Bottom-Up Proteomics: Advancements in Sample Preparation
Abstract
1. Introduction
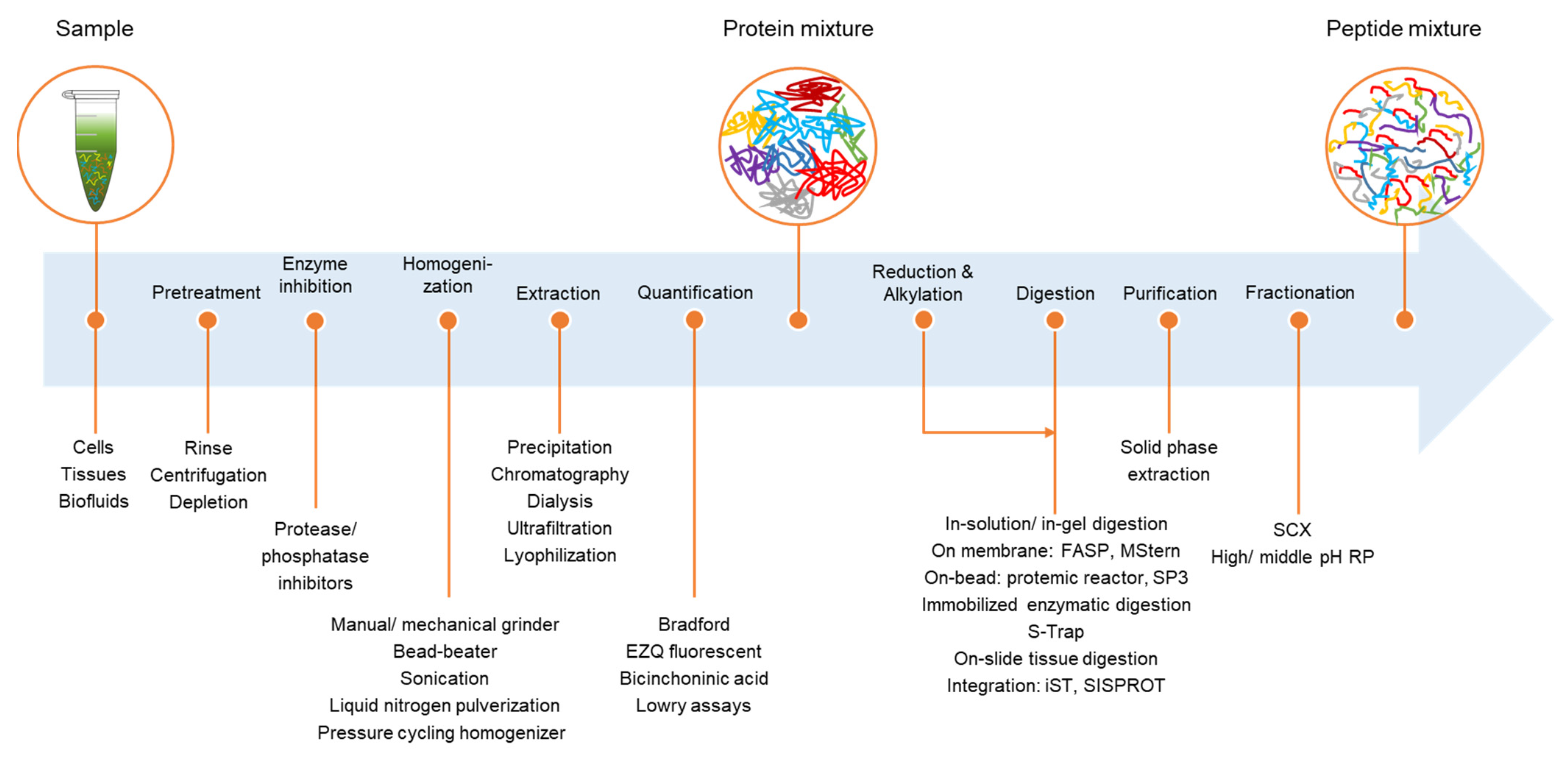
2. From Biological Samples to Proteins
2.1. Cells
2.2. Biological Fluids
2.3. Tissues
2.4. Protein Quantification
3. From Proteins to Peptides
3.1. Protein Digestion
3.1.1. In-Solution and In-Gel Digestion
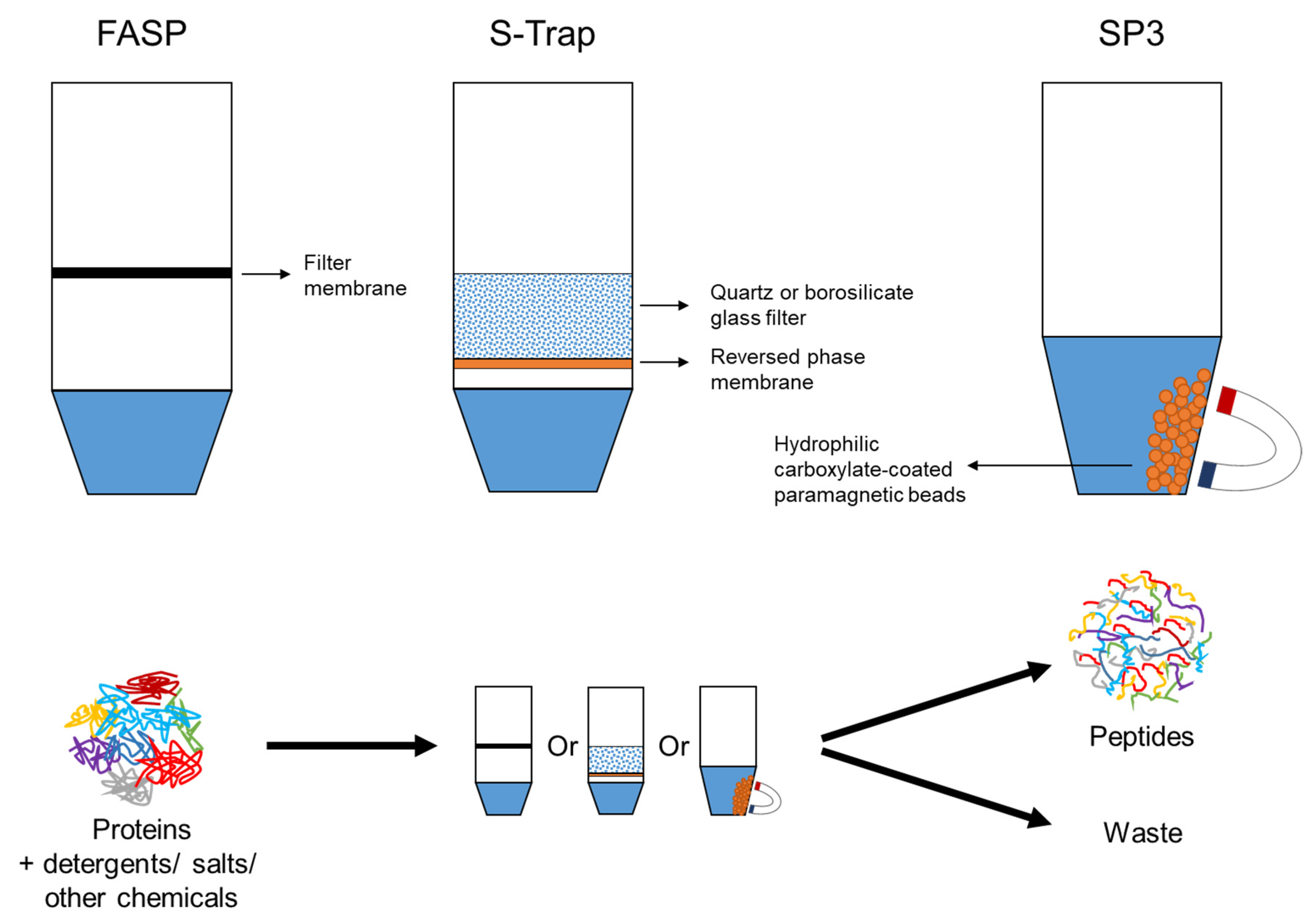
3.1.2. On-Membrane Digestion: FASP and MStern
3.1.3. Bead-Based Methods: Proteomic Reactor and SP3
3.1.4. Immobilized Enzymatic Digestion
3.1.5. Suspension-Trapping Method
3.1.6. On-Slide Digestion
3.1.7. Enzymes for Digestion
3.1.8. Enrichment of Post-Translational Modifications
3.2. Peptide Purification
3.3. Peptide Fractionation
4. Integration of Protein Digestion, Peptide Clean-Up, and Fractionation
5. Authors’ Outlook and Concluding Remarks
| Method | Feature | Ref. | |
|---|---|---|---|
| ISD | Urea-based ISD | Proteins are not separated from contaminants during reduction, alkylation, and digestion; not applicable for detergent-containing samples; sample loss; large starting amount (>100 µg protein); long digestion time; low throughput and reproducibility | [97] |
| SCASP | Use cyclodextrin to remove SDS before digestion | [98] | |
| Simultaneous lysis, reduction, and alkylation | Use TCEP and CAA; reduce time and sample loss | [99] | |
| SPEED | Use pure TFA to dissolve cells and tissues without strong detergent; use tris(hydroxymethyl)-aminomethane to neutralize sample and precipitate proteins; reduce time and sample loss | [101] | |
| In-gel digestion | Use polyacrylamide gel electrophoresis to separate proteins; cut, digest, and analyze gel spots separately; low throughput and reproducibility | [53] | |
| On-membrane digestion | FASP | Use a membrane (3000 or 10,000 Da) to separate proteins from detergents and contaminants; on-membrane digestion; tolerant to strong detergent; long centrifugal time; reduced performance with samples of low protein amount (<20 μg) | [54,102,106] |
| N-Glyco-FASP | Use lectins to enrich and PNGase F to deglycosylate N-glycopeptides | [161] | |
| MED-FASP | Use multiple enzymes for digestion | [162] | |
| iFASP | Combine FASP with TMT or iTRAQ | [107] | |
| eFASP | Use 0.2% deoxycholic acid instead of urea; increase efficiency of trypsin digestion | [108] | |
| Express eFASP | Use TCEP and 4-vinylpyridine for simultaneous reduction and alkylation | [108] | |
| MicroFASP | Use a filter with surface area of ∼0.1 mm2 to process low amount samples; applicable to samples of 100 cells or 1 μg protein | [112] | |
| μFASP | Use 96-well plates with small filter area (∼0.8 mm2) to process low amount samples; applicable to samples of 0.4 μg protein | [113] | |
| MStern | Use a membrane with 0.45 µm pore size to reduce processing time; high number of missed cleaved proteins | [114] | |
| fa-SPEED | Use pure TFA (similar to SPEED); use acetone to facilitate the protein aggregation; use 0.2 µm spin filter to reduce centrifugation time; quick hands-on time (~22 min, excluding digestion) | [101] | |
| Bead-based digestion | Proteomic reactor | Use SCX or SAX beads to bind proteins; all steps are performed in a small volume (~50 nL) of a capillary; applicable to samples of low protein amount (<10 μg) | [24,116] |
| SP3 | Use hydrophilic carboxylate-coated paramagnetic beads to bind proteins; use a magnetic rack to separate proteins from contaminants; applicable to low protein amount (~100 ng); bead clumping and aggregation | [56,121] | |
| USP3 | Use TFA to hydrolyze DNA and RNA; use TCEP and CAA; reduce time | [124] | |
| C4-tip | Use C4 RP resin tip to entrap proteins; use 30% acetonitrile in digestion buffer to increase peptide recovery and reduce missed cleavage percentage | [125] | |
| IMERs | Flow-through devices contain entrapped enzymes; less reagent consumption; fast reaction rate; applicable to low protein amount (<1 μg); integration with fractionation and LC-MS/MS; complex instrumental setups | [127,128] | |
| S-Trap | Use methanol to precipitate proteins; use a quartz or borosilicate glass depth filter to trap proteins; quick centrifugal time (~1 min per step, total process time ~20–30 min); integration with RP fractionation | [55,115] | |
| On-slide digestion | Applicable to FFPE and fresh frozen tissue slides; suitable for MALDI IMS | [143,144] | |
| Integrated methods | iST | Combine C18 membrane (filter) and SCX or SAX disks (fractionation); all steps are performed in a device; sample loss due to C18 material-protein binding; applicable to low protein amount (~1 µg) | [99,106] |
| Micro-FASP + RPLC | Combine micro-FASP and C18 microreactor (sample preparation and fractionation); chemical solution volumes ~5 µL | [176] | |
| RCPR | Use SCX column for cell loading, protein reduction, alkylation, and digestion; combine the SCX column and a C18 RP column for 2D-LC | [177,178] | |
| SISPROT | Integrate SCX or SAX or SCX+SAX beads with C18 disks in a pipet tip for digestion and fractionation (1 or 2 dimensions); applicable to low protein amount (~1 µL serum) | [23,32,34] | |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nesvizhskii, A.I.; Vitek, O.; Aebersold, R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 2007, 4, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Bludau, I.; Aebersold, R. Proteomic and interactomic insights into the molecular basis of cell functional diversity. Nat. Rev. Mol. Cell Biol. 2020, 21, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Agarwal, A.; Young, J.N.; Owji, S.; Luu, Y.; Poplausky, D.; Yassky, D.; Estrada, Y.; Ungar, J.; Krueger, J.G.; et al. Proteomic profiling of a patient with cutaneous melanoma metastasis regression following topical contact sensitizer diphencyprone and immune checkpoint inhibitor treatment. Sci. Rep. 2022, 12, 22364. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Shin, H.A.; Duong, V.-A.; Lee, H.; Lew, H. The Role of Extracellular Vesicles in Optic Nerve Injury: Neuroprotection and Mitochondrial Homeostasis. Cells 2022, 11, 3720. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, C.; Wang, B.; Xu, F.; Ma, F.; Qu, Y.; Jiang, D.; Li, K.; Feng, J.; Tian, S.; et al. Proteomic characterization of gastric cancer response to chemotherapy and targeted therapy reveals potential therapeutic strategies. Nat. Commun. 2022, 13, 5723. [Google Scholar] [CrossRef] [PubMed]
