Differential Expression and Clinical Relevance of C-X-C Motif Chemokine Receptor 4 (CXCR4) in Renal Cell Carcinomas, Benign Renal Tumors, and Metastases
Abstract
1. Introduction
2. Results
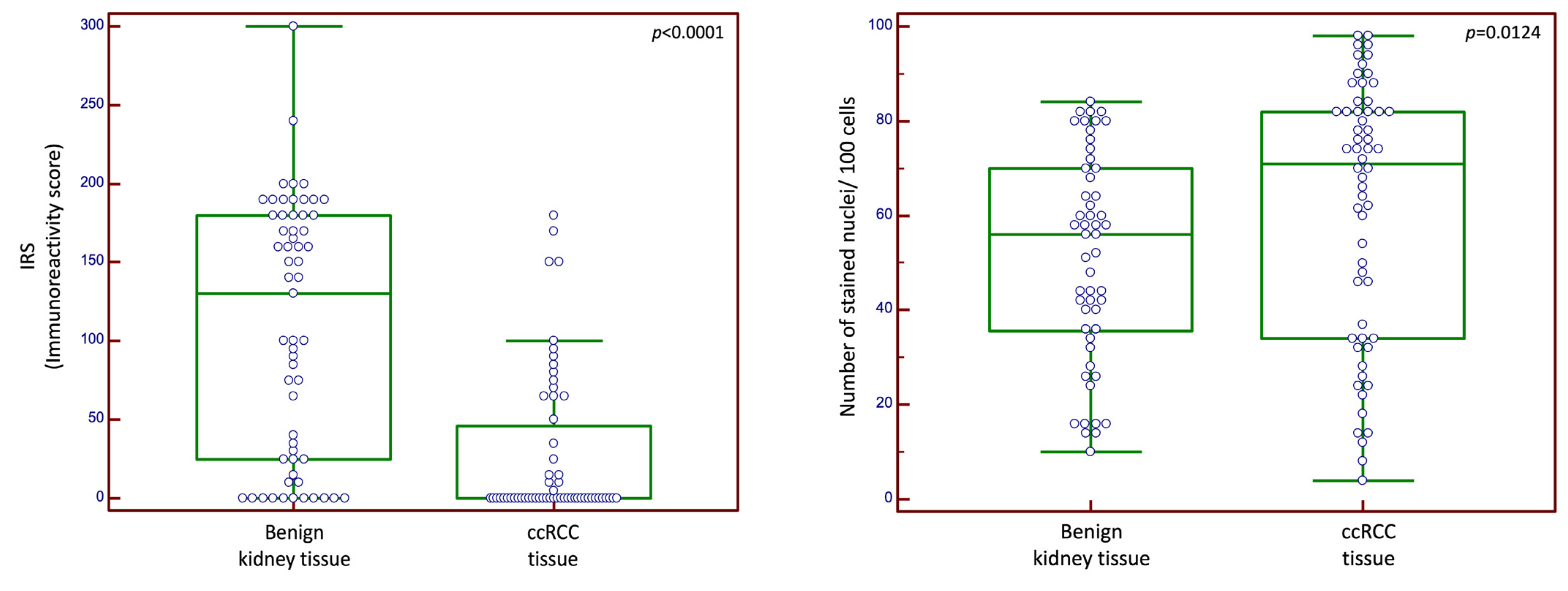
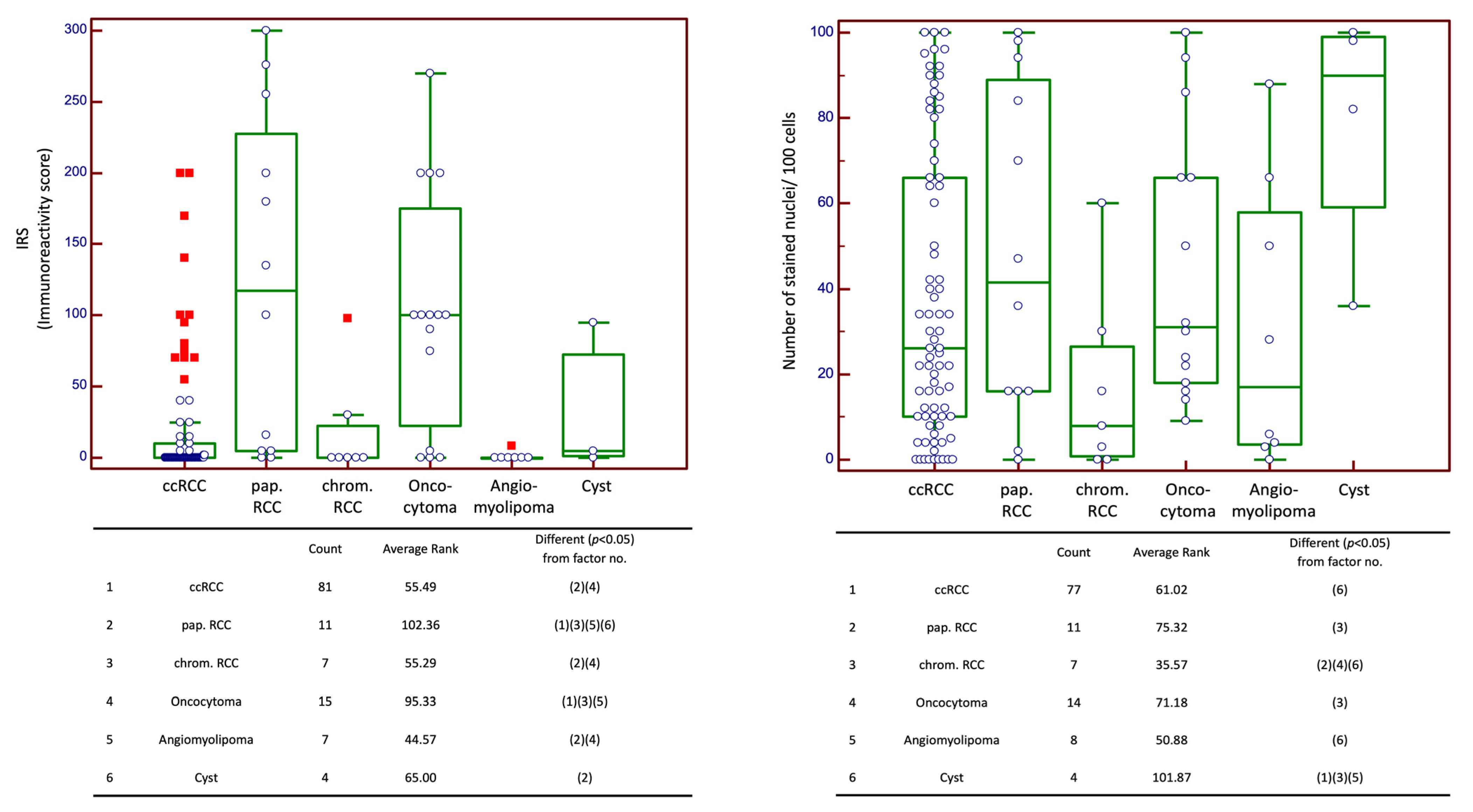
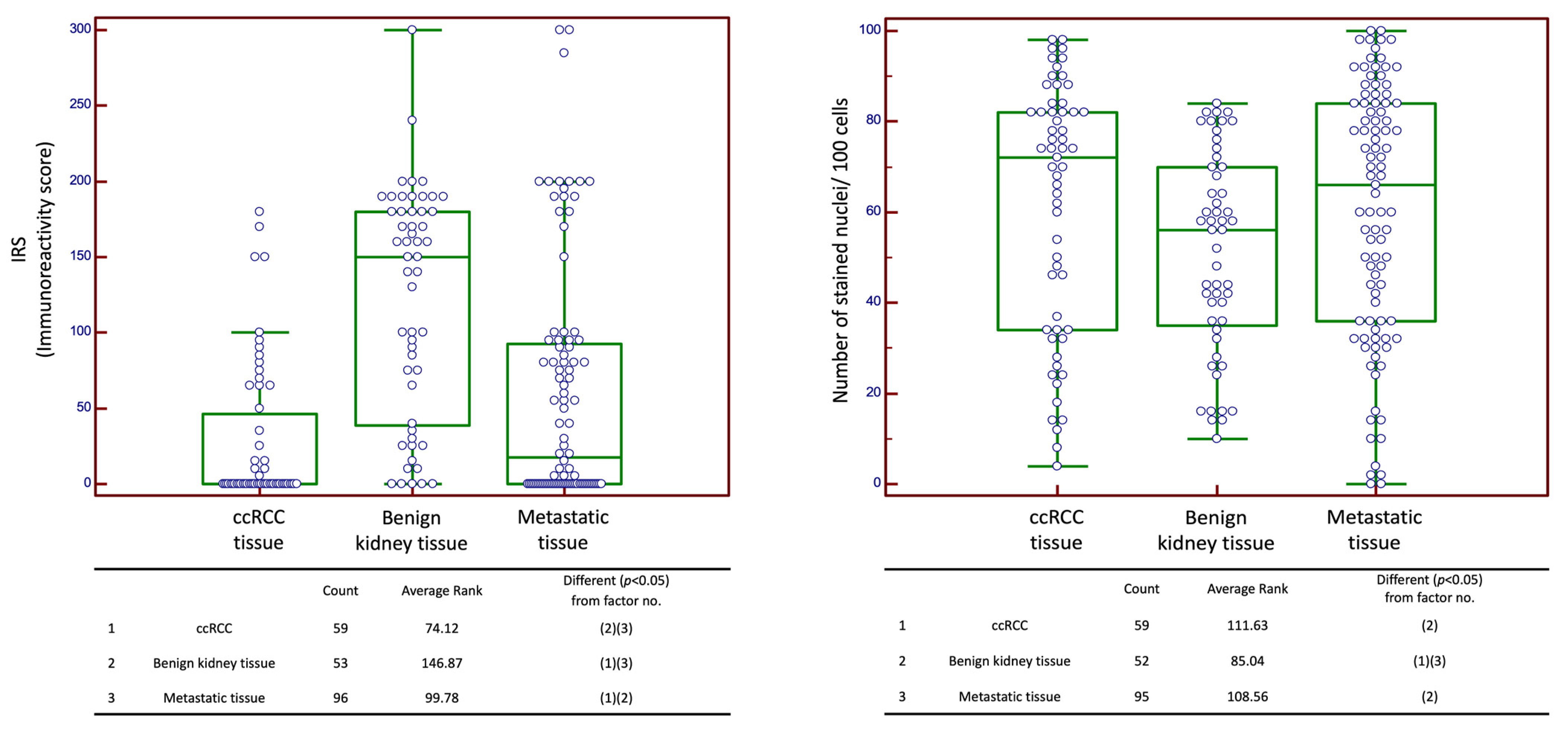
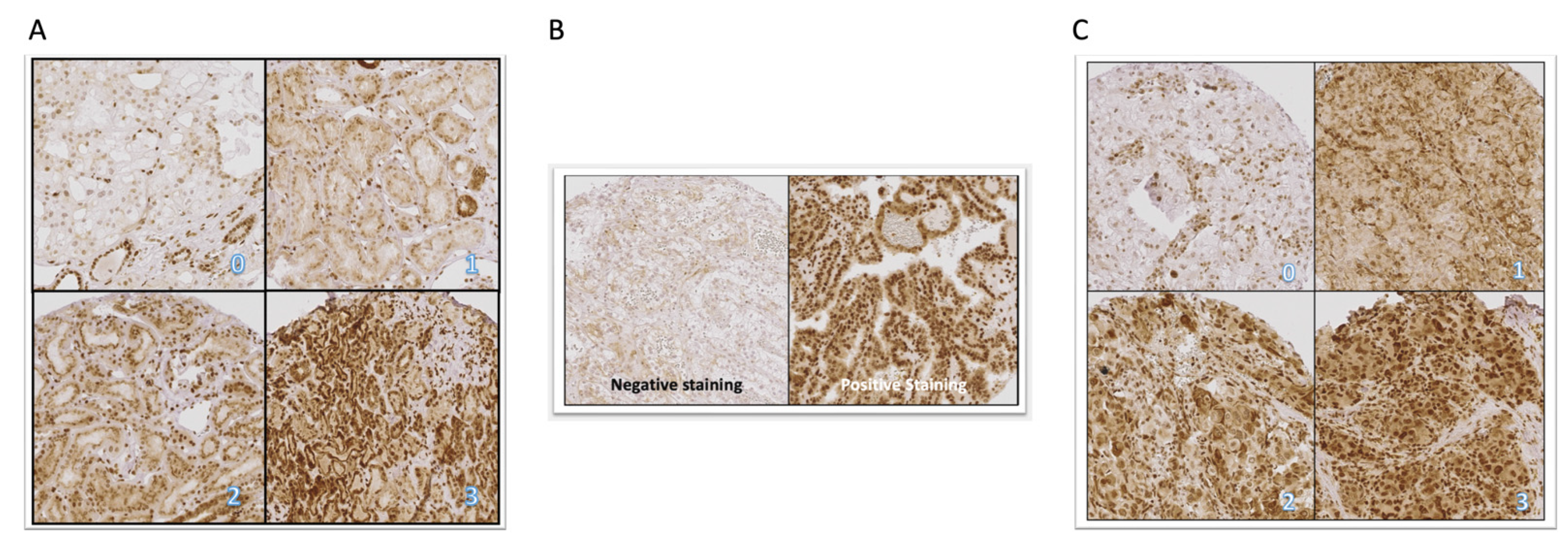
2.1. Expression of CXCR4
2.2. CXCR4 Expression in Correlation with Clinicopathological Data in Cohort n1
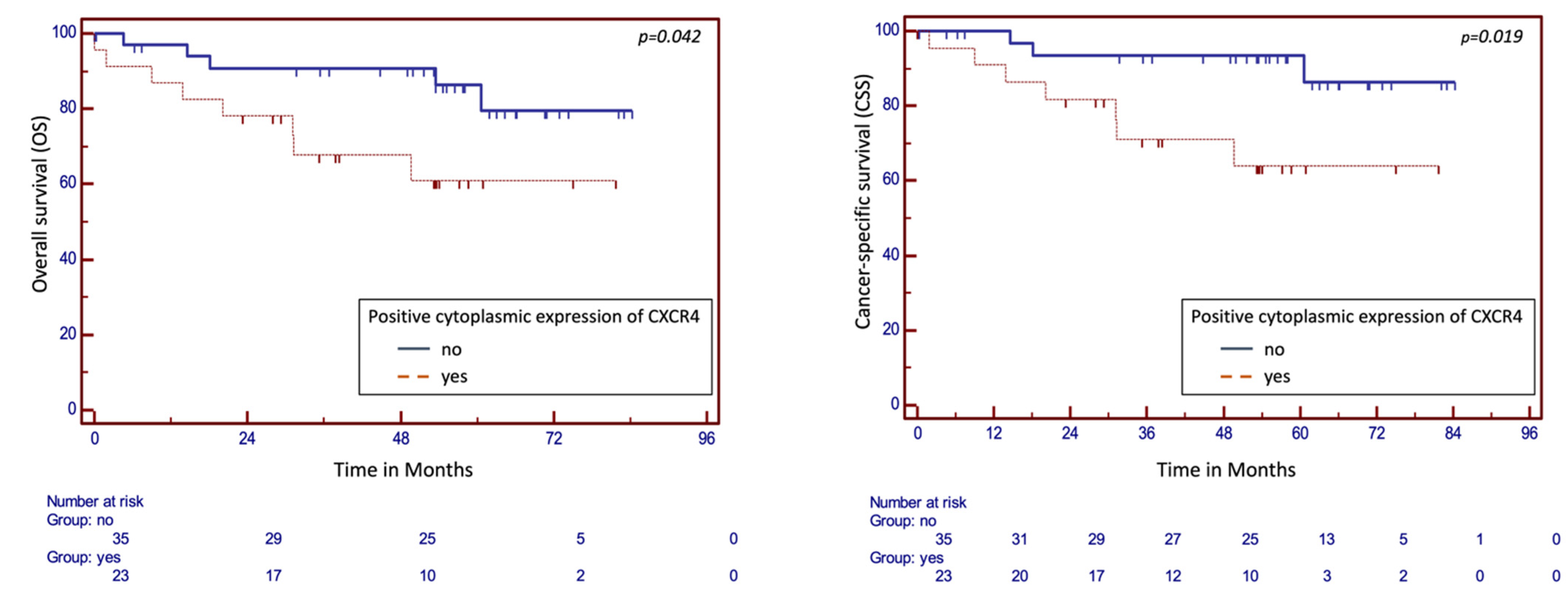
2.3. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Cohorts
4.2. TMA and Immunohistochemistry
4.3. Semi-Quantitative Evaluation of TMAs
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Rot, A.; von Andrian, U.H. Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 2004, 22, 891–928. [Google Scholar] [CrossRef]
- Luster, A.D. Chemokines—Chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 1998, 338, 436–445. [Google Scholar] [CrossRef]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef]
- Teixidó, J.; Martínez-Moreno, M.; Díaz-Martínez, M.; Sevilla-Movilla, S. The good and bad faces of the CXCR4 chemokine receptor. Int. J. Biochem. Cell Biol. 2018, 95, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cao, H.B.; Li, W.J.; Zhao, L. The CXCL12 (SDF-1)/CXCR4 chemokine axis: Oncogenic properties, molecular targeting, and synthetic and natural product CXCR4 inhibitors for cancer therapy. Chin. J. Nat. Med. 2018, 16, 801–810. [Google Scholar] [CrossRef]
