PPARβ/δ Ligands Regulate Oxidative Status and Inflammatory Response in Inflamed Corpus Luteum—An In Vitro Study
Abstract
1. Introduction
2. Results
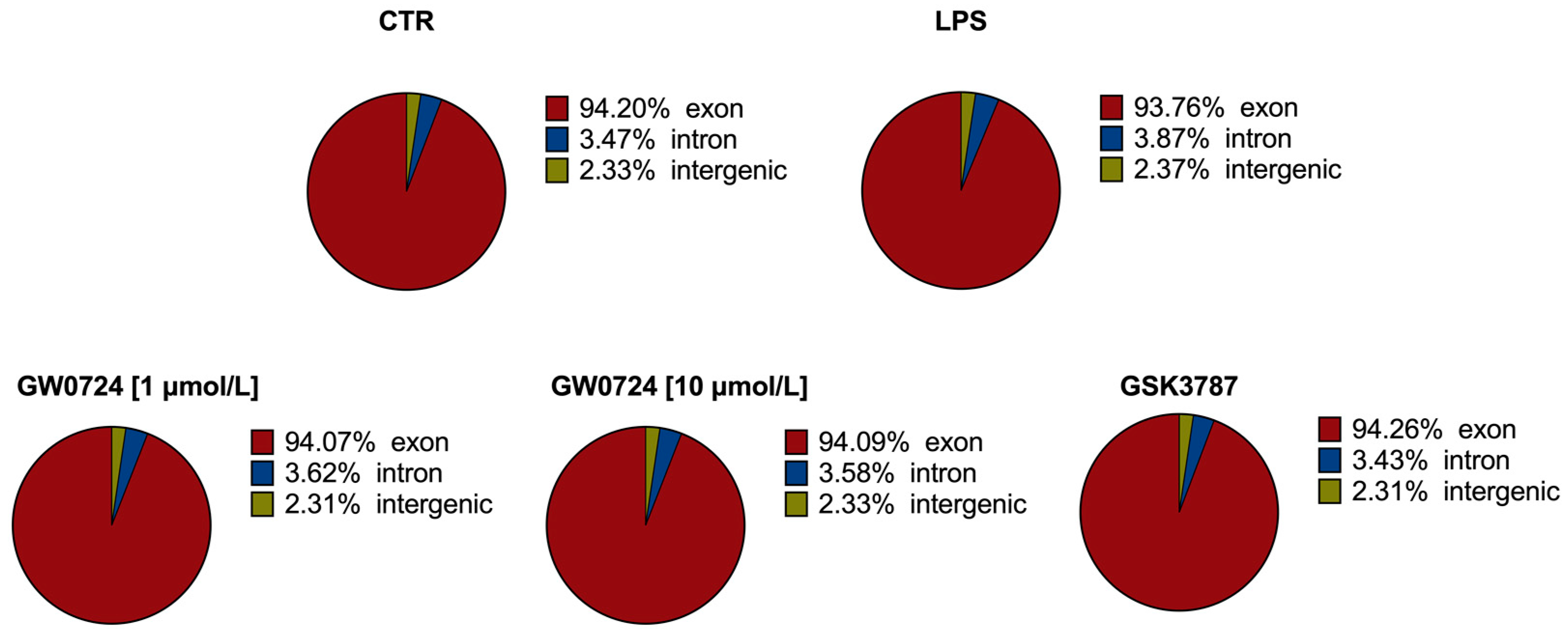
2.1. Statistics of RNA Sequencing
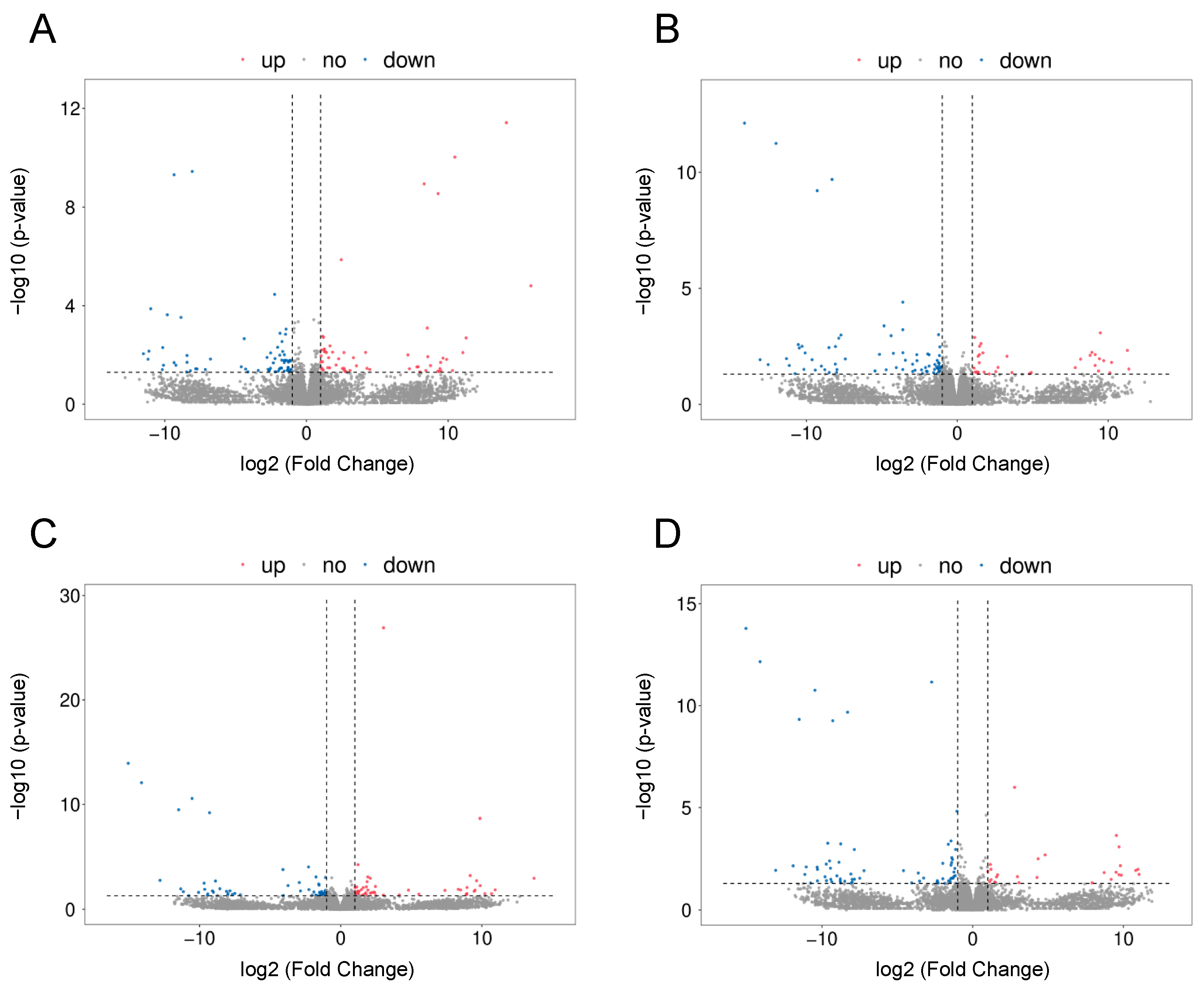
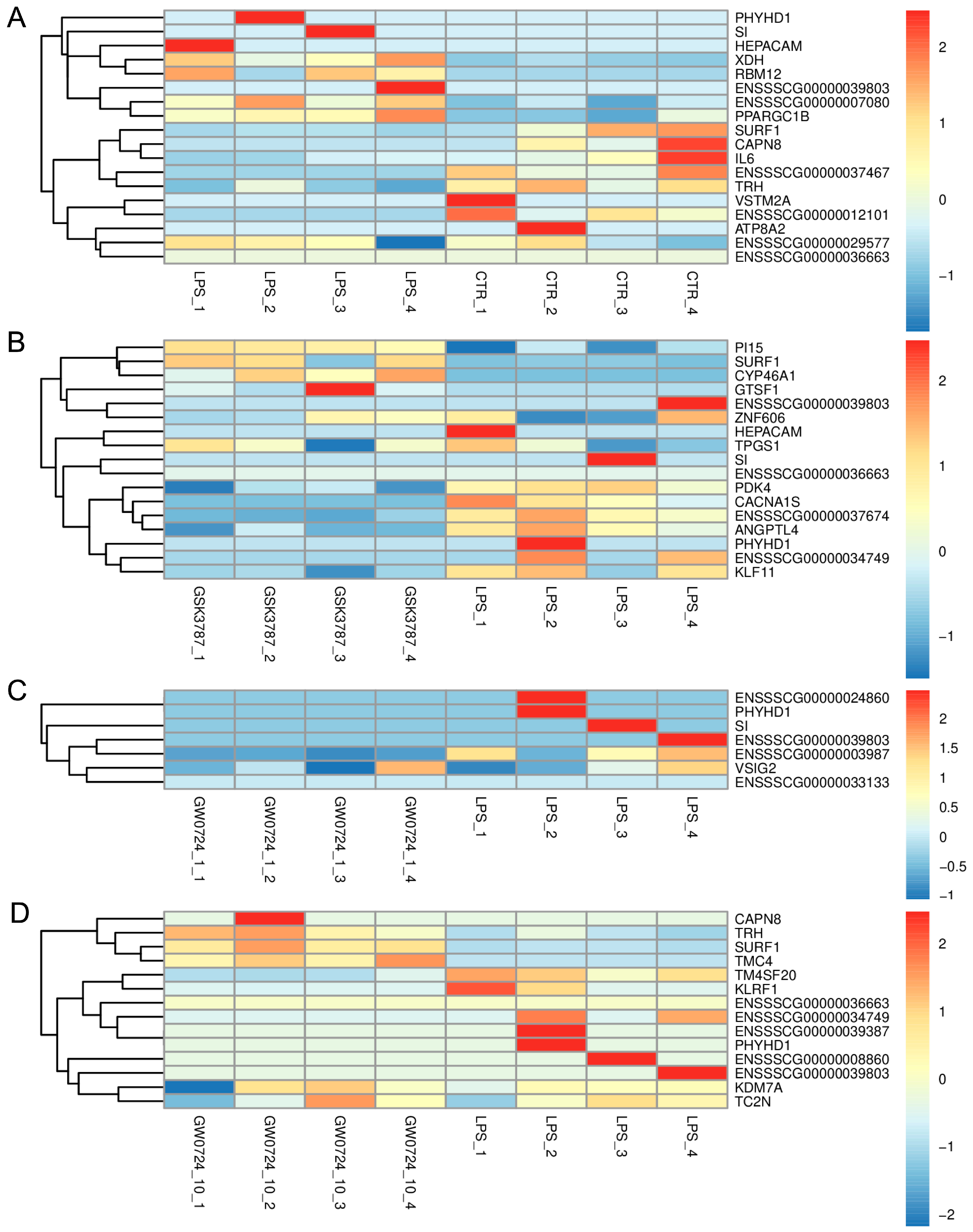
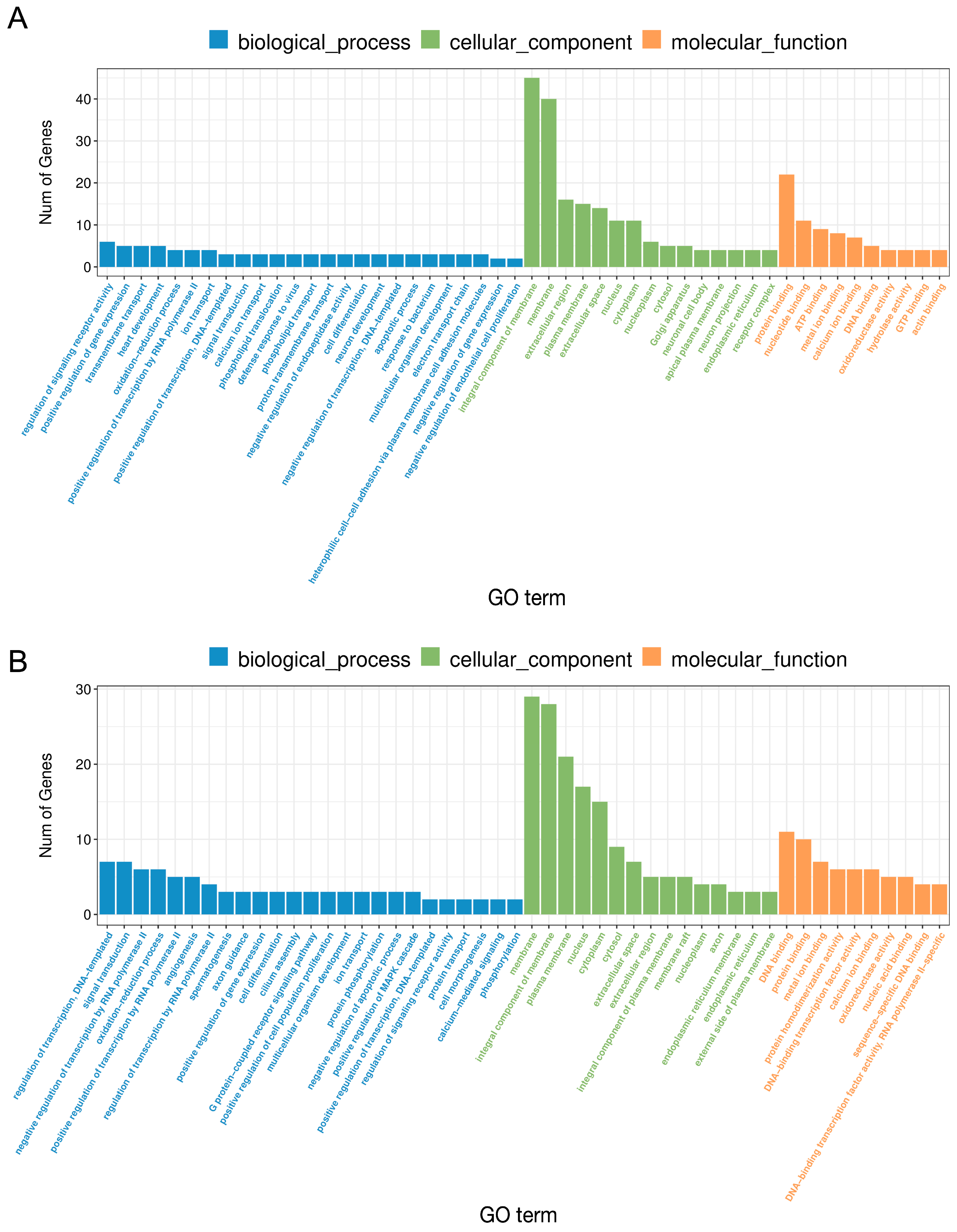
2.2. The Effect of LPS on Differential Gene Expression in the Corpus Luteum
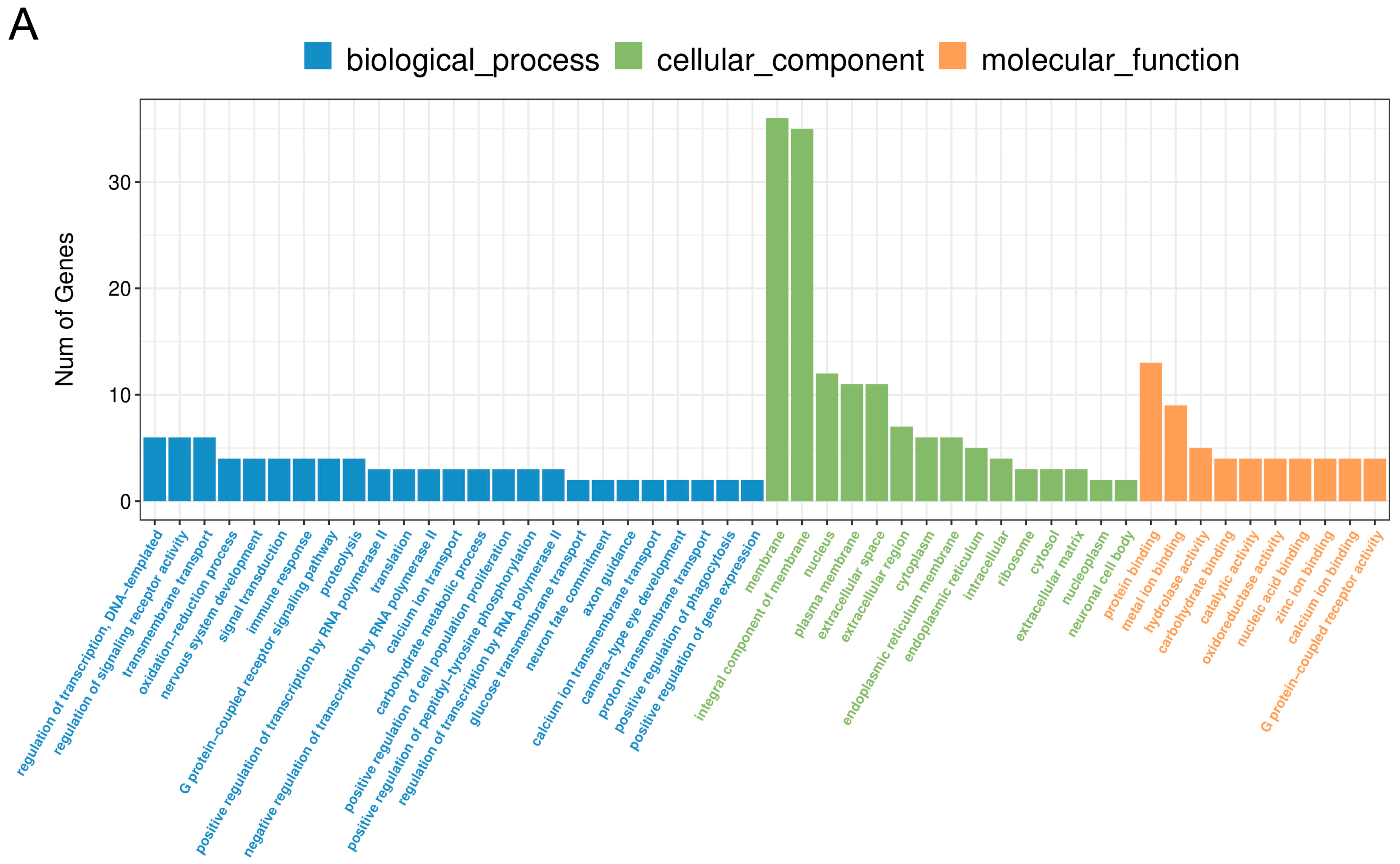
2.3. The Effect of PPARβ/δ Agonist on Differential Gene Expression in the Corpus Luteum
2.4. Comparative Analysis between Two Doses of PPARβ/δ Agonist
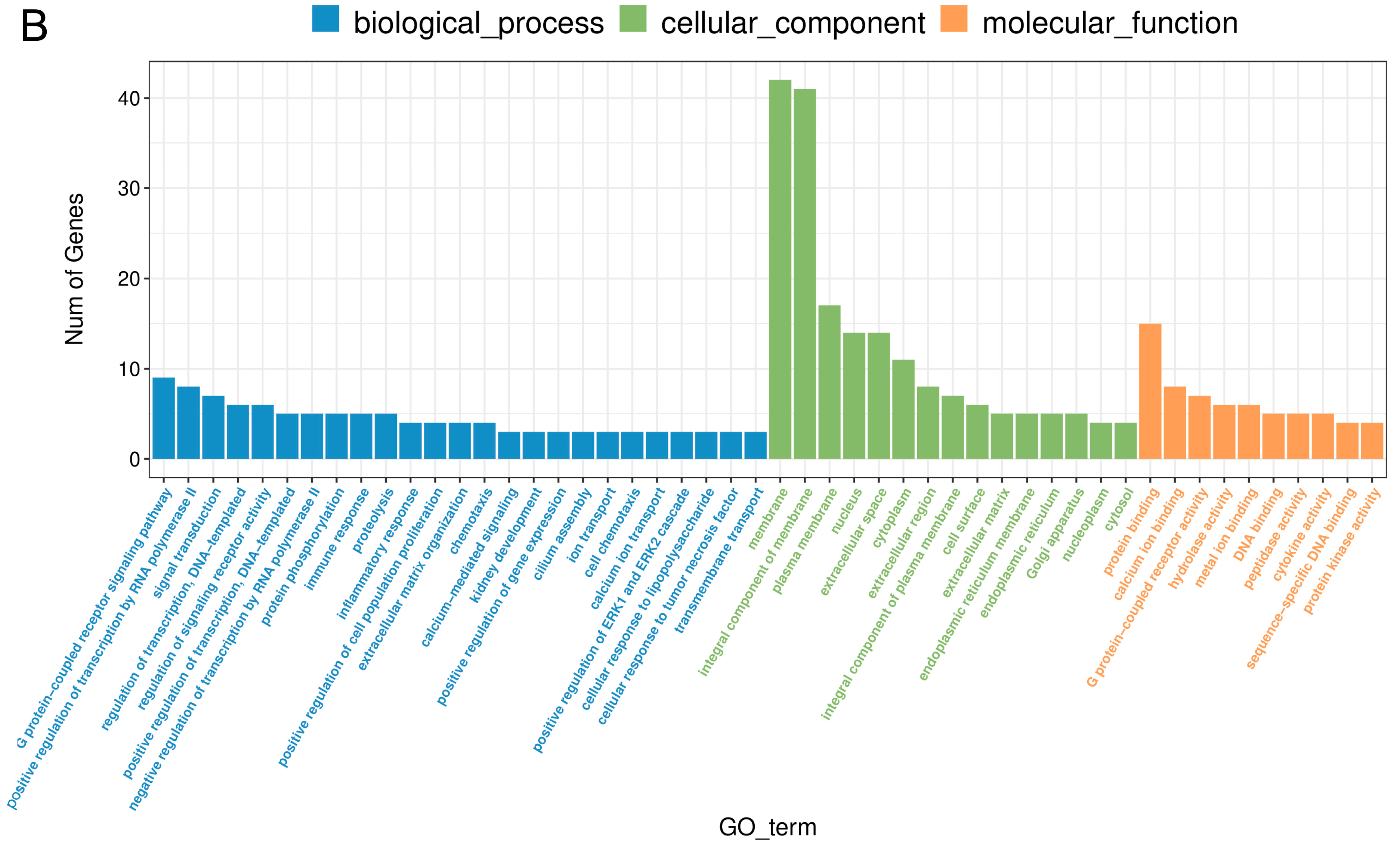
2.5. The Effect of PPARβ/δ Antagonist on Differential Gene Expression in the Corpus Luteum
2.6. Real-Time PCR Analysis
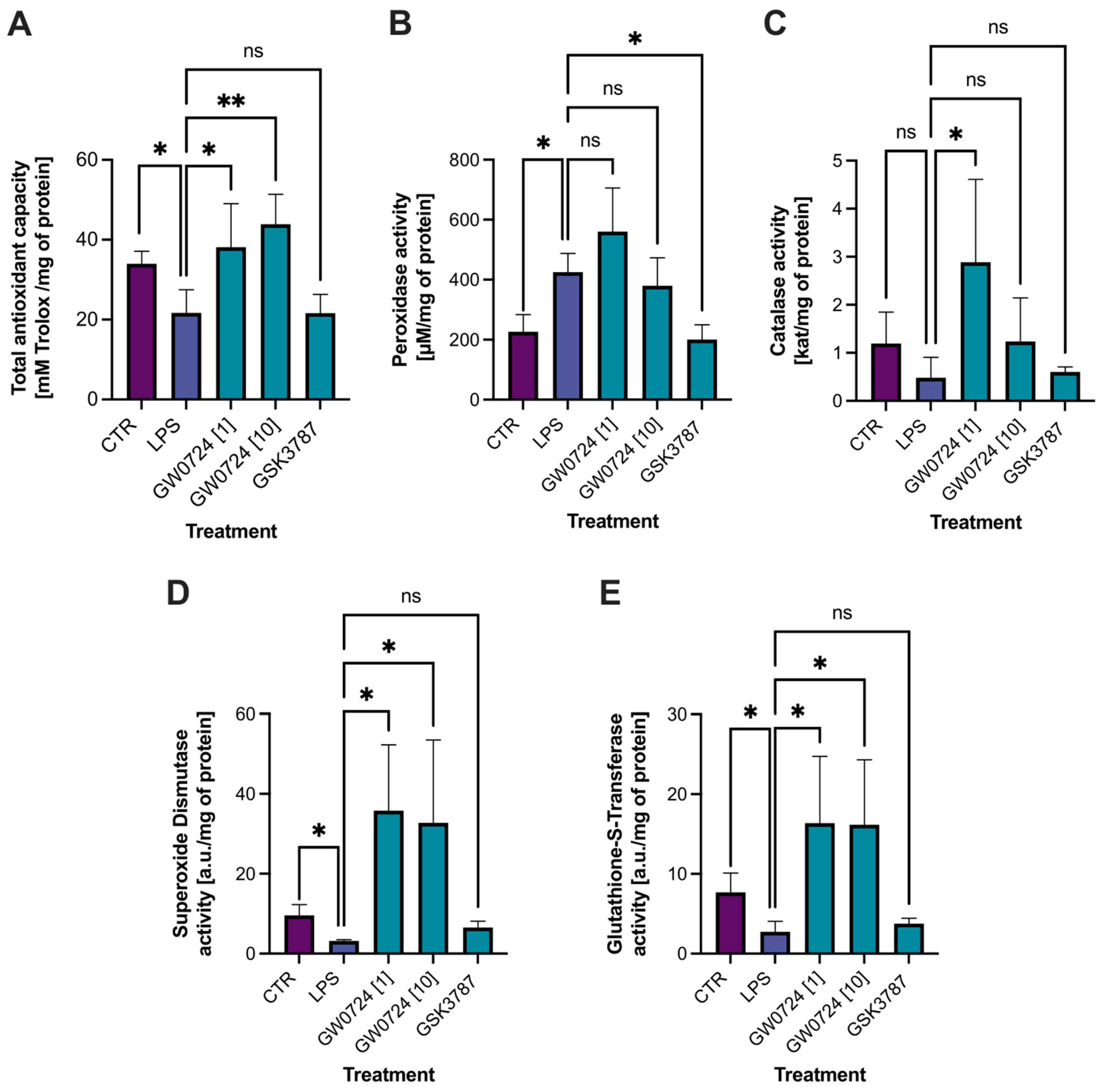
2.7. Biochemical Analyses
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. In Vitro Experiment
4.3. RNA Isolation, Library Preparation and Sequencing Procedure
4.4. Transcript Assembly and Analysis of Differentially Expressed Genes
4.5. Real-Time PCR
4.6. Biochemical Analyses
4.6.1. Tissue Extract Preparation for Biochemical Analyses
4.6.2. Antioxidant Capacity
4.6.3. Peroxidase Activity
4.6.4. Catalase Activity
4.6.5. Superoxide Dismutase Activity
4.6.6. Glutathione S-Transferase Activity
4.6.7. Statistical Analysis for Biochemical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmid-Schönbein, G.W. Analysis of Inflammation. Annu. Rev. Biomed. Eng. 2006, 8, 93–151. [Google Scholar] [CrossRef] [PubMed]
- Herath, S.; Fischer, D.P.; Werling, D.; Williams, E.J.; Lilly, S.T.; Dobson, H.; Bryant, C.E.; Sheldon, I.M. Expression and Function of Toll-Like Receptor 4 in the Endometrial Cells of the Uterus. Endocrinology 2006, 147, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Cronin, J.G.; Turner, M.L.; Goetze, L.; Bryant, C.E.; Sheldon, I.M. Toll-Like Receptor 4 and MYD88-Dependent Signaling Mechanisms of the Innate Immune System Are Essential for the Response to Lipopolysaccharide by Epithelial and Stromal Cells of the Bovine Endometrium1. Biol. Reprod. 2012, 86, 51. [Google Scholar] [CrossRef] [PubMed]
