New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production
Abstract
1. Introduction
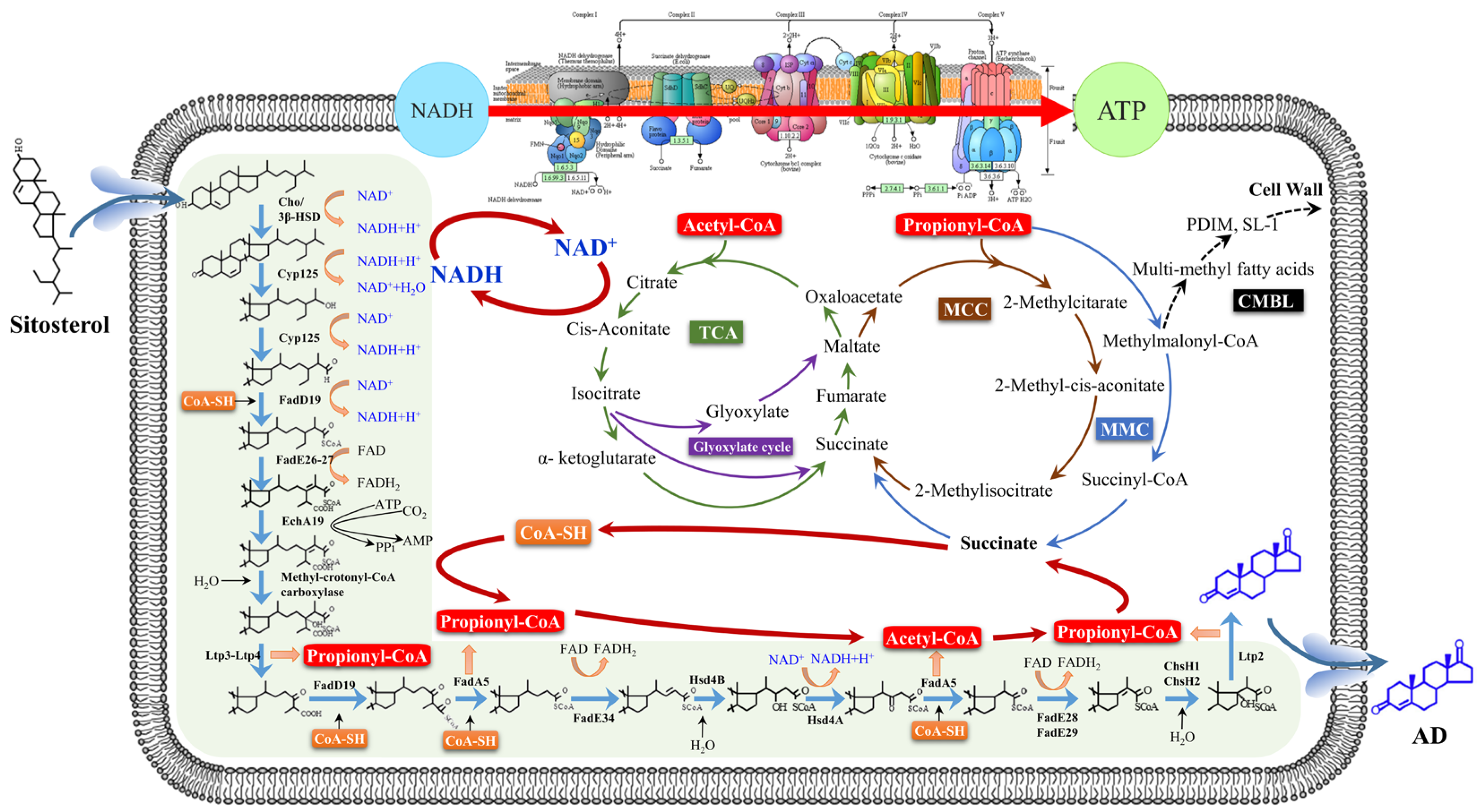
2. Core Metabolic Pathway of Steroid (CMS) in M. neoaurum
3. Non-Core Metabolic Pathway of Steroids in M. neoaurum
3.1. Phytosterol Uptake
3.2. Phytosterol-Side Chain Degradation and Coenzyme I Metabolism
3.3. Phytosterol-Side Chain Degradation and Propionyl-CoA Metabolism
3.4. Phytosterol-Side Chain Degradation and Reactive Oxygen Species
3.5. Phytosterol-Side Chain Degradation and Energy Metabolism
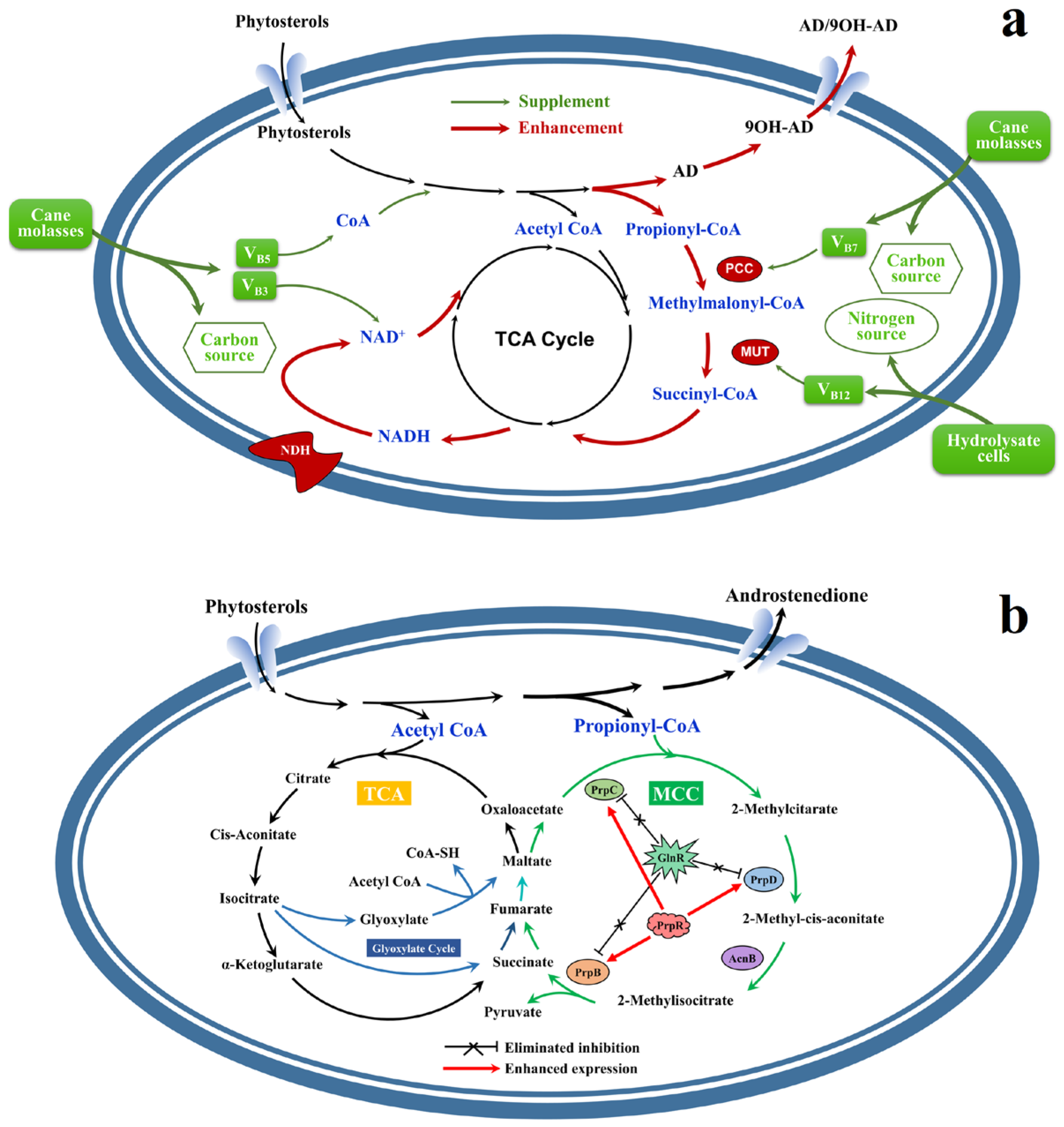
4. Rational Construction of Recombinant M. neoaurum Based on NCMS
4.1. Phytosterol Uptake Pathway Modification
4.2. Metabolic Regulation Based on Coenzyme I
4.3. Metabolic Regulation Based on Propionyl-CoA
4.4. Regulation Strategy of ROS Level
4.5. Energy Metabolism Regulation Based on ATP
5. Enhancement of C-19 Steroid Yield by Improving the Medium Composition and Fermentation Biotechnology
5.1. Optimization of Transformation Medium
5.2. Selection and Optimization of Carbon and Nitrogen Sources
5.3. Construction and Improvement of Fermentation Technology
6. Comparison of the Influence of Various Cofactor Regulation Strategies on AD Production
7. Concentrations and Future Considerations
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, A.; Zhang, X.; Li, Y.; Wang, Z.; Lv, Y.; Liu, J.; Alam, M.A.; Xiong, W.; Xu, J. Mycolicibacterium cell factory for the production of steroid-based drug intermediates. Biotechnol. Adv. 2021, 53, 107860. [Google Scholar] [CrossRef] [PubMed]
- Seth, D.; Kamat, D. Intranasal steroid therapy for allergic rhinitis. Pediatr. Ann. 2019, 48, e43–e48. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cabezon, L.; Galán, B.; García, J.L. New insights on steroid biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Malaviya, A.; Gomes, J. Androstenedione production by biotransformation of phytosterols. Bioresour. Technol. 2008, 99, 6725–6737. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Shen, Y.; Qin, N.; Wang, Y.; Su, L.; Wang, M. Genetic differences in ksdD influence on the ADD/AD ratio of Myco-bacterium neoaurum. J. Ind. Microbiol. Biotechnol. 2015, 42, 507–513. [Google Scholar] [CrossRef]
- Nunes, V.O.; Vanzellotti, N.D.C.; Fraga, J.L.; Pessoa, F.L.P.; Ferreira, T.F.; Amaral, P.F.F. Biotransformation of phytosterols into androstenedione—A technological prospecting study. Molecules 2022, 27, 3164. [Google Scholar] [CrossRef]
- Rodríguez-García, A.; Fernández-Alegre, E.; Morales, A.; Sola-Landa, A.; Lorraine, J.; Macdonald, S.; Dovbnya, D.; Smith, M.C.M.; Donova, M.; Barreiro, C. Complete genome sequence of ‘Mycobacterium neoaurum’ NRRL B-3805, an androstenedione (AD) producer for industrial biotransformation of sterols. J. Biotechnol. 2016, 224, 64–65. [Google Scholar] [CrossRef]
- Peng, H.; Wang, Y.; Jiang, K.; Chen, X.; Zhang, W.; Zhang, Y.; Deng, Z.; Qu, X. A dual role reductase from phytosterols ca-tabolism enables the efficient production of valuable steroid precursors. Angew. Chemie. Int. Ed. 2020, 60, 5414–5420. [Google Scholar] [CrossRef]
- Bragin, E.Y.; Shtratnikova, V.Y.; Dovbnya, D.V.; Schelkunov, M.I.; Pekov, Y.A.; Malakho, S.G.; Egorova, O.V.; Ivashina, T.V.; Sokolov, S.L.; Ashapkin, V.V.; et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. Strains. J. Steroid Biochem. Mol. Biol. 2013, 138, 41–53. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, X.; Wang, Y.; Han, Y.; Sun, J.; Shi, J.; Zhang, B. Identification, function, and application of 3-ketosteroid Δ1-dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons. Microb. Cell Fact. 2018, 17, 77. [Google Scholar] [CrossRef]
- Yao, K.; Xu, L.Q.; Wang, F.Q.; Wei, D.Z. Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab. Eng. 2014, 24, 181–191. [Google Scholar] [CrossRef]
- Shao, M.; Zhang, X.; Rao, Z.; Xu, M.; Yang, T.; Li, H.; Xu, Z.; Yang, S. A mutant form of 3-ketosteroid-Δ1-dehydrogenase gives altered androst-1,4-diene-3, 17-dione/androst-4-ene-3,17-dione molar ratios in steroid biotransformations by Mycobacterium neoaurum ST-095. J. Ind. Microbiol. Biotechnol. 2016, 43, 691–701. [Google Scholar] [CrossRef]