- Pedde, R.D.; Li, H.; Borchers, C.H.; Akbari, M. Microfluidic-Mass Spectrometry Interfaces for Translational Proteomics. Trends Biotechnol. 2017, 35, 954–970. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Paša-Tolić, L. High-Throughput Proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef]
- Alexovič, M.; Urban, P.L.; Tabani, H.; Sabo, J. Recent advances in robotic protein sample preparation for clinical analysis and other biomedical applications. Clin. Chim. Acta 2020, 507, 104–116. [Google Scholar] [CrossRef]
- Kuras, M.; Betancourt, L.H.; Rezeli, M.; Rodriguez, J.; Szasz, M.; Zhou, Q.; Miliotis, T.; Andersson, R.; Marko-Varga, G. Assessing Automated Sample Preparation Technologies for High-Throughput Proteomics of Frozen Well Characterized Tissues from Swedish Biobanks. J. Proteome Res. 2019, 18, 548–556. [Google Scholar] [CrossRef]
- Fu, Q.; Kowalski, M.P.; Mastali, M.; Parker, S.J.; Sobhani, K.; van den Broek, I.; Hunter, C.L.; Van Eyk, J.E. Highly Reproducible Automated Proteomics Sample Preparation Workflow for Quantitative Mass Spectrometry. J. Proteome Res. 2018, 17, 420–428. [Google Scholar] [CrossRef]
- Brodbelt, J.S. Deciphering combinatorial post-translational modifications by top-down mass spectrometry. Curr. Opin. Chem. Biol. 2022, 70, 102180. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.H.; Wang, Y.S. Advances in high-resolution mass spectrometry techniques for analysis of high mass-to-charge ions. Mass Spectrom. Rev. 2022. early review. [Google Scholar] [CrossRef] [PubMed]
- McCool, E.N.; Lubeckyj, R.A.; Shen, X.; Chen, D.; Kou, Q.; Liu, X.; Sun, L. Deep Top-Down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia coli Proteome. Anal. Chem. 2018, 90, 5529–5533. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.M.; Smith, L.M. Overview and considerations in bottom-up proteomics. Analyst 2023, 148, 475–486. [Google Scholar] [CrossRef]
- Duong, V.-A.; Park, J.-M.; Lee, H. Review of Three-Dimensional Liquid Chromatography Platforms for Bottom-Up Proteomics. Int. J. Mol. Sci. 2020, 21, 1524. [Google Scholar] [CrossRef]
- Duong, V.-A.; Park, J.-M.; Lim, H.-J.; Lee, H. Proteomics in Forensic Analysis: Applications for Human Samples. Appl. Sci. 2021, 11, 3393. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, L.; Zhang, Y. Chromatographic separation of peptides and proteins for characterization of proteomes. Chem. Commun. 2023, 59, 270–281. [Google Scholar] [CrossRef]
- Nickerson, J.L.; Baghalabadi, V.; Rajendran, S.R.C.K.; Jakubec, P.J.; Said, H.; McMillen, T.S.; Dang, Z.; Doucette, A.A. Recent advances in top-down proteome sample processing ahead of MS analysis. Mass Spectrom. Rev. 2021, 42, 457–495. [Google Scholar] [CrossRef]
- Woldmar, N.; Schwendenwein, A.; Kuras, M.; Szeitz, B.; Boettiger, K.; Tisza, A.; László, V.; Reiniger, L.; Bagó, A.G.; Szállási, Z.; et al. Proteomic analysis of brain metastatic lung adenocarcinoma reveals intertumoral heterogeneity and specific alterations associated with the timing of brain metastases. ESMO Open. 2023, 8, 100741. [Google Scholar] [CrossRef]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Shotgun Proteomics as a Powerful Tool for the Study of the Proteomes of Plants, Their Pathogens, and Plant–Pathogen Interactions. Proteomes 2022, 10, 5. [Google Scholar] [CrossRef]
- Lenčo, J.; Jadeja, S.; Naplekov, D.K.; Krokhin, O.V.; Khalikova, M.A.; Chocholouš, P.; Urban, J.; Broeckhoven, K.; Nováková, L.; Švec, F. Reversed-Phase Liquid Chromatography of Peptides for Bottom-Up Proteomics: A Tutorial. J. Proteome Res. 2022, 21, 2846–2892. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.-Z.; Li, N.; Wang, Y.-T.; Liu, N.; Guo, M.-Q.; Sun, B.-q.; Zhou, H.; Liu, L.; Wu, J.-L. Acid/Salt/pH Gradient Improved Resolution and Sensitivity in Proteomics Study Using 2D SCX-RP LC–MS. J. Proteome Res. 2017, 16, 3470–3475. [Google Scholar] [CrossRef]
- Chen, W.; Wang, S.; Adhikari, S.; Deng, Z.; Wang, L.; Chen, L.; Ke, M.; Yang, P.; Tian, R. Simple and Integrated Spintip-Based Technology Applied for Deep Proteome Profiling. Anal. Chem. 2016, 88, 4864–4871. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Hou, W.; Lambert, J.-P.; Tian, R.; Figeys, D. Analysis of low-abundance proteins using the proteomic reactor with pH fractionation. Talanta 2010, 80, 1526–1531. [Google Scholar] [CrossRef]
- Lee, H.-J.; Na, K.; Kwon, M.-S.; Park, T.; Kim, K.S.; Kim, H.; Paik, Y.-K. A new versatile peptide-based size exclusion chromatography platform for global profiling and quantitation of candidate biomarkers in hepatocellular carcinoma specimens. Proteomics 2011, 11, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Garbis, S.D.; Roumeliotis, T.I.; Tyritzis, S.I.; Zorpas, K.M.; Pavlakis, K.; Constantinides, C.A. A Novel Multidimensional Protein Identification Technology Approach Combining Protein Size Exclusion Prefractionation, Peptide Zwitterion−Ion Hydrophilic Interaction Chromatography, and Nano-Ultraperformance RP Chromatography/nESI-MS2 for the in-Depth Analysis of the Serum Proteome and Phosphoproteome: Application to Clinical Sera Derived from Humans with Benign Prostate Hyperplasia. Anal. Chem. 2011, 83, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, S.; Boersema, P.J.; Heck, A.J.R.; Mohammed, S. Zwitterionic Hydrophilic Interaction Liquid Chromatography (ZIC-HILIC and ZIC-cHILIC) Provide High Resolution Separation and Increase Sensitivity in Proteome Analysis. Anal. Chem. 2011, 83, 3440–3447. [Google Scholar] [CrossRef]