- Lavon, Y.; Leitner, G.; Goshen, T.; Braw-Tal, R.; Jacoby, S.; Wolfenson, D. Exposure to Endotoxin during Estrus Alters the Timing of Ovulation and Hormonal Concentrations in Cows. Theriogenology 2008, 70, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Lüttgenau, J.; Herzog, K.; Strüve, K.; Latter, S.; Boos, A.; Bruckmaier, R.M.; Bollwein, H.; Kowalewski, M.P. LPS-Mediated Effects and Spatio-Temporal Expression of TLR2 and TLR4 in the Bovine Corpus Luteum. Reproduction 2016, 151, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.J.; Herath, S.; England, G.C.W.; Dobson, H.; Bryant, C.E.; Sheldon, I.M. Effect of Escherichia Coli Infection of the Bovine Uterus from the Whole Animal to the Cell. Animal 2008, 2, 1153–1157. [Google Scholar] [CrossRef]
- Daniel, J.A.; Abrams, M.S.; DeSouza, L.; Wagner, C.G.; Whitlock, B.K.; Sartin, J.L. Endotoxin Inhibition of Luteinizing Hormone in Sheep. Domest. Anim. Endocrinol. 2003, 25, 13–19. [Google Scholar] [CrossRef]
- Herzog, K.; Strüve, K.; Kastelic, J.P.; Piechotta, M.; Ulbrich, S.E.; Pfarrer, C.; Shirasuna, K.; Shimizu, T.; Miyamoto, A.; Bollwein, H. Escherichia Coli Lipopolysaccharide Administration Transiently Suppresses Luteal Structure and Function in Diestrous Cows. Reproduction 2012, 144, 467–476. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The Pig: A Model for Human Infectious Diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Takada, I.; Makishima, M. Peroxisome Proliferator-Activated Receptor Agonists and Antagonists: A Patent Review (2014-Present). Expert Opin. Pat. 2020, 30, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zerani, M.; Polisca, A.; Boiti, C.; Maranesi, M. Current Knowledge on the Multifactorial Regulation of Corpora Lutea Lifespan: The Rabbit Model. Animals 2021, 11, 296. [Google Scholar] [CrossRef] [PubMed]
- Parillo, F.; Maranesi, M.; Brecchia, G.; Gobbetti, A.; Boiti, C.; Zerani, M. In Vivo Chronic and In Vitro Acute Effects of Di(2-Ethylhexyl) Phthalate on Pseudopregnant Rabbit Corpora Lutea: Possible Involvement of Peroxisome Proliferator-Activated Receptor Gamma1. Biol. Reprod. 2014, 90, 41. [Google Scholar] [CrossRef]