- Xiong, L.-B.; Liu, H.-H.; Xu, L.-Q.; Sun, W.-J.; Wang, F.-Q.; Wei, D.-Z. Improving the production of 22-hydroxy-23,24-bisnorchol-4-ene-3-one from sterols in Mycobacterium neoaurum by increasing cell permeability and modifying multiple genes. Microb. Cell Factories 2017, 16, 1–10. [Google Scholar] [CrossRef]
- Lobastova, T.G.; Khomutov, S.M.; Shutov, A.A.; Donova, M.V. Microbiological synthesis of stereoisomeric 7(α/β)-hydroxytestololactones and 7(α/β)-hydroxytestolactones. Appl. Microbiol. Biotechnol. 2019, 103, 4967–4976. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Zhang, D.; Wu, Q.; Zhu, D.; Ma, Y. Characterization of new recombinant 3-ketosteroid-Δ1-dehydrogenases for the biotransformation of steroids. Appl. Microbiol. Biotechnol. 2017, 101, 6049–6060. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, H.; Zhu, L.; Zhang, W.; You, S.; Qi, W.; Qian, J.; Su, R.; He, Z. A combined strategy of metabolic pathway regulation and two-step bioprocess for improved 4-androstene-3,17-dione production with an engineered Mycobacterium neoaurum. Biochem. Eng. J. 2020, 164, 107789. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and comparative genomic studies robustly support division of the genus mycobacterium into an emended genus mycobacterium and four novel genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2016, 66, 1603–1606. [Google Scholar] [CrossRef]
- Malaviya, A.; Gomes, J. Rapid screening and isolation of a fungus for sitosterol to androstenedione biotransformation. Appl. Biochem. Biotechnol. 2009, 158, 374–386. [Google Scholar] [CrossRef]
- Lin, Y.; Song, X.; Fu, J.; Lin, J.; Qu, Y. Microbial transformation of phytosterol in corn flour and soybean flour to 4-androstene-3,17-dione by Fusarium moniliforme sheld. Bioresour. Technol. 2009, 100, 1864–1867. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, G.; Ge, F.; Li, W.; Zeng, L.; Cao, W. Efficient biotransformation of cholesterol to androsta-1,4-diene-3,17-dione by a newly isolated actinomycete Gordonia neofelifaecis. World J. Microbiol. Biotechnol. 2011, 27, 759–765. [Google Scholar] [CrossRef]
- Chaudhari, P.N.; Chaudhari, B.L.; Chincholkar, S.B. Cholesterol biotransformation to androsta-1,4-diene-3,17-dione by growing cells of Chryseobacterium gleum. Biotechnol. Lett. 2010, 32, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V.; Egorova, O. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012, 94, 1423–1447. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Wang, F.-Q.; Zhang, H.-C.; Wei, D.-Z. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab. Eng. 2013, 15, 75–87. [Google Scholar] [CrossRef]
- Uhía, I.; Galán, B.; Morales, V.; García, J.L. Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2155. Environ. Microbiol. 2011, 13, 943–959. [Google Scholar] [CrossRef]
- Horinouchi, M.; Kurita, T.; Hayashi, T.; Kudo, T. Steroid degradation genes in Comamonas testosteroni TA441: Isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100kb steroid degradation gene hot spot. J. Steroid Biochem. Mol. Biol. 2010, 122, 253–263. [Google Scholar] [CrossRef]
- Ivashina, T.V.; Nikolayeva, V.M.; Dovbnya, D.V.; Donova, M.V. Cholesterol oxidase ChoD is not a critical enzyme accounting for oxidation of sterols to 3-keto-4-ene steroids in fast-growing Mycobacterium sp. VKM Ac-1815D. J. Steroid Biochem. Mol. Biol. 2012, 129, 47–53. [Google Scholar] [CrossRef]
- Kreit, J. Microbial catabolism of sterols: Focus on the enzymes that transform the sterol 3 beta-hydroxy-5-en into 3-keto-4-en. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Lu, R.; Schaefer, C.M.; Nesbitt, N.M.; Kuper, J.; Kisker, C.; Sampson, N.S. Catabolism of the cholesterol side chain in Mycobacterium tuberculosis is controlled by a redox-sensitive thiol switch. ACS Infect. Dis. 2017, 3, 666–675. [Google Scholar] [CrossRef]
- Szentirmai, A. Microbial physiology of sidechain degradation of sterols. J. Ind. Microbiol. Biotechnol. 1990, 6, 101–115. [Google Scholar] [CrossRef]
- Xu, L.-Q.; Liu, Y.-J.; Yao, K.; Liu, H.-H.; Tao, X.-Y.; Wang, F.-Q.; Wei, D.-Z. Unraveling and engineering the production of 23,24-bisnorcholenic steroids in sterol metabolism. Sci. Rep. 2016, 6, 21928. [Google Scholar] [CrossRef]
