,57]. The regulatory relationship between PdMYB2R032/151 and other cofactors or key downstream drought-related genes can be further verified by EMSA (electrophoresis mobility shift assay), ChIP (chromatin immunoprecipitation), and yeast one-hybrid or two-hybrid technologies in the future [56,58].
3.3. Tissue-Specific Expression Pattern of Poplar R2R3-MYB Genes under Drought Stress
Research related to the function of the R2R3-MYB transcription factor has mainly focused on the herbaceous model species A. thaliana, and approximately 80% of functional research on AtR2R3-MYB genes has been reported [11,19,59,60]. Only a few drought-related R2R3-MYB genes, such as PtoMYB142 and PtrMYB94, have been reported in poplar as a woody model species [16,18]. Furthermore, the study characterized the drought-inducible expression patterns of 10 PdMYB2R genes across different tissues in poplar NL895. Despite some differences between the qRT-PR results and P. trichocarpa RNA-seq profiling, the expression levels of these 10 R2R3-MYB genes were significantly changed in three poplar tissues (roots, stems and leaves) in response to drought stress. These PtrMYB2R genes shared a relatively similar expression pattern, but their expression levels differed significantly among the three tissues. This result suggested that these PtrMYB2R genes might play similar regulatory roles under drought stress. The expression of genes is determined by their promoters in plants, and we identified a large number of drought-related elements (e.g., MYB, MYC, ABRE, TATA-box, G-box, W-box, and MBS) in gene promoter regions. In conclusion, PtrMYB2R genes are generally involved in abiotic stress such as drought.
We found that some orthologues of R2R3-MYB genes in Arabidopsis may have similar regulatory roles in poplar under drought stress [26]. For example, Liang et al. (2005) found that overexpression of AtMYB61 (AT1G09540) resulted in decreasing stomatal pore size and affected gas exchange, which could improve the water-use efficiency of plants in water-deficient environments [61]. Homologous genes of AtMYB61, PdMYB2R033 and PdMYB2R060 showed significant changes after drought treatment. This result implicated their potential function in contributing to plant tolerance under drought stress. A large number of abiotic stress-related cis-elements were found in the promoter regions of four genes (AtMYB44, AtMYB73, AtMYB77, and AtMYB70) in the Arabidopsis R2R3-MYB family S22 subgroup. The overexpression of AtMYB44 could enhance the ability of Arabidopsis to resist drought, and loss of the AtMYB73 causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity [14,62]. Three poplar R2R3-MYB genes, PdMYB2R028, PdMYB2R111, and PdMYB2R151, belonged to the R2R3-MYB family subgroup S22 and were also homologous to AtMYB73. The three genes were significantly induced in three poplar organs under drought stress. The results suggested that these R2R3-MYB genes belonging to subgroup S22 might be implicated in the drought-responsive processes of poplar.
The regulatory roles of some poplar R2R3-MYB genes in drought response have been reported. For example, recent studies have demonstrated that PtrMYB121 (Potri.002G185900) and PtoMYB170 (Potri.005G001600) can promote the accumulation of lignin and cellulose, and PtoMYB170 enhances drought tolerance by triggering stomatal closure [17,63]. As a transcriptional activator, PtrMYB94 (Potri.017G082500) was involved in the transcriptional regulation of the expression of ABA and drought stress-related genes (e.g., ABA1, DREB2B, and P5CS2), thereby improving the tolerance of transgenic A. thaliana plants to drought stress [16]. Similar to the results of Fang et al., our qRT-PR results showed that the expression level of PdMYB2R176 (Potri.017G082500) had been increased four times in the leaves of the poplar genotype NL895 at 6 h after PEG6000 treatment compared with the control group (0 h). Taken together, the data suggested that these R2R3-MYB gene members might play a crucial role in drought stress response.