- Zerani, M.; Maranesi, M.; Brecchia, G.; Gobbetti, A.; Boiti, C.; Parillo, F. Evidence for a Luteotropic Role of Peroxisome Proliferator-Activated Receptor Gamma: Expression and In Vitro Effects on Enzymatic and Hormonal Activities in Corpora Lutea of Pseudopregnant Rabbits1. Biol. Reprod. 2013, 88, 62. [Google Scholar] [CrossRef]
- Clark, R.B. The Role of PPARs in Inflammation and Immunity. J. Leukoc. Biol. 2002, 71, 388–400. [Google Scholar] [CrossRef]
- Mierzejewski, K.; Paukszto, Ł.; Kurzyńska, A.; Kunicka, Z.; Jastrzębski, J.P.; Makowczenko, K.G.; Golubska, M.; Bogacka, I. PPARγ Regulates the Expression of Genes Involved in the DNA Damage Response in an Inflamed Endometrium. Sci. Rep. 2022, 12, 4026. [Google Scholar] [CrossRef]
- Mierzejewski, K.; Kurzyńska, A.; Kunicka, Z.; Klepacka, A.; Golubska, M.; Bogacka, I. Peroxisome Proliferator-Activated Receptor Gamma Ligands Regulate the Expression of Inflammatory Mediators in Porcine Endometrium during LPS-Induced Inflammation. Theriogenology 2022, 187, 195–204. [Google Scholar] [CrossRef]
- Liu, Y.; Colby, J.; Zuo, X.; Jaoude, J.; Wei, D.; Shureiqi, I. The Role of PPAR-δ in Metabolism, Inflammation, and Cancer: Many Characters of a Critical Transcription Factor. Int. J. Mol. Sci. 2018, 19, 3339. [Google Scholar] [CrossRef]
- Dunn, S.E.; Bhat, R.; Straus, D.S.; Sobel, R.A.; Axtell, R.; Johnson, A.; Nguyen, K.; Mukundan, L.; Moshkova, M.; Dugas, J.C.; et al. Peroxisome Proliferator–Activated Receptor δ Limits the Expansion of Pathogenic Th Cells during Central Nervous System Autoimmunity. J. Exp. Med. 2010, 207, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Connelly, T.L.; Baer, S.E.; Cooper, J.T.; Bronk, D.A.; Wawrik, B.; Harding, K.; Turk-Kubo, K.A.; Sipler, R.E.; Mills, M.M.; Bronk, D.A.; et al. Printing from Here Is Disabled. Deep Sea Res. 2 Top. Stud. Ocean. 2019, 118, 1–13. [Google Scholar] [CrossRef]
- Matsushita, Y.; Ogawa, D.; Wada, J.; Yamamoto, N.; Shikata, K.; Sato, C.; Tachibana, H.; Toyota, N.; Makino, H. Activation of Peroxisome Proliferator–Activated Receptor δ Inhibits Streptozotocin-Induced Diabetic Nephropathy Through Anti-Inflammatory Mechanisms in Mice. Diabetes 2011, 60, 960–968. [Google Scholar] [CrossRef]