- Akashi, T.; Koizumi, K.; Tsuneyama, K.; Saiki, I.; Takano, Y.; Fuse, H. Chemokine receptor CXCR4 expression and prognosis in patients with metastatic prostate cancer. Cancer Sci. 2008, 99, 539–542. [Google Scholar] [CrossRef]
- Sun, Y.X.; Wang, J.; Shelburne, C.E.; Lopatin, D.E.; Chinnaiyan, A.M.; Rubin, M.A.; Pienta, K.J.; Taichman, R.S. Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J. Cell. Biochem. 2003, 89, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Long, Q.; Guan, B.; Mu, L. Prognostic Value of High CXCR4 Expression in Renal Cell Carcinoma: A System Review and Meta-Analysis. Dis. Markers 2015, 2015, 568980. [Google Scholar] [CrossRef]
- Tang, B.; Tang, F.; Li, Y.; Yuan, S.; Li, B.; Wang, Z.; He, S. Clinicopathological significance of CXCR4 expression in renal cell carcinoma: A meta-analysis. Ann. Surg. Oncol. 2015, 22, 1026–1031. [Google Scholar] [CrossRef]
- Ljungberg, B.; Albiges, L.; Bedke, J.; Bex, A.; Capitanio, U.; Giles, R.; Hora, M.; Klatte, T.; Lam, T.; Marconi, L.; et al. EAU Guidelines; Edn. Presented at the EAU Annual Congress Milan 2021; European Association of Urology: Arnhem, The Netherlands, 2021. [Google Scholar]
- Wang, L.; Wang, L.; Yang, B.; Yang, Q.; Qiao, S.; Wang, Y.; Sun, Y. Strong expression of chemokine receptor CXCR4 by renal cell carcinoma cells correlates with metastasis. Clin. Exp. Metastasis 2009, 26, 1049–1054. [Google Scholar] [CrossRef]