- Cao, J.-L.; Wang, S.-S.; Hu, H.; He, C.-W.; Wan, J.-B.; Su, H.-X.; Wang, Y.-T.; Li, P. Online comprehensive two-dimensional hydrophilic interaction chromatography×reversed-phase liquid chromatography coupled with hybrid linear ion trap Orbitrap mass spectrometry for the analysis of phenolic acids in Salvia miltiorrhiza. J. Chromatogr. A 2018, 1536, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Tsai, C.-F.; Piehowski, P.D.; Wang, Y.; Fillmore, T.L.; Zhao, R.; Moore, R.J.; Zhang, P.; Qian, W.-J.; Smith, R.D.; et al. Automated Nanoflow Two-Dimensional Reversed-Phase Liquid Chromatography System Enables In-Depth Proteome and Phosphoproteome Profiling of Nanoscale Samples. Anal. Chem. 2019, 91, 9707–9715. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Zhu, Y.; Liyu, A.; Liang, Y.; Chen, J.; Piehowski, P.D.; Xu, K.; Zhao, R.; Moore, R.J.; Atkinson, M.A.; et al. Nanowell-mediated two-dimensional liquid chromatography enables deep proteome profiling of <1000 mammalian cells. Chem. Sci. 2018, 9, 6944–6951. [Google Scholar] [CrossRef]
- Zhao, P.; Schulz, T.C.; Sherrer, E.S.; Weatherly, D.B.; Robins, A.J.; Wells, L. The human embryonic stem cell proteome revealed by multidimensional fractionation followed by tandem mass spectrometry. Proteomics 2015, 15, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Lin, L.; Zhou, W.; Chen, W.; Tang, J.; Sun, X.; Huang, P.; Tian, R. Mixed-mode ion exchange-based integrated proteomics technology for fast and deep plasma proteome profiling. J. Chromatogr. A 2018, 1564, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.-A.; Nam, O.; Jin, E.; Park, J.-M.; Lee, H. Discovery of Post-Translational Modifications in Emiliania huxleyi. Molecules 2021, 26, 2027. [Google Scholar] [CrossRef]
- Chen, W.; Adhikari, S.; Chen, L.; Lin, L.; Li, H.; Luo, S.; Yang, P.; Tian, R. 3D-SISPROT: A simple and integrated spintip-based protein digestion and three-dimensional peptide fractionation technology for deep proteome profiling. J. Chromatogr. A 2017, 1498, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.R.; Mohammed, S. Toward a Comprehensive Characterization of a Human Cancer Cell Phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Spicer, V.; Ezzati, P.; Neustaeter, H.; Beavis, R.C.; Wilkins, J.A.; Krokhin, O.V. 3D HPLC-MS with Reversed-Phase Separation Functionality in All Three Dimensions for Large-Scale Bottom-Up Proteomics and Peptide Retention Data Collection. Anal. Chem. 2016, 88, 2847–2855. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Han, X.; Aslanian, A.; Yates, J.R. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef]
- Woods, A.G.; Sokolowska, I.; Ngounou Wetie, A.G.; Channaveerappa, D.; Dupree, E.J.; Jayathirtha, M.; Aslebagh, R.; Wormwood, K.L.; Darie, C.C. Mass Spectrometry for Proteomics-Based Investigation. In Advancements of Mass Spectrometry in Biomedical Research; Woods, A.G., Darie, C.C., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 1–26. [Google Scholar] [CrossRef]
- Schessner, J.P.; Voytik, E.; Bludau, I. A practical guide to interpreting and generating bottom-up proteomics data visualizations. Proteomics 2022, 22, 2100103. [Google Scholar] [CrossRef]
- Craig, R.; Beavis, R.C. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, R.; Yang, Z.; Lee, J.; Liu, Y.; Tian, J.; Qin, X.; Ren, Z.; Ding, H.; Chen, Q.; et al. Comparative Effectiveness and Safety of Oral Phosphodiesterase Type 5 Inhibitors for Erectile Dysfunction: A Systematic Review and Network Meta-analysis. Eur. Urol. 2013, 63, 902–912. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Bern, M.; Kil, Y.J.; Becker, C. Byonic: Advanced Peptide and Protein Identification Software. Curr. Protoc. Bioinform. 2012, 40, 13–20. [Google Scholar] [CrossRef]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Chi, H.; Liu, C.; Yang, H.; Zeng, W.-F.; Wu, L.; Zhou, W.-J.; Wang, R.-M.; Niu, X.-N.; Ding, Y.-H.; Zhang, Y.; et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018, 36, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Danko, K.; Lukasheva, E.; Zhukov, V.A.; Zgoda, V.; Frolov, A. Detergent-Assisted Protein Digestion—On the Way to Avoid the Key Bottleneck of Shotgun Bottom-Up Proteomics. Int. J. Mol. Sci. 2022, 23, 13903. [Google Scholar]
- Merkley, E.D.; Kaiser, B.L.D.; Kreuzer, H. A Proteomics Tutorial. In Applications in Forensic Proteomics: Protein Identification and Profiling; American Chemical Society: Washington, DC, USA, 2019; Volume 1339, pp. 9–28. [Google Scholar]
- Yang, Z.; Sun, L. Recent technical progress in sample preparation and liquid-phase separation-mass spectrometry for proteomic analysis of mass-limited samples. Anal. Methods 2021, 13, 1214–1225. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. PROTEOMICS 2014, 14, 1000–1006. [Google Scholar] [CrossRef]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef]
- Alexovič, M.; Sabo, J.; Longuespée, R. Microproteomic sample preparation. Proteomics 2021, 21, 2000318. [Google Scholar] [CrossRef]
- Zhu, Y.; Piehowski, P.D.; Zhao, R.; Chen, J.; Shen, Y.; Moore, R.J.; Shukla, A.K.; Petyuk, V.A.; Campbell-Thompson, M.; Mathews, C.E.; et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 2018, 9, 882. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Huang, M.; Wang, X.-K.; Zhu, Y.; Li, J.-S.; Wong, C.C.L.; Fang, Q. Nanoliter-Scale Oil-Air-Droplet Chip-Based Single Cell Proteomic Analysis. Anal. Chem. 2018, 90, 5430–5438. [Google Scholar] [CrossRef]