- Luz-Crawford, P.; Ipseiz, N.; Espinosa-Carrasco, G.; Caicedo, A.; Tejedor, G.; Toupet, K.; Loriau, J.; Scholtysek, C.; Stoll, C.; Khoury, M.; et al. PPARβ/δ Directs the Therapeutic Potential of Mesenchymal Stem Cells in Arthritis. Ann. Rheum. Dis. 2016, 75, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, S.; Cai, L.; Ma, W.; Shi, Z. Lipopolysaccharide and Heat Stress Impair the Estradiol Biosynthesis in Granulosa Cells via Increase of HSP70 and Inhibition of Smad3 Phosphorylation and Nuclear Translocation. Cell Signal. 2017, 30, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Moravek, M.B.; Ward, E.A.; Lebovic, D.I. Thiazolidinediones as Therapy for Endometriosis: A Case Series. Gynecol. Obs. Invest. 2009, 68, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Talukder, S.; Kerrisk, K.L.; Gabai, G.; Celi, P. Role of Oxidant–Antioxidant Balance in Reproduction of Domestic Animals. Anim. Prod. Sci. 2017, 57, 1588. [Google Scholar] [CrossRef]
- Okamoto, K.; Kusano, T.; Nishino, T. Chemical Nature and Reaction Mechanisms of the Molybdenum Cofactor of Xanthine Oxidoreductase. Curr. Pharm. Des. 2013, 19, 2606–2614. [Google Scholar] [CrossRef] [PubMed]
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine Oxidoreductase-Derived Reactive Species: Physiological and Pathological Effects. Oxid. Med. Cell Longev. 2016, 2016, 3527579. [Google Scholar] [CrossRef]
- Brichac, J.; Ho, K.K.; Honzatko, A.; Wang, R.; Lu, X.; Weiner, H.; Picklo, M.J. Enantioselective Oxidation of Trans -4-Hydroxy-2-Nonenal Is Aldehyde Dehydrogenase Isozyme and Mg2+ Dependent. Chem. Res. Toxicol. 2007, 20, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Marchitti, S.A.; Brocker, C.; Orlicky, D.J.; Vasiliou, V. Molecular Characterization, Expression Analysis, and Role of ALDH3B1 in the Cellular Protection against Oxidative Stress. Free Radic. Biol. Med. 2010, 49, 1432–1443. [Google Scholar] [CrossRef]