- Zhao, F.L.; Guo, W. Expression of stromal derived factor-1 (SDF-1) and chemokine receptor (CXCR4) in bone metastasis of renal carcinoma. Mol. Biol. Rep. 2011, 38, 1039–1045. [Google Scholar] [CrossRef]
- Nengroo, M.A.; Khan, M.A.; Verma, A.; Datta, D. Demystifying the CXCR4 conundrum in cancer biology: Beyond the surface signaling paradigm. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188790. [Google Scholar] [CrossRef]
- Busillo, J.M.; Benovic, J.L. Regulation of CXCR4 signaling. Biochim. Biophys. Acta 2007, 1768, 952–963. [Google Scholar] [CrossRef]
- Zuo, J.; Wen, M.; Li, S.; Lv, X.; Wang, L.; Ai, X.; Lei, M. Overexpression of CXCR4 promotes invasion and migration of non-small cell lung cancer via EGFR and MMP-9. Oncol. Lett. 2017, 14, 7513–7521. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Mezzapelle, R. The Chemokine Receptor CXCR4 in Cell Proliferation and Tissue Regeneration. Front. Immunol. 2020, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guo, L.; Zhao, H.; Zhao, J.; Weng, H.; Zhao, B. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget 2015, 6, 5022–5040. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.G.; Kozin, S.V.; Kirkpatrick, N.D.; Xu, L.; Fukumura, D.; Jain, R.K. CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway inhibition: An emerging sensitizer for anticancer therapies? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 2074–2080. [Google Scholar] [CrossRef] [PubMed]