- Shao, X.; Wang, X.; Guan, S.; Lin, H.; Yan, G.; Gao, M.; Deng, C.; Zhang, X. Integrated Proteome Analysis Device for Fast Single-Cell Protein Profiling. Anal. Chem. 2018, 90, 14003–14010. [Google Scholar] [CrossRef]
- Lamanna, J.; Scott, E.Y.; Edwards, H.S.; Chamberlain, M.D.; Dryden, M.D.M.; Peng, J.; Mair, B.; Lee, A.; Chan, C.; Sklavounos, A.A.; et al. Digital microfluidic isolation of single cells for -Omics. Nat. Commun. 2020, 11, 5632. [Google Scholar] [CrossRef]
- Senavirathna, L.; Ma, C.; Chen, R.; Pan, S. Spectral Library-Based Single-Cell Proteomics Resolves Cellular Heterogeneity. Cells 2022, 11, 2450. [Google Scholar] [CrossRef]
- Kwon, D.; Park, J.-M.; Duong, V.-A.; Hong, S.-J.; Cho, B.-K.; Lee, C.-G.; Choi, H.-K.; Kim, D.-M.; Lee, H. Comparative Proteomic Profiling of Marine and Freshwater Synechocystis Strains Using Liquid Chromatography-Tandem Mass Spectrometry. J. Mar. Sci. Eng. 2020, 8, 790. [Google Scholar] [CrossRef]
- Havanapan, P.-o.; Thongboonkerd, V. Are Protease Inhibitors Required for Gel-Based Proteomics of Kidney and Urine? J. Proteome Res. 2009, 8, 3109–3117. [Google Scholar] [CrossRef]
- Thingholm, T.E.; Larsen, M.R.; Ingrell, C.R.; Kassem, M.; Jensen, O.N. TiO2-Based Phosphoproteomic Analysis of the Plasma Membrane and the Effects of Phosphatase Inhibitor Treatment. J. Proteome Res. 2008, 7, 3304–3313. [Google Scholar] [CrossRef]
- Yun, G.; Park, J.-M.; Duong, V.-A.; Mok, J.-H.; Jeon, J.; Nam, O.; Lee, J.; Jin, E.; Lee, H. Proteomic Profiling of Emiliania huxleyi Using a Three-Dimensional Separation Method Combined with Tandem Mass Spectrometry. Molecules 2020, 25, 3028. [Google Scholar] [CrossRef]
- Santa, C.; Anjo, S.I.; Manadas, B. Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach. Proteomics 2016, 16, 1847–1851. [Google Scholar] [CrossRef]
- Evans, D.R.H.; Romero, J.K.; Westoby, M. Chapter 9 Concentration of Proteins and Removal of Solutes. In Methods in Enzymology; Burgess, R.R., Deutscher, M.P., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 97–120. [Google Scholar]
- Koh, K.; Park, M.; Bae, E.S.; Duong, V.-A.; Park, J.-M.; Lee, H.; Lew, H. UBA2 activates Wnt/β-catenin signaling pathway during protection of R28 retinal precursor cells from hypoxia by extracellular vesicles derived from placental mesenchymal stem cells. Stem Cell Res. Ther. 2020, 11, 428. [Google Scholar] [CrossRef]
- Van-An, D.; Jeeyun, A.; Na-Young, H.; Jong-Moon, P.; Jeong-Hun, M.; Tae Wan, K.; Hookeun, L. Proteomic Analysis of the Vitreous Body in Proliferative and Non-Proliferative Diabetic Retinopathy. Curr. Proteom. 2020, 18, 143–152. [Google Scholar] [CrossRef]
- Olszowy, P.; Buszewski, B. Urine sample preparation for proteomic analysis. J. Sep. Sci. 2014, 37, 2920–2928. [Google Scholar] [CrossRef]
- Jesus, J.R.; Santos, H.M.; López-Fernández, H.; Lodeiro, C.; Arruda, M.A.Z.; Capelo, J.L. Ultrasonic-based membrane aided sample preparation of urine proteomes. Talanta 2018, 178, 864–869. [Google Scholar] [CrossRef]
- Cao, X.; Sandberg, A.; Araújo, J.E.; Cvetkovski, F.; Berglund, E.; Eriksson, L.E.; Pernemalm, M. Evaluation of Spin Columns for Human Plasma Depletion to Facilitate MS-Based Proteomics Analysis of Plasma. J. Proteome Res. 2021, 20, 4610–4620. [Google Scholar] [CrossRef]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2013, 42, D959–D965. [Google Scholar] [CrossRef]
- Pringels, L.; Broeckx, V.; Boonen, K.; Landuyt, B.; Schoofs, L. Abundant plasma protein depletion using ammonium sulfate precipitation and Protein A affinity chromatography. J. Chromatogr. B 2018, 1089, 43–59. [Google Scholar] [CrossRef]
- Beer, L.A.; Ky, B.; Barnhart, K.T.; Speicher, D.W. In-Depth, Reproducible Analysis of Human Plasma Using IgY 14 and SuperMix Immunodepletion. In Serum/Plasma Proteomics: Methods and Protocols; Greening, D.W., Simpson, R.J., Eds.; Springer: New York, NY, USA, 2017; pp. 81–101. [Google Scholar] [CrossRef]
- Wu, C.; Duan, J.; Liu, T.; Smith, R.D.; Qian, W.-J. Contributions of immunoaffinity chromatography to deep proteome profiling of human biofluids. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1021, 57–68. [Google Scholar] [CrossRef]
- Blume, J.E.; Manning, W.C.; Troiano, G.; Hornburg, D.; Figa, M.; Hesterberg, L.; Platt, T.L.; Zhao, X.; Cuaresma, R.A.; Everley, P.A.; et al. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat. Commun. 2020, 11, 3662. [Google Scholar] [CrossRef]
- Tu, C.; Rudnick, P.A.; Martinez, M.Y.; Cheek, K.L.; Stein, S.E.; Slebos, R.J.C.; Liebler, D.C. Depletion of abundant plasma proteins and limitations of plasma proteomics. J. Proteome Res. 2010, 9, 4982–4991. [Google Scholar] [CrossRef]
- Oliveira, B.P.; Buzalaf, M.A.R.; Silva, N.C.; Ventura, T.M.O.; Toniolo, J.; Rodrigues, J.A. Saliva proteomic profile of early childhood caries and caries-free children. Acta Odontol. Scand. 2022, 1–11. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Meng, Q.; Zheng, Q.; Wu, J.; Wang, C.; Jia, W.; Figeys, D.; Chang, Y.; Zhou, H. Proteomic analysis of minute amount of colonic biopsies by enteroscopy sampling. Biochem. Biophys. Res. Commun. 2016, 476, 286–292. [Google Scholar] [CrossRef]
- Toni, M.; Angiulli, E.; Miccoli, G.; Cioni, C.; Alleva, E.; Frabetti, F.; Pizzetti, F.; Grassi Scalvini, F.; Nonnis, S.; Negri, A.; et al. Environmental temperature variation affects brain protein expression and cognitive abilities in adult zebrafish (Danio rerio): A proteomic and behavioural study. J. Proteom. 2019, 204, 103396. [Google Scholar] [CrossRef]