4. Materials and Methods
4.1. Genomic Data Retrieval
The genome data of P. trichocarpa (v3.0) and Salix purpurea (v5.1), including genomic DNA sequences, proteins, protein-encoding genes and genome-annotation files (GFF/GTF, general feature format/gene transfer format), were downloaded from the plant genome database Phytozome (https://phytozome.jgi.doe.gov/, accessed on 5 March 2022). We also obtained the genomic information file of Salix suchowensis (GCA_017552425.1) from the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/, accessed on 5 March 2022). In addition, the R2R3-MYB family relevant genomic data of A. thaliana (TAIR10) and Oryza sativa subsp. japonica (v1.0) were downloaded from the Ensembl Plants website (release 53, https://plants.ensembl.org/info/data/ftp/index.html, accessed on 5 March 2022).
4.2. Identification of R2R3-MYB Transcription Factors in Poplar
First, the hidden Markov model (HMM) file of the MYB binding domain (Pfam no. PF00249.31) was retrieved from the Pfam database website (https://pfam.xfam.org/, accessed on 2 May 2022) and used to search MYB repeats in the P. trichocarpa proteins using hmmsearch in the HMMER package (v3.3.2, http://hmmer.org/, accessed on 2 May 2022) with an E-value cutoff of 1 × 10−3. Moreover, the candidate P. trichocarpa R2R3-MYB (PtrMYB2R) proteins were inspected using BLAST+ (v2.9.0) against A. thaliana R2R3-MYB (AthR2R3-MYB) proteins obtained from the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/, accessed on 2 May 2022). The BLASTp hits with an E-value of less than 1 × 10−5 were retained. The proteins obtained by the above two methods are potential R2R3-MYB gene family members. To further confirm the reliability of candidate R2R3-MYB proteins, potential PtrMYB2R sequences were submitted to Pfam (http://pfam.xfam.org/, accessed on 2 May 2022), CDD (https://www.ncbi.nlm.nih.gov/cdd/, accessed on 2 May 2022) and SMART (http://smart.embl-heidelberg.de/, accessed on 2 May 2022) to check the sequences of intact R2 and R3 MYB repeats and to ensure the completeness of the MYB domain. Potential proteins with incomplete conserved MYB domains were eliminated, and the remaining proteins were considered to belong to the poplar R2R3-MYB family.
4.3. Chromosomal Location, Physicochemical Properties and Cis-Acting Element Analysis of R2R3-MYB Genes in Poplar
The chromosomal distribution information of PtrMYB2R genes was extracted from the P. trichocarpa genome annotation file in GFF/GTF format. MapInspect software was used to map the location of R2R3-MYB genes on poplar chromosomes, and they were named according to their corresponding locations on all chromosomes. Then, the physicochemical properties of these PtrMYB2R proteins, such as the isoelectric point (pI), molecular weight (MW), protein length, instability index, aliphatic index and hydropathicity, were calculated using the Python package Biopython (v1.79, https://biopython.org/, accessed on 5 May 2022) and the ProtParam tool on the Expasy website (https://www.expasy.org/resources/protparam, accessed on 5 May 2022). Finally, we predicted the cis-acting elements in the upstream 2,000-bp region of the poplar R2R3-MYB genes using the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 5 May 2022).
4.4. Multiple Sequence Alignment and Phylogenetic Analysis
Multiple sequence alignment (MSA) of PtrMYB2R proteins was carried out using ClustalW (v2.1, http://www.clustal.org/clustal2/, accessed on 5 May 2022) under default parameter settings. The protein substitution model (JTT+F+R8) with the minimum BIC (Bayesian information criterion) score was inferred from the PtrMYB2R MSA files by ModelFinder in IQ-TREE (v1.6.12) [64]. The ML (maximum likelihood) phylogenetic tree was reconstructed using IQ-TREE (v1.6.12) with the optimal substitution model JTT+F+R8 and 1000 bootstrap replicates [65]. The resulting ML tree was visualized using the R package ggtree (v3.2.1) [66].
4.5. PtrMYB2R Gene Expression Analysis
The RNA-seq data of poplar roots, stems and leaves at three-time points were downloaded from the NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra, accessed on 5 May 2022) under BioProject no. PRJEB19784 [67]. The three-time points were composed of an untreated control point, short-term drought (5 days after withholding water), and prolonged drought (5 days after withholding water and 7 days after limited watering). RNA-seq data were checked for quality control using FastQC (v0.11.9), and reads were trimmed with Trimmomatic (v.0.38) [68]. Then, the RNA-seq reads were mapped against the P. trichocarpa (v3.0) genome using the spliced aligner STAR (v2.7.3a) software [69]. The featuresCounts (v2.0.1) software was used to count the numbers of reads that were aligned on the P. trichocarpa genome [70]. Finally, the expression patterns of PtrMYB2Rs in three tissues and under three treatment time points were analyzed with the R package DESeq2 (v1.30.1) [71].