- Hainsworth, J.D.; Reeves, J.A.; Mace, J.R.; Crane, E.J.; Hamid, O.; Stille, J.R.; Flynt, A.; Roberson, S.; Polzer, J.; Arrowsmith, E.R. A Randomized, Open-Label Phase 2 Study of the CXCR4 Inhibitor LY2510924 in Combination with Sunitinib Versus Sunitinib Alone in Patients with Metastatic Renal Cell Carcinoma (RCC). Target. Oncol. 2016, 11, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Nengroo, M.A.; Maheshwari, S.; Singh, A.; Verma, A.; Arya, R.K.; Chaturvedi, P.; Saini, K.K.; Singh, A.K.; Sinha, A.; Meena, S.; et al. CXCR4 intracellular protein promotes drug resistance and tumorigenic potential by inversely regulating the expression of Death Receptor 5. Cell Death Dis. 2021, 12, 464. [Google Scholar] [CrossRef]
- Don-Salu-Hewage, A.S.; Chan, S.Y.; McAndrews, K.M.; Chetram, M.A.; Dawson, M.R.; Bethea, D.A.; Hinton, C.V. Cysteine (C)-x-C receptor 4 undergoes transportin 1-dependent nuclear localization and remains functional at the nucleus of metastatic prostate cancer cells. PLoS ONE 2013, 8, e57194. [Google Scholar] [CrossRef] [PubMed]