- Smith, K.M.; Xu, Y. Tissue sample preparation in bioanalytical assays. Bioanalysis 2012, 4, 741–749. [Google Scholar] [CrossRef]
- Yagi, R.; Masuda, T.; Ogata, S.; Mori, A.; Ito, S.; Ohtsuki, S. Proteomic Evaluation of Plasma Membrane Fraction Prepared from a Mouse Liver and Kidney Using a Bead Homogenizer: Enrichment of Drug-Related Transporter Proteins. Mol. Pharm. 2020, 17, 4101–4113. [Google Scholar] [CrossRef]
- Liu, W.; Xie, L.; He, Y.-H.; Wu, Z.-Y.; Liu, L.-X.; Bai, X.-F.; Deng, D.-X.; Xu, X.-E.; Liao, L.-D.; Lin, W.; et al. Large-scale and high-resolution mass spectrometry-based proteomics profiling defines molecular subtypes of esophageal cancer for therapeutic targeting. Nat. Commun. 2021, 12, 4961. [Google Scholar] [CrossRef] [PubMed]
- Dubacq, S. Performing efficient sample preparation with hard tumor tissue: Precellys® bead-beating homogenizer solution. Nat. Methods 2016, 13, i–iii. [Google Scholar] [CrossRef]
- Prieto, D.A.; Whitely, G.; Johann, D.J.; Blonder, J. Protocol for the Analysis of Laser Capture Microdissected Fresh-Frozen Tissue Homogenates by Silver-Stained 1D SDS-PAGE. In Laser Capture Microdissection: Methods and Protocols; Murray, G.I., Ed.; Springer: New York, NY, USA, 2018; pp. 95–110. [Google Scholar] [CrossRef]
- Mertins, P.; Tang, L.C.; Krug, K.; Clark, D.J.; Gritsenko, M.A.; Chen, L.; Clauser, K.R.; Clauss, T.R.; Shah, P.; Gillette, M.A.; et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat. Protoc. 2018, 13, 1632–1661. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean Roots Grown under Heat Stress Show Global Changes in Their Transcriptional and Proteomic Profiles. Front. Plant Sci. 2016, 7, 517. [Google Scholar] [CrossRef] [PubMed]
- Guyuron, B.; Yohannes, E.; Miller, R.; Chim, H.; Reed, D.; Chance, M.R. Electron Microscopic and Proteomic Comparison of Terminal Branches of the Trigeminal Nerve in Patients with and without Migraine Headaches. Plast. Reconstr. Surg. 2014, 134, 796e–805e. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Xue, Z.; Wu, C.; Sun, R.; Qian, L.; Yue, L.; Ge, W.; Yi, X.; Liu, W.; Chen, C.; et al. High-throughput proteomic sample preparation using pressure cycling technology. Nat. Protoc. 2022, 17, 2307–2325. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Donnell, A.M.; Lewis, S.; Abraham, S.; Subramanian, K.; Figueroa, J.L.; Deepe, G.S.; Vonderheide, A.P. Investigation of an optimal cell lysis method for the study of the zinc metalloproteome of Histoplasma capsulatum. Anal. Bioanal. Chem. 2017, 409, 6163–6172. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Legler, G.; Müller-Platz, C.M.; Mentges-Hettkamp, M.; Pflieger, G.; Jülich, E. On the chemical basis of the Lowry protein determination. Anal. Biochem. 1985, 150, 278–287. [Google Scholar] [CrossRef]
- Oswald, E.S.; Brown, L.M.; Bulinski, J.C.; Hung, C.T. Label-Free Protein Profiling of Adipose-Derived Human Stem Cells under Hyperosmotic Treatment. J. Proteome Res. 2011, 10, 3050–3059. [Google Scholar] [CrossRef]
- Yu, Y.-Q.; Gilar, M.; Lee, P.J.; Bouvier, E.S.P.; Gebler, J.C. Enzyme-Friendly, Mass Spectrometry-Compatible Surfactant for In-Solution Enzymatic Digestion of Proteins. Anal. Chem. 2003, 75, 6023–6028. [Google Scholar] [CrossRef]
- Gan, G.; Xu, X.; Chen, X.; Zhang, X.-F.; Wang, J.; Zhong, C.-Q. SCASP: A Simple and Robust SDS-Aided Sample Preparation Method for Proteomic Research. Mol. Cell. Proteom. 2021, 20, 100051. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Gebreyesus, S.T.; Siyal, A.A.; Kitata, R.B.; Chen, E.S.-W.; Enkhbayar, B.; Angata, T.; Lin, K.-I.; Chen, Y.-J.; Tu, H.-L. Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry. Nat. Commun. 2022, 13, 37. [Google Scholar] [CrossRef]
- Doellinger, J.; Schneider, A.; Hoeller, M.; Lasch, P. Sample Preparation by Easy Extraction and Digestion (SPEED)—A Universal, Rapid, and Detergent-free Protocol for Proteomics Based on Acid Extraction. Mol. Amp; Cell. Proteom. 2020, 19, 209–222. [Google Scholar] [CrossRef]
- Manza, L.L.; Stamer, S.L.; Ham, A.-J.L.; Codreanu, S.G.; Liebler, D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 2005, 5, 1742–1745. [Google Scholar] [CrossRef]
- Wither, M.J.; Hansen, K.C.; Reisz, J.A. Mass Spectrometry-Based Bottom-Up Proteomics: Sample Preparation, LC-MS/MS Analysis, and Database Query Strategies. Curr. Protoc. Protein Sci. 2016, 86, 16.4.1–16.4.20. [Google Scholar] [CrossRef]
- Wiśniewski, J.R. Filter Aided Sample Preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef]
- Davalieva, K.; Kiprijanovska, S.; Dimovski, A.; Rosoklija, G.; Dwork, A.J. Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues. J. Proteom. 2021, 235, 104117. [Google Scholar] [CrossRef]
- Sielaff, M.; Kuharev, J.; Bohn, T.; Hahlbrock, J.; Bopp, T.; Tenzer, S.; Distler, U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017, 16, 4060–4072. [Google Scholar] [CrossRef] [PubMed]
- McDowell, G.S.; Gaun, A.; Steen, H. iFASP: Combining Isobaric Mass Tagging with Filter-Aided Sample Preparation. J. Proteome Res. 2013, 12, 3809–3812. [Google Scholar] [CrossRef]
- Erde, J.; Loo, R.R.O.; Loo, J.A. Enhanced FASP (eFASP) to Increase Proteome Coverage and Sample Recovery for Quantitative Proteomic Experiments. J. Proteome Res. 2014, 13, 1885–1895. [Google Scholar] [CrossRef]
- Yu, Y.; Suh, M.-J.; Sikorski, P.; Kwon, K.; Nelson, K.E.; Pieper, R. Urine Sample Preparation in 96-Well Filter Plates for Quantitative Clinical Proteomics. Anal. Chem. 2014, 86, 5470–5477. [Google Scholar] [CrossRef]
- Deshmukh, A.S.; Steenberg, D.E.; Hostrup, M.; Birk, J.B.; Larsen, J.K.; Santos, A.; Kjøbsted, R.; Hingst, J.R.; Schéele, C.C.; Murgia, M.; et al. Deep muscle-proteomic analysis of freeze-dried human muscle biopsies reveals fiber type-specific adaptations to exercise training. Nat. Commun. 2021, 12, 304. [Google Scholar] [CrossRef]