- He, K.; Sun, H.; Song, H. Engineering phytosterol transport system in Mycobacterium sp. strain MS136 enhances production of 9α-hydroxy-4-androstene-3,17-dione. Biotechnol. Lett. 2018, 40, 673–678. [Google Scholar] [CrossRef]
- Rank, L.; Herring, L.E.; Braunstein, M. Evidence for the mycobacterial Mce4 transporter being a multiprotein complex. J. Bacteriol. 2021, 203. [Google Scholar] [CrossRef]
- Perkowski, E.F.; Miller, B.K.; McCann, J.R.; Sullivan, J.T.; Malik, S.; Allen, I.C.; Godfrey, V.; Hayden, J.D.; Braunstein, M. An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol. Microbiol. 2016, 100, 90–107. [Google Scholar] [CrossRef]
- Nazarova, E.V.; Montague, C.R.; La, T.; Wilburn, K.M.; Sukumar, N.; Lee, W.; Caldwell, S.; Russell, D.G.; VanderVen, B.C. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 2017, 6, 1–22. [Google Scholar] [CrossRef]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef]
- Tahlan, K.; Wilson, R.; Kastrinsky, D.B.; Arora, K.; Nair, V.; Fischer, E.; Barnes, S.W.; Walker, J.R.; Alland, D.; Barry, C.E.; et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2012, 56, 1797–1809. [Google Scholar] [CrossRef]
- Abuhammad, A. Cholesterol metabolism: A potential therapeutic target in Mycobacteria. Br. J. Pharmacol. 2017, 174, 2194–2208. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Xu, Y.; Liu, W.; Zhang, X.; Gong, J.; Xu, Z.; Shi, J. Effects of a nonionic surfactant TX-40 on 9α-hydroxyandrost-4-ene-3,17-dione biosynthesis and physiological properties of Mycobacterium sp. LY-1. Process. Biochem. 2019, 87, 89–94. [Google Scholar] [CrossRef]
- Queiroz, A.; Medina-Cleghorn, D.; Marjanovic, O.; Nomura, D.K.; Riley, L.W. Comparative metabolic profiling of mce1 operon mutant vs. wild-type Mycobacterium tuberculosis strains. Pathog. Dis. 2015, 73, ftv066. [Google Scholar] [CrossRef]
- Su, Z.; Zhang, Z.; Yu, J.; Yuan, C.; Shen, Y.; Wang, J.; Su, L.; Wang, M. Combined enhancement of the propionyl-CoA metabolic pathway for efficient androstenedione production in Mycolicibacterium neoaurum. Microb. Cell Fact. 2022, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sedlaczek, L.; Lisowska, K.; Korycka, M.; Rumijowska, A.; Ziółkowski, A.; Długoński, J. The effect of cell wall components on glycine-enhanced sterol side chain degradation to androstene derivatives by mycobacteria. Appl. Microbiol. Bio-Technol. 1999, 52, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Korkegian, A.; Roberts, D.M.; Blair, R.; Parish, T. Mutations in the essential arabinosyltransferase EmbC lead to alterations in Mycobacterium tuberculosis lipoarabinomannan. J. Biol. Chem. 2014, 289, 35172–35181. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.H.; Harth, G.; Horwitz, M.A.; Eisenberg, D. An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kda major secretory protein (antigen 85B), a mycolyl transferase. J. Mol. Biol. 2001, 307, 671–681. [Google Scholar] [CrossRef]
- Goins, C.M.; Dajnowicz, S.; Smith, M.D.; Parks, J.M.; Ronning, D.R. Mycolyltransferase from Mycobacterium tuberculosis in covalent complex with tetrahydrolipstatin provides insights into antigen 85 catalysis. J. Biol. Chem. 2018, 293, 3651–3662. [Google Scholar] [CrossRef]
- Warrier, T.; Tropis, M.; Werngren, J.; Diehl, A.; Gengenbacher, M.; Schlegel, B.; Schade, M.; Oschkinat, H.; Daffe, M.; Hoffner, S.; et al. Antigen 85C inhibition restricts mycobacterium tuberculosis growth through disruption of cord factor biosynthesis. Antimicrob. Agents Chemother. 2012, 56, 1735–1743. [Google Scholar] [CrossRef]
- Yang, X.; Nesbitt, N.M.; Dubnau, E.; Smith, I.; Sampson, N.S. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 2009, 48, 3819–3821. [Google Scholar] [CrossRef]
- Griffin, J.E.; Pandey, A.K.; Gilmore, S.A.; Mizrahi, V.; Mckinney, J.D.; Bertozzi, C.R.; Sassetti, C.M. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 2012, 19, 218–227. [Google Scholar] [CrossRef]
- Gopinath, K.; Moosa, A.; Mizrahi, V.; Warner, D.F. Vitamin B 12 metabolism in Mycobacterium tuberculosis. Future Microbiol. 2013, 8, 1405–1418. [Google Scholar] [CrossRef]
- Resh, M. Lipid Modification of Proteins: Targeting to Membranes; Elsevier: Amsterdam, The Netherlands, 2013; pp. 736–740. [Google Scholar] [CrossRef]