- Saba, N.F.; Wang, Y.; Fu, H.; Koenig, L.; Khuri, F.R.; Shin, D.M.; Chen, Z.G. Association of Cytoplasmic CXCR4 With Loss of Epithelial Marker and Activation of ERK1/2 and AKT Signaling Pathways in Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2017, 18, e203–e210. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, Y.; Xia, J.; Chen, N.; Wei, Q.; Li, X.; Zhang, P.; Shen, P.F.; Wang, J.; Zeng, H. CXCR4 expression in patients with high-risk locally advanced renal cell carcinoma can independently predict increased risk of disease progression and poor overall survival. Asian Pac. J. Cancer Prev. 2011, 12, 3313–3318. [Google Scholar]
- D’Alterio, C.; Cindolo, L.; Portella, L.; Polimeno, M.; Consales, C.; Riccio, A.; Cioffi, M.; Franco, R.; Chiodini, P.; Cartenì, G.; et al. Differential role of CD133 and CXCR4 in renal cell carcinoma. Cell Cycle 2010, 9, 4492–4500. [Google Scholar] [CrossRef] [PubMed]
- Floranović, M.P.; Petrović, A.R.; Veličković, L.J. Expression of the CXCR4 and CXCR7 in renal cancers; can “the orphan receptor” predict the mortality? Ann. Diagn. Pathol. 2021, 55, 151829. [Google Scholar] [CrossRef] [PubMed]
- Wehler, T.C.; Graf, C.; Biesterfeld, S.; Brenner, W.; Schadt, J.; Gockel, I.; Berger, M.R.; Thüroff, J.W.; Galle, P.R.; Moehler, M.; et al. Strong expression of chemokine receptor CXCR4 by renal cell carcinoma correlates with advanced disease. J. Oncol. 2008, 2008, 626340. [Google Scholar] [CrossRef] [PubMed]
- Atlas, T.H.P. CXCR4. 2022. Available online: https://www.proteinatlas.org/ENSG00000121966-CXCR4 (accessed on 24 October 2022).