- Loroch, S.; Kopczynski, D.; Schneider, A.C.; Schumbrutzki, C.; Feldmann, I.; Panagiotidis, E.; Reinders, Y.; Sakson, R.; Solari, F.A.; Vening, A.; et al. Toward Zero Variance in Proteomics Sample Preparation: Positive-Pressure FASP in 96-Well Format (PF96) Enables Highly Reproducible, Time- and Cost-Efficient Analysis of Sample Cohorts. J. Proteome Res. 2022, 21, 1181–1188. [Google Scholar] [CrossRef]
- Zhang, Z.; Dubiak, K.M.; Huber, P.W.; Dovichi, N.J. Miniaturized Filter-Aided Sample Preparation (MICRO-FASP) Method for High Throughput, Ultrasensitive Proteomics Sample Preparation Reveals Proteome Asymmetry in Xenopus laevis Embryos. Anal. Chem. 2020, 92, 5554–5560. [Google Scholar] [CrossRef]
- Sandbaumhüter, F.A.; Nezhyva, M.; Eriksson, O.; Engberg, A.; Kreuger, J.; Andrén, P.E.; Jansson, E.T. Well-Plate μFASP for Proteomic Analysis of Single Pancreatic Islets. J. Proteome Res. 2022, 21, 1167–1174. [Google Scholar] [CrossRef]
- Berger, S.T.; Ahmed, S.; Muntel, J.; Cuevas Polo, N.; Bachur, R.; Kentsis, A.; Steen, J.; Steen, H. MStern Blotting–High Throughput Polyvinylidene Fluoride (PVDF) Membrane-Based Proteomic Sample Preparation for 96-Well Plates*[S]. Mol. Cell. Proteom. 2015, 14, 2814–2823. [Google Scholar] [CrossRef]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef]
- Ethier, M.; Hou, W.; Duewel, H.S.; Figeys, D. The Proteomic Reactor: A Microfluidic Device for Processing Minute Amounts of Protein Prior to Mass Spectrometry Analysis. J. Proteome Res. 2006, 5, 2754–2759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, F.; Wang, Y.; Ning, Z.; Hou, W.; Wright, T.G.; Sundaram, M.; Zhong, S.; Yao, Z.; Figeys, D. Improved Recovery and Identification of Membrane Proteins from Rat Hepatic Cells using a Centrifugal Proteomic Reactor*. Mol. Cell. Proteom. 2011, 10, O111.008425. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Fang, L.; Yang, Y.; Jiang, H.; Yang, H.; Zhang, H.; Zhou, H. Quantitative proteomic analysis reveals the neuroprotective effects of huperzine A for amyloid beta treated neuroblastoma N2a cells. Proteomics 2013, 13, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Ethier, M.; Smith, J.C.; Sheng, Y.; Figeys, D. Multiplexed Proteomic Reactor for the Processing of Proteomic Samples. Anal. Chem. 2007, 79, 39–44. [Google Scholar] [CrossRef]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef]
- Blankenburg, S.; Hentschker, C.; Nagel, A.; Hildebrandt, P.; Michalik, S.; Dittmar, D.; Surmann, K.; Völker, U. Improving Proteome Coverage for Small Sample Amounts: An Advanced Method for Proteomics Approaches with Low Bacterial Cell Numbers. Proteomics 2019, 19, 1900192. [Google Scholar] [CrossRef]
- Paulo, J.A.; Navarrete-Perea, J.; Gygi, S.P. Multiplexed proteome profiling of carbon source perturbations in two yeast species with SL-SP3-TMT. J. Proteom. 2020, 210, 103531. [Google Scholar] [CrossRef]
- Osório, H.; Silva, C.; Ferreira, M.; Gullo, I.; Máximo, V.; Barros, R.; Mendonça, F.; Oliveira, C.; Carneiro, F. Proteomics Analysis of Gastric Cancer Patients with Diabetes Mellitus. J. Clin. Med. 2021, 10, 407. [Google Scholar] [CrossRef]
- Dagley, L.F.; Infusini, G.; Larsen, R.H.; Sandow, J.J.; Webb, A.I. Universal Solid-Phase Protein Preparation (USP3) for Bottom-up and Top-down Proteomics. J. Proteome Res. 2019, 18, 2915–2924. [Google Scholar] [CrossRef]
- Müller, T.; Kalxdorf, M.; Longuespée, R.; Kazdal, D.N.; Stenzinger, A.; Krijgsveld, J. Automated sample preparation with SP3 for low-input clinical proteomics. Mol. Syst. Biol. 2020, 16, e9111. [Google Scholar] [CrossRef]
- Clark, D.J.; Hu, Y.; Schnaubelt, M.; Fu, Y.; Ponce, S.; Chen, S.-Y.; Zhou, Y.; Shah, P.; Zhang, H. Simple Tip-Based Sample Processing Method for Urinary Proteomic Analysis. Anal. Chem. 2019, 91, 5517–5522. [Google Scholar] [CrossRef]
- Wouters, B.; Currivan, S.A.; Abdulhussain, N.; Hankemeier, T.; Schoenmakers, P.J. Immobilized-enzyme reactors integrated into analytical platforms: Recent advances and challenges. TrAC Trends Anal. Chem. 2021, 144, 116419. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Miyazaki, M. Enzyme-immobilized reactors for rapid and efficient sample preparation in MS-based proteomic studies. Proteomics 2013, 13, 457–466. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, S.; Zhao, B.; Weng, Y.; Zhu, X.; Li, S.; Zhang, L.; Zhang, Y. Enzymatic Reactor with Trypsin Immobilized on Graphene Oxide Modified Polymer Microspheres To Achieve Automated Proteome Quantification. Anal. Chem. 2017, 89, 6324–6329. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, L.; Liang, Z.; Shan, Y.; Zhang, Y. Immobilized enzyme reactors in proteomics. TrAC Trends Anal. Chem. 2011, 30, 691–702. [Google Scholar] [CrossRef]
- Nagy, C.; Szabo, R.; Gaspar, A. Microfluidic Immobilized Enzymatic Reactors for Proteomic Analyses—Recent Developments and Trends (2017–2021). Micromachines 2022, 13, 311. [Google Scholar]
- Qu, Y.; Xia, S.; Yuan, H.; Wu, Q.; Li, M.; Zou, L.; Zhang, L.; Liang, Z.; Zhang, Y. Integrated Sample Pretreatment System for N-Linked Glycosylation Site Profiling with Combination of Hydrophilic Interaction Chromatography and PNGase F Immobilized Enzymatic Reactor via a Strong Cation Exchange Precolumn. Anal. Chem. 2011, 83, 7457–7463. [Google Scholar] [CrossRef]
- Wei, Z.; Fan, P.; Jiao, Y.; Wang, Y.; Huang, Y.; Liu, Z. Integrated microfluidic chip for on-line proteome analysis with combination of denaturing and rapid digestion of protein. Anal. Chim. Acta 2020, 1102, 1–10. [Google Scholar] [CrossRef]