4.6. Genome Collinearity Analysis
The collinearity relationship between R2R3-MYB genes in the P. trichocarpa genome poplar was visualized with the R package circlize (v0.4.15) [72]. To further explore the evolutionary mechanism of the poplar R2R3-MYB gene family, interspecific collinearity analysis was performed on five flowering plants, including P. trichocarpa, Salix purpurea, S. suchowensis, A. thaliana, and Orzya sativa. MCScanX (https://github.com/wyp1125/MCScanX, accessed on 6 May 2022) and BLASTP (E-value threshold of 1 × 10−10) were used to identify gene tandem duplication and fragment duplication [73]. The collinear relationship between poplar and the other four species was then visualized using the Python package JCVI (v0.8.12, https://github.com/tanghaibao/jcvi, accessed on 6 May 2022) [74]. Paralogous and orthologous gene pairs were aligned with ClustalW (v2.1), and nonsynonymous (Ka)/synonymous (Ks) for each gene pair was calculated using ParaAT (v2.0) and KaKs-Calculator (v2.0) [75]. The divergence time of duplicated gene pairs was calculated with their Ks values and the synonymous mutation rate (r = 9.1 × 10−9) [76,77].
4.7. Cloning of R2R3-MYB Genes from NL895 Poplar
In this study, the tissue-cultured plantlets of hybrid poplar NL895 (P. deltoides × P. euramericana cv. Nanlin895) were used for cloning 10 selected R2R3-MYB genes. These plantlets were grown on 1/2 Murashige and Skoog (MS) medium under a photoperiod of 16 h of light and 8 h of darkness. The young leaves were harvested from 45-day-old plantlets, frozen immediately in liquid nitrogen, and stored at −80 °C for further RNA isolation.
Total RNA was extracted from poplar NL895 leaves using an RNA extraction kit (Tiangen, Beijing, China), and 1 μg of RNA was reverse-transcribed into cDNA. Then, the high-fidelity PCR enzyme KOD-Plus-Neo (Toyobo, Osaka, Japan) was used for PCR amplification, and the cloning primers in the reaction system are shown in Table S7. The PCR product was verified by 1% agarose gel electrophoresis, and the single and correct band was purified and recovered. The recovered products were connected to pTOP001 Blunt Simple Cloning vector (Genesand, Beijing, China), transformed into E. coli competent cells TSC-C01 (Tsingke, Beijing, China) and cultured overnight at 37 °C. The next day, the monoclonal bacteria were selected for amplification and sent to the biotechnology company (Sangon Biotech, Shanghai, China) for Sanger sequencing.
4.8. Expression Pattern of R2R3 MYB Genes in Poplar NL895 under Drought Stress
The tissue-cultured plantlets of NL895 were grown at an air temperature of 25 °C and 60% relative humidity. NL895 plantlets 45 days old were subjected to water-deficit stress imposed by polyethylene glycol 6000 (PEG6000). The NL895 plantlets with consistent growth conditions were placed in solid 1/2 MS medium containing 10% PEG6000 for 0 h, 1 h, 6 h, 12 h, 24 h (1 day), 72 h (3 days), and 120 h (5 days). Three biological replicates were performed separately for each time point, and each biological replicate was composed of 3~5 leaves. As previously described, total RNA was isolated from the leaf, root and stem tissues of the NL895 plantlets under PEG6000 treatment, and 1 μg of RNA was reverse-transcribed into cDNA. Real-time quantitative PCR (qPCR) was used to detect the expression pattern of these 10 poplar R2R3-MYB genes under drought stress. These qPCR data were normalized and analyzed using the 2-ΔΔCt algorithm [78] with the poplar EF1-α gene (NCBI GenBank no. GQ253565.1) as an internal reference gene. Using the cloned PdMYB2R gene coding sequence (CDS) as the template, qPCR primers were designed on the website of GenScript (https://www.genscript.com/, accessed on 6 June 2022). The primer sequences are shown in Supplemental File Table S8.