- Okuyama Kishima, M.; de Oliveira, C.E.; Banin-Hirata, B.K.; Losi-Guembarovski, R.; Brajão de Oliveira, K.; Amarante, M.K.; Watanabe, M.A. Immunohistochemical expression of CXCR4 on breast cancer and its clinical significance. Anal. Cell. Pathol. 2015, 2015, 891020. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Gao, L.; Yang, Q.; Liu, B.; Wu, Z.; Wang, Y.; Sun, Y. High expression of CXCR4, CXCR7 and SDF-1 predicts poor survival in renal cell carcinoma. World J. Surg. Oncol. 2012, 10, 212. [Google Scholar] [CrossRef]
- Yoshitake, N.; Fukui, H.; Yamagishi, H.; Sekikawa, A.; Fujii, S.; Tomita, S.; Ichikawa, K.; Imura, J.; Hiraishi, H.; Fujimori, T. Expression of SDF-1 alpha and nuclear CXCR4 predicts lymph node metastasis in colorectal cancer. Br. J. Cancer 2008, 98, 1682–1689. [Google Scholar] [CrossRef]
- Cabioglu, N.; Gong, Y.; Islam, R.; Broglio, K.R.; Sneige, N.; Sahin, A.; Gonzalez-Angulo, A.M.; Morandi, P.; Bucana, C.; Hortobagyi, G.N.; et al. Expression of growth factor and chemokine receptors: New insights in the biology of inflammatory breast cancer. Ann. Oncol. 2007, 18, 1021–1029. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, Z.; Liu, B.; Lu, X.; Xiong, Y.; Shi, J.; Li, P.; Chen, J.; Zhang, Z.; Chen, M.; et al. A feed-forward loop between nuclear translocation of CXCR4 and HIF-1α promotes renal cell carcinoma metastasis. Oncogene 2019, 38, 881–895. [Google Scholar] [CrossRef] [PubMed]
- Rasti, A.; Abolhasani, M.; Zanjani, L.S.; Asgari, M.; Mehrazma, M.; Madjd, Z. Reduced expression of CXCR4, a novel renal cancer stem cell marker, is associated with high-grade renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 95–104. [Google Scholar] [CrossRef]
- Werner, R.A.; Kircher, S.; Higuchi, T.; Kircher, M.; Schirbel, A.; Wester, H.J.; Buck, A.K.; Pomper, M.G.; Rowe, S.P.; Lapa, C. CXCR4-Directed Imaging in Solid Tumors. Front. Oncol. 2019, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Xu, L.; Zhu, Y.; Lv, T.; Liu, W.; Liu, Y.; Liu, H.; Chen, L.; Xu, J.; Lin, Z. High CXC chemokine receptor 4 expression is an adverse prognostic factor in patients with clear-cell renal cell carcinoma. Br. J. Cancer 2014, 110, 2261–2268. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Badin, G.; Zhao, A.; Gentil-Perret, A.; Tostain, J.; Péoc’h, M.; Gigante, M. Prognostic value of CXCR4 expression in patients with clear cell renal cell carcinoma. Histol. Histopathol. 2013, 28, 1217–1222. [Google Scholar] [CrossRef]
- Potić Floranović, M.; Ristić Petrović, A.; Veličković, F.; Janković Veličković, L. Expression and prognostic value of CXCL12/CXCR4/CXCR7 axis in clear cell renal cell carcinoma. Clin. Exp. Nephrol. 2021, 25, 1057–1069. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Uhlen, M.E.A. Human Protein Atlas. 2022. Available online: https://www.proteinatlas.org/ENSG00000121966-CXCR4/pathology/renal+cancer/KIRC (accessed on 24 October 2022).
- Wittekind, L.C. International Union against Cancer (UICC): TNM Classification of Malignant Tumors; Wiley-Liss: New York, NY, USA, 2002. [Google Scholar]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Bärlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef]

| n | % | |
|---|---|---|
| Total number of patients | 64 | 100 |
| Male | 37 | 57.8 |
| Female | 27 | 42.2 |
| Age in years | Median 64.2 | Range 35.7–88.0 |
| Pathological stage | ||
| T1a | 18 | 28.1 |
| T1b | 16 | 25.0 |
| T2a | 5 | 7.8 |
| T2b | 1 | 1.6 |
| T3a | 10 | 15.6 |
| T3b | 14 | 21.9 |
| Lymph node status | ||
| N0 | 57 | 89.1 |
| N1 | 7 | 10.9 |
| Distant metastatic status | ||
| M0 | 54 | 84.4 |
| M1 | 9 | 14.1 |
| Mx | 1 | 1.5 |
| R-Status | ||
| R0 | 60 | 93.8 |
| R1 | 4 | 6.2 |
| L-Status | ||
| L0 | 61 | 95.3 |
| L1 | 3 | 4.7 |
| V-Status | ||
| V0 | 46 | 71.9 |
| V1 | 17 | 26.5 |
| V2 | 1 | 1.6 |
| Fuhrman grading | ||
| G1 | 14 | 21.8% |
| G2 | 41 | 64.1% |
| G3/G4 | 9 | 14.1% |