- Zhang, S.; Yuan, H.; Zhao, B.; Zhang, L.; Zhang, Y. Integrated platform with combination of on-line protein digestion, isotope dimethyl labeling and multidimensional peptide separation for high-throughput proteome quantification. Anal. Chim. Acta 2018, 1000, 172–179. [Google Scholar] [CrossRef]
- Duong, V.-A.; Park, J.-M.; Lee, H. A review of suspension trapping digestion method in bottom-up proteomics. J. Sep. Sci. 2022, 45, 3150–3168. [Google Scholar] [CrossRef]
- Zougman, A.; Wilson, J.P.; Roberts, L.D.; Banks, R.E. Detergent-Free Simultaneous Sample Preparation Method for Proteomics and Metabolomics. J. Proteome Res. 2020, 19, 2838–2844. [Google Scholar] [CrossRef] [PubMed]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Optimisation of Sporosori Purification and Protein Extraction Techniques for the Biotrophic Protozoan Plant Pathogen Spongospora subterranea. Molecules 2020, 25, 3109. [Google Scholar] [CrossRef] [PubMed]
- Baniasad, M.; Kim, Y.; Shaffer, M.; Sabag-Daigle, A.; Leleiwi, I.; Daly, R.A.; Ahmer, B.M.M.; Wrighton, K.C.; Wysocki, V.H. Optimization of proteomics sample preparation for identification of host and bacterial proteins in mouse feces. Anal. Bioanal. Chem. 2022, 414, 2317–2331. [Google Scholar] [CrossRef]
- Hayoun, K.; Gouveia, D.; Grenga, L.; Pible, O.; Armengaud, J.; Alpha-Bazin, B. Evaluation of Sample Preparation Methods for Fast Proteotyping of Microorganisms by Tandem Mass Spectrometry. Front. Microbiol. 2019, 10, 1985. [Google Scholar] [CrossRef]
- Mikulášek, K.; Konečná, H.; Potěšil, D.; Holánková, R.; Havliš, J.; Zdráhal, Z. SP3 Protocol for Proteomic Plant Sample Preparation Prior LC-MS/MS. Front. Plant Sci. 2021, 12, 635550. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Caterino, M.; Cevenini, A.; Jung, V.; Chhuon, C.; Lipecka, J.; Fedele, R.; Guerrera, I.C.; Ruoppolo, M. Dataset of a comparative proteomics experiment in a methylmalonyl-CoA mutase knockout HEK 293 cell model. Data Brief 2020, 33, 106453. [Google Scholar] [CrossRef]
- Wojtkiewicz, M.; Berg Luecke, L.; Kelly, M.I.; Gundry, R.L. Facile Preparation of Peptides for Mass Spectrometry Analysis in Bottom-Up Proteomics Workflows. Curr. Protoc. 2021, 1, e85. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, R.; Sethi, M.K.; Zaia, J. On-slide tissue digestion for mass spectrometry based glycomic and proteomic profiling. MethodsX 2019, 6, 2329–2347. [Google Scholar] [CrossRef]
- Judd, A.M.; Gutierrez, D.B.; Moore, J.L.; Patterson, N.H.; Yang, J.; Romer, C.E.; Norris, J.L.; Caprioli, R.M. A recommended and verified procedure for in situ tryptic digestion of formalin-fixed paraffin-embedded tissues for analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry. J. Mass Spectrom. 2019, 54, 716–727. [Google Scholar] [CrossRef]
- Olsen, J.V.; Ong, S.-E.; Mann, M. Trypsin Cleaves Exclusively C-terminal to Arginine and Lysine Residues*. Mol. Cell. Proteom. 2004, 3, 608–614. [Google Scholar] [CrossRef]
- Tyers, M.; Mann, M. From genomics to proteomics. Nature 2003, 422, 193–197. [Google Scholar] [CrossRef]
- Giansanti, P.; Tsiatsiani, L.; Low, T.Y.; Heck, A.J.R. Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat. Protoc. 2016, 11, 993–1006. [Google Scholar] [CrossRef]
- Li, W.; Li, F.; Zhang, X.; Lin, H.-K.; Xu, C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct. Target. Ther. 2021, 6, 422. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Pieroni, L.; Iavarone, F.; Olianas, A.; Greco, V.; Desiderio, C.; Martelli, C.; Manconi, B.; Sanna, M.T.; Messana, I.; Castagnola, M.; et al. Enrichments of post-translational modifications in proteomic studies. J. Sep. Sci. 2020, 43, 313–336. [Google Scholar] [CrossRef]
- Humphrey, S.J.; James, D.E.; Mann, M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef]
- Fíla, J.; Honys, D. Enrichment techniques employed in phosphoproteomics. Amino Acids 2012, 43, 1025–1047. [Google Scholar] [CrossRef]
- Qiu, W.; Evans, C.A.; Landels, A.; Pham, T.K.; Wright, P.C. Phosphopeptide enrichment for phosphoproteomic analysis—A tutorial and review of novel materials. Anal. Chim. Acta 2020, 1129, 158–180. [Google Scholar] [CrossRef]
- Ahn, Y.H.; Kim, J.Y.; Yoo, J.S. Quantitative mass spectrometric analysis of glycoproteins combined with enrichment methods. Mass Spectrom. Rev. 2015, 34, 148–165. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.-j.; Martin, D.B.; Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003, 21, 660–666. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.; Ha, M.Y.; Lee, E.K.; Choo, J. Immobilization of aminophenylboronic acid on magnetic beads for the direct determination of glycoproteins by matrix assisted laser desorption ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16, 1456–1460. [Google Scholar] [CrossRef]
- Tang, J.; Liu, Y.; Qi, D.; Yao, G.; Deng, C.; Zhang, X. On-plate-selective enrichment of glycopeptides using boronic acid-modified gold nanoparticles for direct MALDI-QIT-TOF MS analysis. Proteomics 2009, 9, 5046–5055. [Google Scholar] [CrossRef]
- Lin, Z.A.; Pang, J.L.; Lin, Y.; Huang, H.; Cai, Z.W.; Zhang, L.; Chen, G.N. Preparation and evaluation of a phenylboronate affinity monolith for selective capture of glycoproteins by capillary liquid chromatography. Analyst 2011, 136, 3281–3288. [Google Scholar] [CrossRef]
- Heo, S.-H.; Lee, S.-J.; Ryoo, H.-M.; Park, J.-Y.; Cho, J.-Y. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics 2007, 7, 4292–4302. [Google Scholar] [CrossRef]
- Choi, E.; Loo, D.; Dennis, J.W.; O’Leary, C.A.; Hill, M.M. High-throughput lectin magnetic bead array-coupled tandem mass spectrometry for glycoprotein biomarker discovery. Electrophoresis 2011, 32, 3564–3575. [Google Scholar] [CrossRef]