- Mishra, D.P.; Dhali, A. Endotoxin Induces Luteal Cell Apoptosis through the Mitochondrial Pathway. Prostaglandins Other Lipid. Mediat. 2007, 83, 75–88. [Google Scholar] [CrossRef]
- Yoon, S.; Hong, M.; Wilson, I.A. An Unusual Dimeric Structure and Assembly for TLR4 Regulator RP105–MD-1. Nat. Struct. Mol. Biol. 2011, 18, 1028–1035. [Google Scholar] [CrossRef]
- Yi, L.; Shen, H.; Zhao, M.; Shao, P.; Liu, C.; Cui, J.; Wang, J.; Wang, C.; Guo, N.; Kang, L.; et al. Inflammation-Mediated SOD-2 Upregulation Contributes to Epithelial-Mesenchymal Transition and Migration of Tumor Cells in Aflatoxin G1-Induced Lung Adenocarcinoma. Sci. Rep. 2017, 7, 7953. [Google Scholar] [CrossRef]
- Flynn, J.M.; Melov, S. SOD2 in Mitochondrial Dysfunction and Neurodegeneration. Free Radic. Biol. Med. 2013, 62, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Al-Gubory, K.H.; Garrel, C.; Faure, P.; Sugino, N. Roles of Antioxidant Enzymes in Corpus Luteum Rescue from Reactive Oxygen Species-Induced Oxidative Stress. Reprod. Biomed. Online 2012, 25, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Schwanhäusser, B.; Wolf, J.; Selbach, M.; Busse, D. Synthesis and Degradation Jointly Determine the Responsiveness of the Cellular Proteome. BioEssays 2013, 35, 597–601. [Google Scholar] [CrossRef]
- Gry, M.; Rimini, R.; Strömberg, S.; Asplund, A.; Pontén, F.; Uhlén, M.; Nilsson, P. Correlations between RNA and Protein Expression Profiles in 23 Human Cell Lines. BMC Genom. 2009, 10, 365. [Google Scholar] [CrossRef] [PubMed]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Kim, W.-J.; Kang, Y.-J.; Koh, E.-M.; Ahn, K.-S.; Cha, H.-S.; Lee, W.-H. LIGHT Is Involved in the Pathogenesis of Rheumatoid Arthritis by Inducing the Expression of Pro-Inflammatory Cytokines and MMP-9 in Macrophages. Immunology 2005, 114, 272–279. [Google Scholar] [CrossRef]
- Shaikh, R.B.; Santee, S.; Granger, S.W.; Butrovich, K.; Cheung, T.; Kronenberg, M.; Cheroutre, H.; Ware, C.F. Constitutive Expression of LIGHT on T Cells Leads to Lymphocyte Activation, Inflammation, and Tissue Destruction. J. Immunol. 2001, 167, 6330–6337. [Google Scholar] [CrossRef]