- Ouellet, H.; Johnston, J.B.; de Montellano, P.R.O. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef]
- Puckett, S.; Trujillo, C.; Wang, Z.; Eoh, H.; Ioerger, T.R.; Krieger, I.; Sacchettini, J.; Schnappinger, D.; Rhee, K.Y.; Ehrt, S. Glyox-ylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, E2225–E2232. [Google Scholar] [CrossRef]
- Lyonnet, B.B.; Diacovich, L.; Cabruja, M.; Bardou, F.; Quémard, A.; Gago, G.; Gramajo, H. Pleiotropic effect of AccD5 and AccE5 depletion in acyl-coenzyme A carboxylase activity and in lipid biosynthesis in mycobacteria. PLoS ONE 2014, 9, e99853. [Google Scholar] [CrossRef]
- Eoh, H.; Rhee, K.Y. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc. Natl. Acad. Sci. USA 2014, 111, 4976–4981. [Google Scholar] [CrossRef]
- Singh, P.; Sinha, R.; Tyagi, G.; Sharma, N.K.; Saini, N.K.; Chandolia, A.; Prasad, A.K.; Varma-Basil, M.; Bose, M. PDIM and SL1 accumulation in Mycobacterium tuberculosis is associated with mce4A expression. Gene 2018, 642, 178–187. [Google Scholar] [CrossRef]
- Claes, W.A.; Puhler, A.; Kalinowski, J. Identification of two prpDBC gene clusters in Corynebacterium glutamicum and their involvement in propionate degradation via the 2-methylcitrate cycle. J. Bacteriol. 2002, 184, 2728–2739. [Google Scholar] [CrossRef]
- Textor, S.; Wendisch, V.F.; de Graaf, A.; Müller, U.; Linder, M.I.; Linder, D.; Buckel, W. Propionate oxidation in Escherichia coli: Evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 1997, 168, 428–436. [Google Scholar] [CrossRef]
- Borisov, V.; Siletsky, S.; Nastasi, M.; Forte, E. ROS defense systems and terminal oxidases in bacteria. Antioxidants 2021, 10, 839. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Sampaio-Marques, B.; Barbosa, C.; Rodrigues, F.; Costa, V.; Mendes-Faia, A.; Ludovico, P.; Leao, C. Ac-cumulation of non-superoxide anion reactive oxygen species mediates nitrogen-limited alcoholic fermentation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 7918–7924. [Google Scholar] [CrossRef]
- Kerscher, S.; Dröse, S.; Zickermann, V.; Brandt, U. The three families of respiratory NADH dehydrogenases. Results Probl. Cell Differ. 2008, 45, 185–222. [Google Scholar] [CrossRef]
- Muller, F.L.; Liu, Y.; Abdul-Ghani, M.A.; Lustgarten, M.S.; Bhattacharya, A.; Jang, Y.C.; Van Remmen, H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem. J. 2008, 409, 491–499. [Google Scholar] [CrossRef]
- Kussmaul, L.; Hirst, J. The mechanism of superoxide production by NADH: Ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 7607–7612. [Google Scholar] [CrossRef] [PubMed]
- Knuuti, J.; Belevich, G.; Sharma, V.; Bloch, D.A.; Verkhovskaya, M. A single amino acid residue controls ROS production in the respiratory complex I from Escherichia coli. Mol. Microbiol. 2013, 90, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.-J.; Wang, L.; Liu, H.-H.; Liu, Y.-J.; Ren, Y.-H.; Wang, F.-Q.; Wei, D.-Z. Characterization and engineering control of the effects of reactive oxygen species on the conversion of sterols to steroid synthons in Mycobacterium neoaurum. Metab. Eng. 2019, 56, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Richard-Greenblatt, M.; Bach, H.; Adamson, J.; Peña-Diaz, S.; Li, W.; Steyn, A.J.; Av-Gay, Y. Regulation of ergothioneine biosynthesis and its effect on Mycobacterium tuberculosis growth and infectivity. J. Biol. Chem. 2015, 290, 23064–23076. [Google Scholar] [CrossRef]
- Servillo, L.; Castaldo, D.; Casale, R.; D’Onofrio, N.; Giovane, A.; Cautela, D.; Balestrieri, M.L. An uncommon redox behavior sheds light on the cellular antioxidant properties of ergothioneine. Free. Radic. Biol. Med. 2015, 79, 228–236. [Google Scholar] [CrossRef]
- Si, M.; Zhao, C.; Zhang, B.; Wei, D.; Chen, K.; Yang, X.; Xiao, H.; Shen, X. Overexpression of mycothiol disulfide reductase enhances Corynebacterium glutamicum robustness by modulating cellular redox homeostasis and antioxidant proteins under oxidative stress. Sci. Rep. 2016, 6, 29491. [Google Scholar] [CrossRef]
- Nakajima, S.; Satoh, Y.; Yanashima, K.; Matsui, T.; Dairi, T. Ergothioneine protects Streptomyces coelicolor A3(2) from oxidative stresses. J. Biosci. Bioeng. 2015, 120, 294–298. [Google Scholar] [CrossRef]