- Zielinska, D.F.; Gnad, F.; Wiśniewski, J.R.; Mann, M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Mann, M. Consecutive Proteolytic Digestion in an Enzyme Reactor Increases Depth of Proteomic and Phosphoproteomic Analysis. Anal. Chem. 2012, 84, 2631–2637. [Google Scholar] [CrossRef]
- Aksnes, H.; Van Damme, P.; Goris, M.; Starheim, K.K.; Marie, M.; Støve, S.I.; Hoel, C.; Kalvik, T.V.; Hole, K.; Glomnes, N.; et al. An Organellar Nα-Acetyltransferase, Naa60, Acetylates Cytosolic N Termini of Transmembrane Proteins and Maintains Golgi Integrity. Cell Rep. 2015, 10, 1362–1374. [Google Scholar] [CrossRef]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H. Comparison of Two Solid-Phase Extraction (SPE) Methods for the Identification and Quantification of Porcine Retinal Protein Markers by LC-MS/MS. Int. J. Mol. Sci. 2018, 19, 3847. [Google Scholar] [CrossRef]
- Shen, Y.; Tolić, N.; Masselon, C.; Paša-Tolić, L.; Camp, D.G.; Hixson, K.K.; Zhao, R.; Anderson, G.A.; Smith, R.D. Ultrasensitive Proteomics Using High-Efficiency On-Line Micro-SPE-NanoLC-NanoESI MS and MS/MS. Anal. Chem. 2004, 76, 144–154. [Google Scholar] [CrossRef]
- Bladergroen, M.R.; van der Burgt, Y.E.M. Solid-phase extraction strategies to surmount body fluid sample complexity in high-throughput mass spectrometry-based proteomics. J. Anal. Methods Chem. 2015, 2015, 250131. [Google Scholar] [CrossRef]
- Chen, D.; Shen, X.; Sun, L. Strong cation exchange-reversed phase liquid chromatography-capillary zone electrophoresis-tandem mass spectrometry platform with high peak capacity for deep bottom-up proteomics. Anal. Chim. Acta 2018, 1012, 1–9. [Google Scholar] [CrossRef]
- Boichenko, A.P.; Govorukhina, N.; van der Zee, A.G.J.; Bischoff, R. Multidimensional separation of tryptic peptides from human serum proteins using reversed-phase, strong cation exchange, weak anion exchange, and fused-core fluorinated stationary phases. J. Sep. Sci. 2013, 36, 3463–3470. [Google Scholar] [CrossRef]
- Betancourt, L.H.; De Bock, P.-J.; Staes, A.; Timmerman, E.; Perez-Riverol, Y.; Sanchez, A.; Besada, V.; Gonzalez, L.J.; Vandekerckhove, J.; Gevaert, K. SCX charge state selective separation of tryptic peptides combined with 2D-RP-HPLC allows for detailed proteome mapping. J. Proteom. 2013, 91, 164–171. [Google Scholar] [CrossRef]
- Xu, B.; Wang, F.; Song, C.; Sun, Z.; Cheng, K.; Tan, Y.; Wang, H.; Zou, H. Large-Scale Proteome Quantification of Hepatocellular Carcinoma Tissues by a Three-Dimensional Liquid Chromatography Strategy Integrated with Sample Preparation. J. Proteome Res. 2014, 13, 3645–3654. [Google Scholar] [CrossRef]
- Ye, X.; Tang, J.; Mao, Y.; Lu, X.; Yang, Y.; Chen, W.; Zhang, X.; Xu, R.; Tian, R. Integrated proteomics sample preparation and fractionation: Method development and applications. TrAC Trends Anal. Chem. 2019, 120, 115667. [Google Scholar] [CrossRef]
- Ishihama, Y.; Rappsilber, J.; Mann, M. Modular Stop and Go Extraction Tips with Stacked Disks for Parallel and Multidimensional Peptide Fractionation in Proteomics. J. Proteome Res. 2006, 5, 988–994. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Adachi, J.; Hashiguchi, K.; Nagano, M.; Sato, M.; Sato, A.; Fukamizu, K.; Ishihama, Y.; Tomonaga, T. Improved Proteome and Phosphoproteome Analysis on a Cation Exchanger by a Combined Acid and Salt Gradient. Anal. Chem. 2016, 88, 7899–7903. [Google Scholar] [CrossRef]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef]
- Zhang, Z.; Dovichi, N.J. Seamlessly Integrated Miniaturized Filter-Aided Sample Preparation Method to Fractionation Techniques for Fast, Loss-Less, and In-Depth Proteomics Analysis of 1 μg of Cell Lysates at Low Cost. Anal. Chem. 2022, 94, 10135–10141. [Google Scholar] [CrossRef]
- Tian, R.; Wang, S.; Elisma, F.; Li, L.; Zhou, H.; Wang, L.; Figeys, D. Rare Cell Proteomic Reactor Applied to Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-based Quantitative Proteomics Study of Human Embryonic Stem Cell Differentiation. Mol. Cell. Proteom. 2011, 10, S1–S10. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, S.; He, S.; Liu, C.; Fu, C.; Tang, M.; Liu, C.; Sun, Y.; Lam, H.; Liu, Z.; et al. Fully integrated on-line strategy for highly sensitive proteome profiling of 10–500 mammalian cells. Analyst 2023, 148, 120–127. [Google Scholar] [CrossRef]
- Lin, L.; Zheng, J.; Yu, Q.; Chen, W.; Xing, J.; Chen, C.; Tian, R. High throughput and accurate serum proteome profiling by integrated sample preparation technology and single-run data independent mass spectrometry analysis. J. Proteom. 2018, 174, 9–16. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Tian, R. An integrated strategy for highly sensitive phosphoproteome analysis from low micrograms of protein samples. Analyst 2018, 143, 3693–3701. [Google Scholar] [CrossRef]
- Gao, W.; Li, H.; Liu, L.; Huang, P.; Wang, Z.; Chen, W.; Ye, M.; Yu, X.; Tian, R. An integrated strategy for high-sensitive and multi-level glycoproteome analysis from low micrograms of protein samples. J. Chromatogr. A 2019, 1600, 46–54. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duong, V.-A.; Lee, H. Bottom-Up Proteomics: Advancements in Sample Preparation. Int. J. Mol. Sci. 2023, 24, 5350. https://doi.org/10.3390/ijms24065350
Duong V-A, Lee H. Bottom-Up Proteomics: Advancements in Sample Preparation. International Journal of Molecular Sciences. 2023; 24(6):5350. https://doi.org/10.3390/ijms24065350
Chicago/Turabian StyleDuong, Van-An, and Hookeun Lee. 2023. "Bottom-Up Proteomics: Advancements in Sample Preparation" International Journal of Molecular Sciences 24, no. 6: 5350. https://doi.org/10.3390/ijms24065350
APA StyleDuong, V.-A., & Lee, H. (2023). Bottom-Up Proteomics: Advancements in Sample Preparation. International Journal of Molecular Sciences, 24(6), 5350. https://doi.org/10.3390/ijms24065350