4.9. Subcellular Localization of the R2R3-MYB Transcription Factor in Poplar
The subcellular localization of PMYB2R proteins was predicted using three online tools, including BUSCA (http://busca.biocomp.unibo.it/, accessed on 20 June 2022), Cello (http://cello.life.nctu.edu.tw/) and WoLFPSORT (https://wolfpsort.hgc.jp/, accessed on 20 June 2022). To ensure the reliability of the prediction results, we combined the three prediction results to determine the final subcellular localization of each R2R3-MYB gene.
The PEG-mediated transformation of poplar NL895 protoplast was performed according to the method described by Tan et al. [79]. Two poplar R2R3-MYB proteins were selected as the target genes to verify the subcellular localization. Specifically, the CDS of PdMYB2R032 and PdMYB2R151 without a stop codon was inserted into the vector p2GWF7.0 for fusion expression of the target gene and green fluorescent protein (GFP). D53-mCherry, a nuclear localization protein with a red fluorescent label, was used as a positive control [80]. First, poplar leaves were chopped into tiny pieces, and protoplasts were obtained by enzymatic hydrolysis with pectinase (Sigma, Saint Louis, MO, USA) and cellulase (Sigma, Saint Louis, MO, USA). Then, the plasmid containing the target gene was transformed into poplar NL895 protoplasts. Finally, the subcellular localization information of the two target proteins was captured under an AxioScope A1 fluorescence microscope (Zeiss, Oberkochen, Germany).
4.10. Transcriptional Activation Assay
The full-length of PdMYB2R032 and PdMYB2R151 cDNA were cloned into the pGBKT7 plasmid (VT006, Coolaber, Beijing, China). The two fusion plasmids, including pGBKT7-PdMYB2R032 and pGBKT7-PdMYB2R151, were transformed into yeast cells AH109 (CC300, Coolaber, Beijing, China). In the study, empty pGBKT7 vector and pGBKT7-53 vector (VT007, Coolaber, Beijing, China) were used as negative and positive controls, respectively. Finally, the transformed yeast cells were streaked and cultured on SD medium (SD/-Trp and SD/-Trp/-Ade/-His) at 29 °C for 48 h.
5. Conclusions
In this research, a total of 210 members belonging to the R2R3-MYB gene family were identified in the entire P. trichocarpa nuclear genome and were phylogenetically divided into 23 subgroups. A total of 207 R2R3-MYB genes were relatively unevenly distributed on the 19 P. trichocarpa chromosomes. A large number of whole-genome duplications and a few tandem duplication events were found in these R2R3-MYB genes, suggesting that whole-genome duplication might have made a large contribution to the rapid expansion of the poplar R2R3-MYB family. There were a large number of drought stress-related cis-acting elements in the promoter regions of R2R3-MYB genes, suggesting that the R2R3-MYB genes might be involved in the response to drought stress. Almost all poplar R2R3-MYB transcription factors were predicted to be localized in the nucleus, and the nuclear positioning of two R2R3-MYB proteins was validated in a subcellular localization analysis in poplar NL895 protoplasts. In addition, RNA-seq profiling and qRT-PCR analysis showed that the expression levels of 10 R2R3-MYB genes changed significantly in different tissues under drought stress. However, how these R2R3-MYB proteins bind to specific cis-acting elements of drought-related target genes to regulate drought response in poplar remains largely unclear. These can be studied in the future by ChIP, EMSA, and yeast one/two-hybrid approaches. This study will lay a foundation for further investigation of the regulatory roles of R2R3-MYB transcription factors in Populus species exposed to water deficit, and provide support for the development of new poplar genotypes with elevated drought tolerance.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms24065389/s1.
Author Contributions
S.Z. and M.H. conceived the project. S.Z. designed the entire experiment. S.Z. and X.Z. carried out the gene family analysis. X.Z., H.W. and Y.C. conducted the molecular experiments. X.Z. prepared this manuscript, and S.Z. revised it. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by grants from the Fourteen Five-Year National Science and Technology Support Program (2021YFD2201200), Natural Science Foundation of Jiangsu Province (BK20150879), and Qinglan Project of the Jiangsu Education Department. The funding bodies did not have a role in the design of the study, data collection, analysis, interpretation of data, writing the manuscript, or the decision to publish.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Table.