- Luo, Z.; Zeng, W.; Du, G.; Chen, J.; Zhou, J. Enhanced pyruvate production in Candida glabrata by engineering ATP futile cycle system. ACS Synth. Biol. 2019, 8, 787–795. [Google Scholar] [CrossRef]
- Johnson, K.M.; Cleary, J.; Fierke, C.A.; Opipari, A.W.; Glick, G.D. Mechanistic basis for therapeutic targeting of the mito-chondrial F 1 F o-ATPase. ACS Chem. Biol. 2006, 1, 304–308. [Google Scholar] [CrossRef]
- Boecker, S.; Zahoor, A.; Schramm, T.; Link, H.; Klamt, S. Broadening the scope of enforced ATP wasting as a tool for metabolic engineering in Escherichia coli. Biotechnol. J. 2019, 14, 1800438. [Google Scholar] [CrossRef]
- Liu, J.; Kandasamy, V.; Würtz, A.; Jensen, P.R.; Solem, C. Stimulation of acetoin production in metabolically engineered Lactococcus lactis by increasing ATP demand. Appl. Microbiol. Biotechnol. 2016, 100, 9509–9517. [Google Scholar] [CrossRef]
- Chao, Y.P.; Liao, J.C. Metabolic responses to substrate futile cycling in Escherichia coli. J. Biol. Chem. 1994, 269, 5122–5126. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8106492 (accessed on 7 January 2023). [CrossRef]
- Hädicke, O.; Bettenbrock, K.; Klamt, S. Enforced ATP futile cycling increases specific productivity and yield of anaerobic lactate production in Escherichia coli. Biotechnol. Bioeng. 2015, 112, 2195–2199. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Cui, Z.; Liang, Q.; Qi, Q. Enhancement of succinate yield by manipulating NADH/NAD+ ratio and ATP generation. Appl. Microbiol. Biotechnol. 2017, 101, 3153–3161. [Google Scholar] [CrossRef]
- Chubukov, V.; Gerosa, L.; Kochanowski, K.; Sauer, U. Coordination of microbial metabolism. Nat. Rev. Genet. 2014, 12, 327–340. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Yao, Y.; Xu, Q.; Shi, H.; Wang, K.; Feng, W.; Shen, Y. Coexpression of VHb and MceG genes in Mycobacterium sp. strain LZ2 enhances androstenone production via immobilized repeated batch fermentation. Bioresour. Technol. 2021, 342, 125965. [Google Scholar] [CrossRef]
- Xiong, L.-B.; Liu, H.-H.; Song, X.-W.; Meng, X.-G.; Liu, X.-Z.; Ji, Y.-Q.; Wang, F.-Q.; Wei, D.-Z. Improving the biotransformation of phytosterols to 9α-hydroxy-4-androstene-3,17-dione by deleting embC associated with the assembly of cell envelope in Mycobacterium neoaurum. J. Biotechnol. 2020, 323, 341–346. [Google Scholar] [CrossRef]
- Xiong, L.-B.; Liu, H.-H.; Song, L.; Dong, M.-M.; Ke, J.; Liu, Y.-J.; Liu, K.; Zhao, M.; Wang, F.-Q.; Wei, D.-Z. Improving the bio-transformation efficiency of soybean phytosterols in Mycolicibacterium neoaurum by the combined deletion of fbpC3 and embC in cell envelope synthesis. Synth. Syst. Biotechnol. 2022, 7, 453–459. [Google Scholar] [CrossRef]
- Xiong, L.-B.; Liu, H.-H.; Zhao, M.; Liu, Y.-J.; Song, L.; Xie, Z.-Y.; Xu, Y.-X.; Wang, F.-Q.; Wei, D.-Z. Enhancing the bioconversion of phytosterols to steroidal intermediates by the deficiency of kasB in the cell wall synthesis of Mycobacterium neoaurum. Microb. Cell Factories 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Su, L.; Shen, Y.; Gao, T.; Cui, L.; Luo, J.; Wang, M. Regulation of NAD (H) pool by overexpression of nicotinic acid phosphoribosyltransferase for AD (D) production in Mycobacterium neoaurum. Lect. Notes Electr. Eng. 2018, 444, 357–364. [Google Scholar]
- Su, L.; Shen, Y.; Zhang, W.; Gao, T.; Shang, Z.; Wang, M. Cofactor engineering to regulate NAD+/NADH ratio with its ap-plication to phytosterols biotransformation. Microb. Cell Fact. 2017, 16, 182. [Google Scholar] [CrossRef] [PubMed]
- Harbut, M.B.; Yang, B.; Liu, R.; Yano, T.; Vilchèze, C.; Cheng, B.; Lockner, J.; Guo, H.; Yu, C.; Franzblau, S.G.; et al. Small molecules targeting mycobacterium tuberculosis type II NADH dehydrogenase exhibit antimycobacterial activity. Angew. Chemie. Int. Ed. 2018, 57, 3478–3482. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, Y.; Shen, Y.; Zhang, X.; Xu, S.; Shang, Z.; Xia, M.; Wang, M. Efficient production of androstenedione by re-peated batch fermentation in waste cooking oil media through regulating NAD+/NADH ratio and strengthening cell vitality of Mycobacterium neoaurum. Bioresour. Technol. 2019, 279, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Zhao, Y.; Liu, Y.; Yang, T.; Xu, M.; Zhang, X.; Rao, Z. Intracellular environment improvement of Mycobacterium neoaurum for enhancing androst-1,4-diene-3,17-dione production by manipulating NADH and reactive oxygen species levels. Molecules 2019, 24, 3841. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, R.; Wu, Y.; Wang, D.; Wei, D. Nitrate metabolism decreases the steroidal alcohol byproduct compared with ammonium in biotransformation of phytosterol to androstenedione by Mycobacterium neoaurum. Appl. Biochem. Biotechnol. 2020, 190, 1553–1560. [Google Scholar] [CrossRef]