- Otterdal, K.; Haukeland, J.W.; Yndestad, A.; Dahl, T.B.; Holm, S.; Segers, F.M.; Gladhaug, I.P.; Konopski, Z.; Damås, J.K.; Halvorsen, B.; et al. Increased Serum Levels of LIGHT/TNFSF14 in Nonalcoholic Fatty Liver Disease: Possible Role in Hepatic Inflammation. Clin. Transl. Gastroenterol. 2015, 6, e95. [Google Scholar] [CrossRef]
- Hatziagelaki, E.; Pergialiotis, V.; Kannenberg, J.M.; Trakakis, E.; Tsiavou, A.; Markgraf, D.F.; Carstensen-Kirberg, M.; Pacini, G.; Roden, M.; Dimitriadis, G.; et al. Association between Biomarkers of Low-Grade Inflammation and Sex Hormones in Women with Polycystic Ovary Syndrome. Exp. Clin. Endocrinol. Diabetes 2020, 128, 723–730. [Google Scholar] [CrossRef]
- Tiranti, V.; Hoertnagel, K.; Carrozzo, R.; Galimberti, C.; Munaro, M.; Granatiero, M.; Zelante, L.; Gasparini, P.; Marzella, R.; Rocchi, M.; et al. Mutations of SURF-1 in Leigh Disease Associated with Cytochrome c Oxidase Deficiency. Am. J. Hum. Genet. 1998, 63, 1609–1621. [Google Scholar] [CrossRef]
- De Deken, X.; Corvilain, B.; Dumont, J.E.; Miot, F. Roles of DUOX-Mediated Hydrogen Peroxide in Metabolism, Host Defense, and Signaling. Antioxid. Redox. Signal. 2014, 20, 2776–2793. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, S.; Till, A.; Sina, C.; Arlt, A.; Grasberger, H.; Schreiber, S.; Rosenstiel, P. DUOX2-Derived Reactive Oxygen Species Are Effectors of NOD2-Mediated Antibacterial Responses. J. Cell. Sci. 2009, 122, 3522–3530. [Google Scholar] [CrossRef]
- Wu, Y.; Antony, S.; Meitzler, J.L.; Doroshow, J.H. Molecular Mechanisms Underlying Chronic Inflammation-Associated Cancers. Cancer Lett. 2014, 345, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, Y.; Hu, C.; Wang, Y.; Yan, Z.; Li, Z.; Wu, R. Mitochondria and Oxidative Stress in Ovarian Endometriosis. Free Radic. Biol. Med. 2019, 136, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Scarlett, D.-J.G.; Herst, P.M.; Berridge, M.V. Multiple Proteins with Single Activities or a Single Protein with Multiple Activities: The Conundrum of Cell Surface NADH Oxidoreductases. Biochim. Et. Biophys. Acta BBA-Bioenerg. 2005, 1708, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Li, R.; Kim, B.; Palvolgyi, R.; Ho, T.; Yang, Q.-Z.; Xu, J.; Szeto, W.L.; Honda, H.; Berliner, J.A. Ox-PAPC Activation of PMET System Increases Expression of Heme Oxygenase-1 in Human Aortic Endothelial Cell. J. Lipid. Res. 2009, 50, 265–274. [Google Scholar] [CrossRef]