Acknowledgments
We would like to thank Huixin Pan at Nanjing Forestry University and Xinye Zhang at Hubei Forestry Academy for help in technical guidance.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Haberstroh, S.; Werner, C. The role of species interactions for forest resilience to drought. Plant Biol. 2022, 24, 1098–1107. [Google Scholar] [CrossRef]
- Salmon, Y.; Dietrich, L.; Sevanto, S.; Hölttä, T.; Dannoura, M.; Epron, D. Drought impacts on tree phloem: From cell-level responses to ecological significance. Tree Physiol. 2019, 39, 173–191. [Google Scholar] [CrossRef]
- Qiu, J. China faces up to groundwater crisis. Nature 2010, 466, 308. [Google Scholar] [CrossRef]
- Wu, K.; Qu, Y.; Rong, H.; Han, X.; Tian, Y.; Xu, L. Identification and Expression Analysis of the Populus trichocarpa GASA-Gene Family. Int. J. Mol. Sci. 2022, 23, 1507. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Ma, J.; Wan, D.; Duan, B.; Bai, X.; Bai, Q.; Chen, N.; Ma, T. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 2019, 17, 451–460. [Google Scholar] [CrossRef]
- Yao, T.; Zhang, J.; Xie, M.; Yuan, G.; Tschaplinski, T.J.; Muchero, W.; Chen, J.G. Transcriptional Regulation of Drought Response in Arabidopsis and Woody Plants. Front. Plant Sci. 2020, 11, 572137. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Liu, Y.; Shang, X.; Fang, S. Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. Int. J. Mol. Sci. 2022, 23, 3429. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 2015, 56, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Cheong, J.J. AtMYB44 interacts with TOPLESS-RELATED corepressors to suppress protein phosphatase 2C gene transcription. Biochem. Biophys. Res. Commun. 2018, 507, 437–442. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, X.; Wang, H.; Tang, X.; Liu, C.; Yin, H.; Ye, S.; Jiang, Y.; Duan, Y.; Luo, K. The poplar R2R3 MYB transcription factor PtrMYB94 coordinates with abscisic acid signaling to improve drought tolerance in plants. Tree Physiol. 2020, 40, 46–59. [Google Scholar] [CrossRef]
- Xu, C.; Fu, X.; Liu, R.; Guo, L.; Ran, L.; Li, C.; Tian, Q.; Jiao, B.; Wang, B.; Luo, K. PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 2017, 37, 1713–1726. [Google Scholar] [CrossRef]
- Song, Q.; Kong, L.; Yang, X.; Jiao, B.; Hu, J.; Zhang, Z.; Xu, C.; Luo, K. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis. Tree Physiol. 2022, 42, 2133–2147. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Arce-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef]
- Soler, M.; Camargo, E.L.; Carocha, V.; Cassan-Wang, H.; San Clemente, H.; Savelli, B.; Hefer, C.A.; Paiva, J.A.; Myburg, A.A.; Grima-Pettenati, J. The Eucalyptus grandis R2R3-MYB transcription factor family: Evidence for woody growth-related evolution and function. New Phytol. 2015, 206, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Tang, B.; Dai, X.; Xie, L.; Liu, F.; Zou, X. Genome-Wide Identification and Capsaicinoid Biosynthesis-Related Expression Analysis of the R2R3-MYB Gene Family in Capsicum annuum L. Front. Genet. 2020, 11, 598183. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Yao, S.; Chen, P.; Wang, D.; Agassin, R.H.; Hou, Y.; Zhang, C.; Ji, K. Transcriptome Identification of R2R3-MYB Gene Family Members in Pinus massoniana and PmMYB4 Response to Drought Stress. Forests 2023, 14, 410. [Google Scholar] [CrossRef]