| Sarcomatoid features | ||
| Yes | 4 | 6.2 |
| No | 60 | 93.8 |
| Tumor necrosis | ||
| Yes | 9 | 14.1 |
| No | 55 | 85.9 |
| Primary tumor size in cm | Median 5.5 | Range 1.4–16.0 |
| FU time (months) from date of diagnosis of primary ccRCC | Mean 47.02 Median 53.34 | 95% CI (mean) 42.03–52.02 Range (Median) 0.00–87.72 |
| N2: Cohort of Various Histological Entities | ||||||
|---|---|---|---|---|---|---|
| n | % | |||||
| Total number of patients | 146 | 100 | ||||
| Male | 90 | 61.64 | ||||
| Female | 56 | 38.36 | ||||
| Age in years | Median in years 64.08 | Range 24.30–89.82 | ||||
| Malignant tumors within this cohort | ||||||
| ccRCC | Pap. RCC | Chrom. RCC | ||||
| n | % | n | % | n | % | |
| Number of Patients | 87 | 59.59 | 14 | 9.59 | 7 | 4.79 |
| Male | 53 | 36.30 | 11 | 7.53 | 4 | 2.74 |
| Female | 34 | 23.29 | 3 | 2.05 | 3 | 2.05 |
| Age in years | Median 65.08 | Range 27.66–89.82 | Median 63.38 | Range 37.72–81.11 | Median 46.54 | Range 44.70–81.15 |
| Pathological Stage | ||||||
| T1a | 42 | 28.77 | 6 | 4.11 | 2 | 1.37 |
| T1b | 18 | 12.33 | 3 | 2.05 | 3 | 2.05 |
| T2a | 4 | 2.74 | 2 | 1.37 | 2 | 1.37 |
| T2b | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| T3a | 20 | 13.70 | 2 | 1.37 | 0 | 0.00 |
| T3b | 2 | 1.37 | 1 | 0.68 | 0 | 0.00 |
| T3c | 1 | 0.68 | 0 | 0.00 | 0 | 0.00 |
| T4 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Lymph node status | ||||||
| N0 | 79 | 54.11 | 12 | 8.22 | 7 | 4.79 |
| N1 | 6 | 4.11 | 2 | 1.37 | 0 | 0.00 |
| Nx | 2 | 1.37 | 0 | 0.00 | 0 | 0.00 |
| Distant metastatic status | ||||||
| M0 | 74 | 50.68 | 1 | 0.68 | 7 | 4.79 |
| M1 | 11 | 7.53 | 13 | 8.90 | 0 | 0.00 |
| Mx | 2 | 1.37 | 0 | 0.00 | 0 | 0.00 |
| R-Status | ||||||
| R0 | 77 | 52.74 | 11 | 7.53 | 7 | 4.79 |
| R1 | 7 | 4.79 | 2 | 1.37 | 0 | 0.00 |
| RX | 3 | 2.05 | 1 | 0.68 | 0 | 0.00 |
| L-Status | ||||||
| L0 | 85 | 58.22 | 11 | 7.53 | 7 | 4.79 |
| L1 | 2 | 1.37 | 3 | 2.05 | 0 | 0.00 |
| V-Status | ||||||
| V0 | 72 | 49.32 | 13 | 8.90 | 7 | 4.79 |
| V1 | 11 | 7.53 | 0 | 0.00 | 0 | 0.00 |
| V2 | 4 | 2.74 | 1 | 0.68 | 0 | 0.00 |
| Fuhrman grading | ||||||
| G1 | 25 | 17.12 | 2 | 1.37 | 1 | 0.68 |
| G2 | 48 | 32.88 | 9 | 6.16 | 5 | 3.42 |
| G3-4 | 13 | 8.90 | 2 | 1.37 | 1 | 0.68 |
| Gx | 1 | 0.68 | 1 | 0.68 | 0 | 0.00 |
| Sarcomatoid features | ||||||
| Yes | 3 | 2.05 | 1 | 0.68 | 0 | 0.00 |
| No | 84 | 57.53 | 13 | 8.90 | 7 | 4.79 |
| Tumor Necrosis | ||||||
| Yes | 21 | 14.38 | 6 | 4.11 | 1 | 0.68 |
| No | 66 | 45.21 | 8 | 5.48 | 6 | 4.11 |
| Primary tumor size in cm | Median 3.5 | Range 0.5–15.00 | Median 3.9 | Range 1.5–12.00 | Median 4.3 | Range 2.00–7.80 |
| Benign tumors within this cohort | ||||||
| Oncocytoma | Angiomyolipoma | Cyst | ||||
| n | % | n | % | n | % | |
| Number of patients | 16 | 10.96 | 10 | 6.85 | 7 | 4.79 |
| Male | 9 | 6.16 | 3 | 2.05 | 6 | 4.11 |
| Female | 7 | 4.79 | 7 | 4.79 | 1 | 0.68 |
| Age in years | Median 62.56 | Range 51.38–77.86 | Median 60.88 | Range 24.30–74.13 | Median 61.28 | Range 32.56–78.82 |
| Unknown tumors within this cohort | ||||||
| n | % | |||||
| Number of patients | 5 | 3.42 | ||||
| Male | 3 | 2.05 | ||||
| Female | 2 | 1.37 | ||||
| Age in years | Median 60.4 | Range 34.32–74.85 | ||||
| N3: Metastases cohort | ||
|---|---|---|
| n | % | |
| Total number of patients with met. RCC | 92 | 100% |
| Total number of metastases | 137 | 100% |
| Histology of primary tumor | ||
| ccRCC | 83 | 90.22% |
| Pap. RCC | 3 | 3.26% |
| Chrom. RCC | 2 | 2.17% |
| Unknown | 4 | 4.35% |
| Metastasis | ||
| metachronous | 62 | 67.39% |
| synchronous | 27 | 29.35% |
| unknown | 3 | 3.26% |
| Metastatic site | ||
| Lung | 35 | 25.55% |
| Soft tissue | 21 | 15.33% |
| Bone | 18 | 13.14% |
| Lymph node | 15 | 10.95% |
| Adrenal gland | 11 | 8.03% |
| Pancreas | 6 | 4.38% |
| Liver | 5 | 3.65% |
| Local recurrence | 4 | 2.92% |
| Diaphragm/ Muscle | 3 | 2.19% |
| Pleura | 2 | 1.46% |
| Testicle | 2 | 1.46% |