- Lawlor, K.E.; Campbell, I.K.; Metcalf, D.; O’Donnell, K.; van Nieuwenhuijze, A.; Roberts, A.W.; Wicks, I.P. Critical Role for Granulocyte Colony-Stimulating Factor in Inflammatory Arthritis. Proc. Natl. Acad. Sci. USA 2004, 101, 11398–11403. [Google Scholar] [CrossRef]
- Welser-Alves, J.V.; Milner, R. Microglia Are the Major Source of TNF-α and TGF-Β1 in Postnatal Glial Cultures; Regulation by Cytokines, Lipopolysaccharide, and Vitronectin. Neurochem. Int. 2013, 63, 47–53. [Google Scholar] [CrossRef]
- Tsuruta, Y.; Park, Y.-J.; Siegal, G.P.; Liu, G.; Abraham, E. Involvement of Vitronectin in Lipopolysaccaride-Induced Acute Lung Injury. J. Immunol. 2007, 179, 7079–7086. [Google Scholar] [CrossRef]
- Bae, H.-B.; Zmijewski, J.W.; Deshane, J.S.; Zhi, D.; Thompson, L.C.; Peterson, C.B.; Chaplin, D.D.; Abraham, E. Vitronectin Inhibits Neutrophil Apoptosis through Activation of Integrin-Associated Signaling Pathways. Am. J. Respir. Cell. Mol. Biol. 2012, 46, 790–796. [Google Scholar] [CrossRef]
- Park, Y.-J.; Liu, G.; Lorne, E.F.; Zhao, X.; Wang, J.; Tsuruta, Y.; Zmijewski, J.; Abraham, E. PAI-1 Inhibits Neutrophil Efferocytosis. Proc. Natl. Acad. Sci. USA 2008, 105, 11784–11789. [Google Scholar] [CrossRef]
- Budagian, V.; Bulanova, E.; Paus, R.; Bulfonepaus, S. IL-15/IL-15 Receptor Biology: A Guided Tour through an Expanding Universe. Cytokine Growth Factor Rev. 2006, 17, 259–280. [Google Scholar] [CrossRef]
- Hiromatsu, T.; Yajima, T.; Matsuguchi, T.; Nishimura, H.; Wajjwalku, W.; Arai, T.; Nimura, Y.; Yoshikai, Y. Overexpression of Interleukin-15 Protects against Escherichia Coli –Induced Shock Accompanied by Inhibition of Tumor Necrosis Factor–α–Induced Apoptosis. J. Infect. Dis. 2003, 187, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Divanovic, S.; Trompette, A.; Atabani, S.F.; Madan, R.; Golenbock, D.T.; Visintin, A.; Finberg, R.W.; Tarakhovsky, A.; Vogel, S.N.; Belkaid, Y.; et al. Negative Regulation of Toll-like Receptor 4 Signaling by the Toll-like Receptor Homolog RP105. Nat. Immunol. 