- Yin, Y.; Guo, C.; Shi, H.; Zhao, J.; Ma, F.; An, W.; He, X.; Luo, Q.; Cao, Y.; Zhan, X. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Five Solanaceae Species and Identification of Members Regulating Carotenoid Biosynthesis in Wolfberry. Int. J. Mol. Sci. 2022, 23, 2259. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Wang, Z.; Tang, X.; Yu, L.; Qi, G.; Wang, D.; Yan, X.; Kong, Y.; Zhou, G. R2R3-MYB gene pairs in Populus: Evolution and contribution to secondary wall formation and flowering time. J. Exp. Bot. 2014, 65, 4255–4269. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, Z.; Guo, Q.; Yao, W.; Liu, H.; Zhou, B.; Jiang, T. Characterization of the Poplar R2R3-MYB Gene Family and Over-Expression of PsnMYB108 Confers Salt Tolerance in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 571881. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Guo, T.; Guo, B.; Chen, Z.; An, X. Comprehensive analysis of the R2R3-MYB transcription factor gene family in Populus trichocarpa. Ind. Crops Prod. 2021, 168, 7. [Google Scholar] [CrossRef]
- Zhou, F.; Chen, Y.; Wu, H.; Yin, T. Genome-Wide Comparative Analysis of R2R3 MYB Gene Family in Populus and Salix and Identification of Male Flower Bud Development-Related Genes. Front. Plant Sci. 2021, 12, 721558. [Google Scholar] [CrossRef]
- Singh, P.; Mukhopadhyay, K. Comprehensive molecular dissection of TIFY Transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat (Triticum aestivum L.). Sci. Rep. 2021, 11, 9739. [Google Scholar] [CrossRef]
- Zeng, D.; Dai, L.J.; Li, X.; Li, W.; Qu, G.Z.; Li, S. Genome-Wide Identification of the ERF Transcription Factor Family for Structure Analysis, Expression Pattern, and Response to Drought Stress in Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2023, 24, 3697. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, W.; Xia, P.; Yin, J.; Chen, H.; Li, W.; Ma, D. Genome-Wide Identification Transcriptional Expression Analysis of E2F-DP Transcription Factor Family in Wheat. Plant Mol. Biol. Report. 2022, 40, 339–358. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Seidl, V.; Seiboth, B.; Karaffa, L.; Kubicek, C.P. The fungal STRE-element-binding protein Seb1 is involved but not essential for glycerol dehydrogenase (gld1) gene expression and glycerol accumulation in Trichoderma atroviride during osmotic stress. Fungal Genet. Biol. 2004, 41, 1132–1140. [Google Scholar] [CrossRef]
- Liang, J.; Zhang, H.; Yi, L.; Tang, Y.; Long, H.; Yu, M.; Deng, G. Identification of HvLRX, a new dehydration and light responsive gene in Tibetan hulless barley (Hordeum vulgare var. nudum). Genes Genom. 2021, 43, 1445–1461. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M. Survey of Promoters Cis-elements and Genes Co- expression of Oxidative Defense Pathway in Arabidopsis thaliana L. Plant. Int. J. Agric. Crop Sci. 2012, 4, 1021–1025. [Google Scholar]
- Goldsbrough, A.P.; Albrecht, H.; Stratford, R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 1993, 3, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.K.; Rao, G.Y. Insights into the Diversification and Evolution of R2R3-MYB Transcription Factors in Plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB Transcription Factors—Functions outside the DNA-Binding Domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wall, P.K.; Leebens-Mack, J.H.; Lindsay, B.G.; Soltis, D.E.; Doyle, J.J.; Soltis, P.S.; Carlson, J.E.; Arumuganathan, K.; Barakat, A.; et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006, 16, 738–749. [Google Scholar] [CrossRef]
- Berlin, S.; Lagercrantz, U.; von Arnold, S.; Ost, T.; Rönnberg-Wästljung, A.C. High-density linkage mapping and evolution of paralogs and orthologs in Salix and Populus. BMC Genom. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- González, M.; Carrasco, B.; Salazar, E. Genome-wide identification and characterization of R2R3MYB family in Rosaceae. Genom. Data 2016, 9, 50–57. [Google Scholar] [CrossRef]
- Luan, X.; Xu, W.; Zhang, J.; Shen, T.; Chen, C.; Xi, M.; Zhong, Y.; Xu, M. Genome-Scale Identification, Classification, and Expression Profiling of MYB Transcription Factor Genes in Cinnamomum camphora. Int. J. Mol. Sci. 2022, 23, 14279. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef]