| Small intestine | 2 | 1.46% |
| Skin | 1 | 0.73% |
| Larynx | 1 | 0.73% |
| Sympathetic trunk | 1 | 0.73% |
| Mesentery | 1 | 0.73% |
| Peritoneum | 1 | 0.73% |
| Spleen | 1 | 0.73% |
| Brain | 1 | 0.73% |
| Unknown | 6 | 4.38% |
| n1: ccRCC Cohort | T-Stage (T > T2) | N-Stage (N+) | Lymphovascular Invasion (L+) | Vascular Invasion (V+) | Grading (G > G2) | Sarcomatoid Differentiation | Necrosis |
|---|---|---|---|---|---|---|---|
| Positive cytoplasmic CXCR4 expression | p = 0.5809 | p = 0.2370 | p = 0.0097 | p = 0.1298 | p = 0.6861 | p = 0.6142 | p = 0.0634 |
| Positive nuclear CXCR4 expression | p = 0.2204 | p = 0.7859 | p = 0.2411 | p = 0.5148 | p = 0.8787 | p = 0.4132 | p = 0.9616 |
| Cancer-Specific Survival (CSS) | Overall Survival (OS) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Covariate | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||||
| p | HR | 95% CI | p | HR | 95% CI | p | HR | 95% CI | p | HR | 95% CI | |
| Clinical Stage ≥ T2 | 0.0093 | 7.6407 | 1.6629 to 35.1085 | 0.1386 | 4.3516 | 0.6278 to 30.1617 | 0.0046 | 6.3617 | 1.7835–22.6920 | 0.1071 | 3.9410 | 0.7497 to 20.7186 |
| Sarcomatoid features | 0.0003 | 13.9040 | 3.4158 to 56.5963 | 0.8367 | 0.7732 | 0.0678 to 8.8149 | 0.0014 | 9.0238 | 2.3498–34.6527 | 0.8128 | 1.3192 | 0.1348 to 12.9154 |
| N+ | <0.0001 | 15.5824 | 4.6962 to 51.7044 | 0.1364 | 6.3868 | 0.5636 to 72.3807 | <0.0001 | 9.1089 | 3.1287–26.5202 | 0.4155 | 2.3116 | 0.3107 to 17.1968 |
| Venous invasion V+ | 0.0092 | 3.2142 | 1.3412 to 7.70 | 0.7326 | 0.7371 | 0.1292 to 4.2035 | 0.0152 | 2.6846 | 1.2140–5.9367 | 0.5763 | 0.6658 | 0.1610 to 2.7536 |
| Lymphovascular invasion L+ | <0.0001 | 35.9364 | 7.0777 to 182.4646 | 0.5824 | 2.1766 | 0.1380 to 34.3246 | <0.0001 | 19.0835 | 4.4745–81.3896 | 0.4900 | 2.8076 | 0.1520 to 51.8496 |
| Fuhrman grading G > 2 | 0.0004 | 8.9889 | 2.6983 to 29.9454 | 0.4745 | 2.0447 | 0.2909 to 14.3739 | 0.0026 | 5.5545 | 1.8337–16.8248 | 0.6026 | 1.6717 | 0.2440 to 11.4528 |
| Cytoplasmic expression CXCR4 | 0.0317 | 4.4826 | 1.1483 to 17.4993 | 0.1386 | 4.3516 | 0.6278 to 30.1617 | 0.0457 | 3.0454 | 1.0272–9.0290 | 0.5233 | 1.6381 | 0.3626 to 7.4002 |
| Nuclear expression CXCR4 | 0.5092 | 1.5775 | 0.4103 to 6.0648 | 0.5529 | 1.8713 | 0.2388–14.6646 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maas, M.; Kurcz, A.; Hennenlotter, J.; Scharpf, M.; Fend, F.; Walz, S.; Stühler, V.; Todenhöfer, T.; Stenzl, A.; Bedke, J.; et al. Differential Expression and Clinical Relevance of C-X-C Motif Chemokine Receptor 4 (CXCR4) in Renal Cell Carcinomas, Benign Renal Tumors, and Metastases. Int. J. Mol. Sci. 2023, 24, 5227. https://doi.org/10.3390/ijms24065227
Maas M, Kurcz A, Hennenlotter J, Scharpf M, Fend F, Walz S, Stühler V, Todenhöfer T, Stenzl A, Bedke J, et al. Differential Expression and Clinical Relevance of C-X-C Motif Chemokine Receptor 4 (CXCR4) in Renal Cell Carcinomas, Benign Renal Tumors, and Metastases. International Journal of Molecular Sciences. 2023; 24(6):5227. https://doi.org/10.3390/ijms24065227
Chicago/Turabian StyleMaas, Moritz, Aymone Kurcz, Jörg Hennenlotter, Marcus Scharpf, Falko Fend, Simon Walz, Viktoria Stühler, Tilman Todenhöfer, Arnulf Stenzl, Jens Bedke, and et al. 2023. "Differential Expression and Clinical Relevance of C-X-C Motif Chemokine Receptor 4 (CXCR4) in Renal Cell Carcinomas, Benign Renal Tumors, and Metastases" International Journal of Molecular Sciences 24, no. 6: 5227. https://doi.org/10.3390/ijms24065227
APA StyleMaas, M., Kurcz, A., Hennenlotter, J., Scharpf, M., Fend, F., Walz, S., Stühler, V., Todenhöfer, T., Stenzl, A., Bedke, J., & Rausch, S. (2023). Differential Expression and Clinical Relevance of C-X-C Motif Chemokine Receptor 4 (CXCR4) in Renal Cell Carcinomas, Benign Renal Tumors, and Metastases. International Journal of Molecular Sciences, 24(6), 5227. https://doi.org/10.3390/ijms24065227