- Leonardi, R.; Zhang, Y.-M.; Rock, C.O.; Jackowski, S. Coenzyme A: Back in action. Prog. Lipid Res. 2005, 44, 125–153. [Google Scholar] [CrossRef]
- Upton, A.M.; McKinney, J.D. Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology 2007, 153, 3973–3982. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, Y.; Wei, L.; Hu, F.; Hua, Q. Effects of the methylmalonyl-CoA metabolic pathway on ansamitocin production in Actinosynnema pretiosum. Appl. Biochem. Biotechnol. 2017, 181, 1167–1178. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Shen, Y.; Zhang, X.; Zhang, Z.; Xu, S.; Luo, J.; Xia, M.; Wang, M. Economical production of androstenedi-one and 9A-hydroxyandrostenedione using untreated cane molasses by recombinant mycobacteria. Bioresour. Technol. 2019, 290, 121750. [Google Scholar] [CrossRef]
- Masiewicz, P.; Brzostek, A.; Wolański, M.; Dziadek, J.; Zakrzewska-Czerwińska, J. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS ONE 2012, 7, e43651. [Google Scholar] [CrossRef]
- Liu, X.-X.; Shen, M.-J.; Liu, W.-B.; Ye, B.-C. GlnR-mediated regulation of short-chain fatty acid assimilation in Mycobacterium smegmatis. Front. Microbiol. 2018, 9, 1311. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, X.; Wang, X.; Wang, L.; Xia, M.; Luo, J.; Shen, Y.; Wang, M. Improving phytosterol biotransformation at low nitrogen levels by enhancing the methylcitrate cycle with transcriptional regulators PrpR and GlnR of Mycobacterium neoaurum. Microb. Cell Factories 2020, 19, 1–11. [Google Scholar] [CrossRef]
- Sena, F.V.; Sousa, F.M.; Oliveira, A.S.F.; Soares, C.M.; Catarino, T.; Pereira, M.M. Regulation of the mechanism of type-II NADH: Quinone oxidoreductase from S. aureus. Redox Biol. 2018, 16, 209–214. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Shen, Y.; Zhang, X.; Zan, Z.; Xia, M.; Luo, J.; Wang, M. Efficient repeated batch production of androstenedione using untreated cane molasses by Mycobacterium neoaurum driven by ATP futile cycle. Bioresour. Technol. 2020, 309, 123307. [Google Scholar] [CrossRef]
- Chen, K.-C.; Wey, H.-C. Dissolution-enzyme kinetics of 11β-hydroxylation of cortexolone by Curvularia lunata. Enzym. Microb. Technol. 1990, 12, 616–621. [Google Scholar] [CrossRef]
- Cook, G.M.; Hards, K.; Vilchèze, C.; Hartman, T.; Berney, M. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2014, 2, 804–811. [Google Scholar] [CrossRef]
- Sun, W.-J.; Liu, Y.-J.; Liu, H.; Ma, J.; Ren, Y.; Wang, F.; Wei, D. Enhanced conversion of sterols to steroid synthons by augmenting the peptidoglycan synthesis gene pbpB in Mycobacterium neoaurum. J. Basic Microbiol. 2019, 59, 924–935. [Google Scholar] [CrossRef]
- Li, H.; Fu, Z.; Li, H.; Zhang, X.; Shi, J.; Xu, Z. Enhanced biotransformation of dehydroepiandrosterone to 3β,7α,15α-trihydroxy-5-androsten-17-one with Gibberella intermedia CA3-1 by natural oils addition. J. Ind. Microbiol. Biotechnol. 2014, 41, 1497–1504. [Google Scholar] [CrossRef]
- Mancilla, R.A.; Little, C.; Amoroso, A. Efficient bioconversion of high concentration phytosterol microdispersion to 4-Androstene-3,17-dione (AD) by Mycobacterium sp. B3805. Appl. Biochem. Biotechnol. 2017, 185, 494–506. [Google Scholar] [CrossRef]