- Peng, X.; Liu, H.; Wang, D.; Shen, S. Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016, 17, 251. [Google Scholar] [CrossRef]
- Yang, X.; Guo, T.; Li, J.; Chen, Z.; Guo, B.; An, X. Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int. J. Biol. Macromol. 2021, 191, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhu, S.; Zhu, L.; Wang, D.; Liu, Y.; Liu, S.; Zhang, J.; Hao, Z.; Lu, Y.; Cheng, T.; et al. Characterization of the Liriodendron Chinense MYB Gene Family and Its Role in Abiotic Stress Response. Front. Plant Sci. 2021, 12, 641280. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef]
- Wei, Q.; Chen, R.; Wei, X.; Liu, Y.; Zhao, S.; Yin, X.; Xie, T. Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genom. 2020, 21, 792. [Google Scholar] [CrossRef]
- Oh, I.H.; Reddy, E.P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999, 18, 3017–3033. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Smita, S.; Katiyar, A.; Chinnusamy, V.; Pandey, D.M.; Bansal, K.C. Transcriptional Regulatory Network Analysis of MYB Transcription Factor Family Genes in Rice. Front. Plant Sci. 2015, 6, 1157. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene Expression and Yeast Two-Hybrid Studies of 1R-MYB Transcription Factor Mediating Drought Stress Response in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2015, 6, 1117. [Google Scholar] [CrossRef]
- Zhang, X.L.; Chen, Y.; Huang, M.R.; Zhu, S. Research progress on drought-responsive transcription factors of Populus. J. Northwest A & F Univ. (Nat. Sci. Ed. ) 2023, 51, 2–14. [Google Scholar]
- Qu, X.; Zou, J.; Wang, J.; Yang, K.; Wang, X.; Le, J. A Rice R2R3-Type MYB Transcription Factor OsFLP Positively Regulates Drought Stress Response via OsNAC. Int. J. Mol. Sci. 2022, 23, 5873. [Google Scholar] [CrossRef]
- Davidson, E.H.; Rast, J.P.; Oliveri, P.; Ransick, A.; Calestani, C.; Yuh, C.H.; Minokawa, T.; Amore, G.; Hinman, V.; Arenas-Mena, C.; et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 2002, 246, 162–190. [Google Scholar] [CrossRef] [PubMed]
- Otim, O. An empirical model of Onecut binding activity at the sea urchin SM50 C-element gene regulatory region. Int. J. Dev. Biol. 2017, 61, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J Plant Physiol 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- Liu, Y.; Man, J.; Wang, Y.; Yuan, C.; Shi, Y.; Liu, B.; Hu, X.; Wu, S.; Zhang, T.; Lian, C. Overexpression of PtrMYB121 Positively Regulates the Formation of Secondary Cell Wall in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 7734. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinformatics 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Hamilton, M.; Dharmawardhana, P.D.; Singh, S.K.; Sullivan, C.; Ben-Hur, A.; Reddy, A.S.N.; Jaiswal, P. Abiotic Stresses Modulate Landscape of Poplar Transcriptome via Alternative Splicing, Differential Intron Retention, and Isoform Ratio Switching. Front. Plant Sci. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 2012, 40, e49. [Google Scholar] [CrossRef]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zhou, S.L.; Ma, H.; Zhang, L.S. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Liu, B.; Wang, L.; Zhang, L.; Hu, J.; Chen, J.; Zheng, H.; Lu, M. Characterization of the Populus Rab family genes and the function of PtRabE1b in salt tolerance. BMC Plant Biol. 2018, 18, 124. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.Y.; Xu, M.; Chen, Y.; Huang, M.R. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tissue Organ Cult. 2013, 114, 11–18. [Google Scholar] [CrossRef]
- Fang, Z.; Ji, Y.; Hu, J.; Guo, R.; Sun, S.; Wang, X. Strigolactones and Brassinosteroids Antagonistically Regulate the Stability of the D53-OsBZR1 Complex to Determine FC1 Expression in Rice Tillering. Mol. Plant 2020, 13, 586–597. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).