- Shen, Y.; Liang, J.; Li, H.; Wang, M. Hydroxypropyl-β-cyclodextrin-mediated alterations in cell permeability, lipid and protein profiles of steroid-transforming Arthrobacter simplex. Appl. Microbiol. Biotechnol. 2014, 99, 387–397. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, M.; Zhang, L.; Ma, Y.; Ma, B.; Zheng, Y.; Liu, H.; Luo, J. Effects of hydroxypropyl-β-cyclodextrin on cell growth, activity, and integrity of steroid-transforming Arthrobacter simplex and Mycobacterium sp. Appl. Microbiol. Biotechnol. 2011, 90, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Shtratnikova, V.Y.; Schelkunov, M.I.; Dovbnya, D.V.; Bragin, E.Y.; Donova, M.V. Effect of methyl-β-cyclodextrin on gene expression in microbial conversion of phytosterol. Appl. Microbiol. Biotechnol. 2017, 101, 4659–4667. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Xu, S.; Shen, Y.; Xia, M.; Ren, X.; Wang, L.; Shang, Z.; Wanga, M. The sterol carrier hydroxypropyl-β-cyclodextrin enhances the metabolism of phytosterols by Mycobacterium neoaurum. Appl. Environ. Microbiol. 2020, 86, e00441-20. [Google Scholar] [CrossRef] [PubMed]
- Gulla, V.; Banerjee, T.; Patil, S. Bioconversion of soysterols to androstenedione by Mycobacterium fortuitum subsp. fortuitum NCIM 5239, a mutant derived from total sterol degrader strain. J. Chem. Technol. Biotechnol. 2010, 85, 1135–1141. [Google Scholar] [CrossRef]
- Su, L.; Shen, Y.; Gao, T.; Luo, J.; Wang, M. Improvement of AD biosynthesis response to enhanced oxygen transfer by oxygen vectors in Mycobacterium neoaurum TCCC 11979. Appl. Biochem. Biotechnol. 2017, 182, 1564–1574. [Google Scholar] [CrossRef]
- Nanou, K.; Roukas, T. Waste cooking oil: A new substrate for carotene production by Blakeslea trispora in submerged fermentation. Bioresour. Technol. 2016, 203, 198–203. [Google Scholar] [CrossRef]
- Kourmentza, C.; Costa, J.; Azevedo, Z.; Servin, C.; Grandfils, C.; De Freitas, V.; Reis, M. Burkholderia thailandensis as a microbial cell factory for the bioconversion of used cooking oil to polyhydroxyalkanoates and rhamnolipids. Bioresour. Technol. 2018, 247, 829–837. [Google Scholar] [CrossRef]
- Li, H.-X.; Lu, Z.-M.; Geng, Y.; Gong, J.-S.; Zhang, X.-J.; Shi, J.-S.; Xu, Z.-H.; Ma, Y.-H. Efficient production of bioactive metabolites from Antrodia camphorata ATCC 200183 by asexual reproduction-based repeated batch fermentation. Bioresour. Technol. 2015, 194, 334–343. [Google Scholar] [CrossRef]
- Reddy, L.V.; Kim, Y.M.; Yun, J.S.; Ryu, H.W.; Wee, Y.J. L-Lactic acid production by combined utilization of agricultural bi-oresources as renewable and economical substrates through batch and repeated-batch fermentation of Enterococcus faecalis RKY1. Bioresour. Technol. 2016, 209, 187–194. [Google Scholar] [CrossRef]
- Han, X.; Song, W.; Liu, G.; Li, Z.; Yang, P.; Qu, Y. Improving cellulase productivity of Penicillium oxalicum RE-10 by repeated fed-batch fermentation strategy. Bioresour. Technol. 2017, 227, 155–163. [Google Scholar] [CrossRef]
- Wijaya, H.; Sasaki, K.; Kahar, P.; Yopi, H.; Kawaguchi, T.; Sazuka, C.; Ogino, B.; Prasetya, A. Kondo, repeated ethanol fer-mentation from membrane-concentrated sweet sorghum juice using the flocculating yeast Saccharomyces cerevisiae F118 strain. Bioresour. Technol. 2018, 265, 542–547. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, X.; Xu, H.; Yao, Z.; Ouyang, P. ε-poly-L-lysine production by immobilized cells of Kitasatospora sp. MY 5-36 in repeated fed-batch cultures. Bioresour. Technol. 2010, 101, 5523–5527. [Google Scholar] [CrossRef]
- Tang, R.; Shen, Y.; Xia, M.; Tu, L.; Luo, J.; Geng, Y.; Gao, T.; Zhou, H.; Zhao, Y.; Wang, M. A highly efficient step-wise biotransformation strategy for direct conversion of phytosterol to boldenone. Bioresour. Technol. 2019, 283, 242–250. [Google Scholar] [CrossRef]
- Chacón, S.J.; Matias, G.; Ezeji, T.C.; Maciel Filho, R.; Mariano, A.P. Three-stage repeated-batch immobilized cell fermentation to produce butanol from non-detoxified sugarcane bagasse hemicellulose hydrolysates. Bioresour. Technol. 2021, 321, 124504. [Google Scholar] [CrossRef]
- Xu, X.; Gao, X.; Feng, J.; Wang, X.; Wei, D. Influence of temperature on nucleus degradation of 4-androstene-3, 17-dione in phytosterol biotransformation by Mycobacterium sp. Lett. Appl. Microbiol. 2015, 61, 63–68. [Google Scholar] [CrossRef]
- Wang, X.; Hua, C.; Xu, X.; Wei, D. Two-step bioprocess for reducing nucleus degradation in phytosterol bioconversion by Mycobacterium neoaurum NwIB-R10(hsd4A). Appl. Biochem. Biotechnol. 2019, 188, 138–146. [Google Scholar] [CrossRef]
- Bin, L.X.; Liu, H.H.; Xu, L.Q.; Wei, D.Z.; Wang, F.Q. Role identification and application of sigD in the transformation of soybean phytosterol to 9α-hydroxy-4-androstene-3,17-dione in Mycobacterium neoaurum. J. Agric. Food Chem. 2017, 65, 626–631. [Google Scholar]
- Liu, K.; Gao, Y.; Li, Z.-H.; Liu, M.; Wang, F.-Q.; Wei, D.-Z. CRISPR-Cas12a assisted precise genome editing of Mycolicibacterium neoaurum. New Biotechnol. 2022, 66, 61–69. [Google Scholar] [CrossRef]
- Olivera, E.R.; Luengo, J.M. Steroids as environmental compounds recalcitrant to degradation: Genetic mechanisms of bacterial biodegradation pathways. Genes 2019, 10, 512. [Google Scholar] [CrossRef]

| Strains | Strategies to Promote Sterol Uptake | Substrate (g/L) | Cosolvent | Fermentation Biotechnology | Main Products | References |
|---|---|---|---|---|---|---|
| Mycobacterium sp. strain MS136 | Enhancement of the Mce4 transport system by over-expressing mceG, yrbE4A and yrbE4B | 13 g/L phytosterol | 70 g/L β-cyclodextrin; 0.2 g/L Tween-80 | Batch fermentation | 6.0 g/L 9OH-AD | [32] |
| Mycobacterium sp. Strain LZ2 | Coexpression of the optimized Vitreoscilla hemoglobin gene and mceG | 10 g/L phytosterol | 25 mM hydroxypropyl-β-cyclodextrin | Immobilized repeated batch fermentation | 6.46 g/L AD | [77] |
| Deletion of kstD1, kstD2 and kstD3 in M. neoaurum ATCC 25795 | Deleted the key gene embC required for the synthesis of lipoarabinomannan from lipomannan | 20 g/L phytosterols | 80 g/L hydrox-ypropyl-β-cyclodextrin | Batch fermentation of resting cell | 9.9 g/L 9-OHAD | [78] |
| Deletion of kstD1, kstD2 and kstD3 in M. neoaurum ATCC 25795 | Deletion fbpC3 (a key factor for the synthesis of m-AG and TDM) and embC | 20 g/L phytosterols | 80 g/L hydrox-ypropyl-β-cyclodextrin | Batch fermentation of resting cell | 11.2 g/L 9-OHAD | [79] |
| Deletion of kstD1, kstD2 and kstD3 in M. neoaurum ATCC 25795 | Deletion kasB (β-ketoacyl-acyl carrier protein synthase gene) | 20 g/L phytosterols | 80 g/L hydrox-ypropyl-β-cyclodextrin | Batch fermentation of resting cell | 10.9 g/L 9-OHAD | [80] |
| Cofactor Regulation Strategies | Strain | Translational Systems | Conversion Rate (%) | Productivity (g/L/d) |
|---|---|---|---|---|
| NDH-II Overexpress | NdhF | None | 23.9 | 0.166 |
| NDH-II Overexpress | NdhF | TOWS | 95.0 | 0.656 |
| NDH-II Overexpress | NdhF | TOWS + Repeated batch | 86.1 | 0.921 |
| MMC Enhance | MNR-Fpcc | None | 25.4 | 0.176 |
| NDH-II Overexpress + MMC Enhance | MNR-Fpcc-Fndh | None | 33.8 | 0.235 |
| NDH-II Overexpress + MMC Enhance | MNR-Fpcc-Fndh | TOWS + Cane molasses + HMC | 96.4 | 0.669 |
| MCC Enhance | MNR-prpR | HP-β-CD | 90.6 | 0.628 |
| GlnR Knockout + MCC Enhance | MNR-prpDBC/ΔglnR | HP-β-CD | 94.3 | 0.654 |
| GlnR Knockout + MCC Enhance | MNR-prpDBC/ΔglnR | Low nitrogen sources + HP-β-CD | 92.8 | 0.644 |
| CAFC | MNR-P3 | HP-β-CD | 81.4 | 0.942 |
| PAFC | MNR-C3 | HP-β-CD | 93.2 | 1.078 |
| PAFC + MMC Enhance | MNR-P3-pccB | HP-β-CD | 94.6 | 1.187 |
| PAFC + MCC Enhance | MNR-P3-prpR | HP-β-CD | 97.3 | 1.222 |
| PAFC + MCC Enhance | MNR-P3-prpR | TOWS + Cane molasses | 97.0 | 1.218 |
| PAFC + MCC Enhance | MNR-P3-prpR | TOWS + Cane molasses + Repeated batch | 94.2 | 1.96 |
| None | MNR | None | 11.5 | 0.067 |
| None | MNR | HP-β-CD | 75.7 | 0.439 |
| None | MNR | TOWS | 72.4 | 0.417 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xiao, P.; Pan, D.; Zhou, X. New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production. Int. J. Mol. Sci. 2023, 24, 5236. https://doi.org/10.3390/ijms24065236
Zhang Y, Xiao P, Pan D, Zhou X. New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production. International Journal of Molecular Sciences. 2023; 24(6):5236. https://doi.org/10.3390/ijms24065236
Chicago/Turabian StyleZhang, Yang, Peiyao Xiao, Delong Pan, and Xiuling Zhou. 2023. "New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production" International Journal of Molecular Sciences 24, no. 6: 5236. https://doi.org/10.3390/ijms24065236
APA StyleZhang, Y., Xiao, P., Pan, D., & Zhou, X. (2023). New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production. International Journal of Molecular Sciences, 24(6), 5236. https://doi.org/10.3390/ijms24065236