2005, 6, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Kaplanski, G. IL-6: A Regulator of the Transition from Neutrophil to Monocyte Recruitment during Inflammation. Trends Immunol. 2003, 24, 25–29. [Google Scholar] [CrossRef]
- Kouno, J.; Nagai, H.; Nagahata, T.; Onda, M.; Yamaguchi, H.; Adachi, K.; Takahashi, H.; Teramoto, A.; Emi, M. Up-Regulation of CC Chemokine, CCL3L1, and Receptors, CCR3, CCR5 in Human Glioblastoma That Promotes Cell Growth. J. Neurooncol. 2004, 70, 301–307. [Google Scholar] [CrossRef]
- Calmon-Hamaty, F.; Combe, B.; Hahne, M.; Morel, J. Lymphotoxin α Revisited: General Features and Implications in Rheumatoid Arthritis. Arthritis Res. 2011, 13, 232. [Google Scholar] [CrossRef]
- Suzuki, M.; Saito-Adachi, M.; Arai, Y.; Fujiwara, Y.; Takai, E.; Shibata, S.; Seki, M.; Rokutan, H.; Maeda, D.; Horie, M.; et al. E74-Like Factor 3 Is a Key Regulator of Epithelial Integrity and Immune Response Genes in Biliary Tract Cancer. Cancer Res. 2021, 81, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, D.C. Mannan-Binding Lectin and Its Role in Innate Immunity. Transfus. Med. 2002, 12, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Royet, J.; Dziarski, R. Peptidoglycan Recognition Proteins: Pleiotropic Sensors and Effectors of Antimicrobial Defences. Nat. Rev. Microbiol. 2007, 5, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J.; Bättig, P.; Bauer, M.; Ruedl, C.; Lässing, U.; Beerli, R.R.; Dietmeier, K.; Ivanova, L.; Pfister, T.; Vogt, L.; et al. CCL19 and CCL21 Induce a Potent Proinflammatory Differentiation Program in Licensed Dendritic Cells. Immunity 2005, 22, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, J.; Huang, A.; Stinson, J.; Heldens, S.; Foster, J.; Dowd, P.; Gurney, A.L.; Wood, W.I. Cloning and Characterization of IL-17B and IL-17C, Two New Members of the IL-17 Cytokine Family. Proc. Natl. Acad. Sci. USA 2000, 97, 773–778. [Google Scholar] [CrossRef]
- Shi, Y.; Ullrich, S.J.; Zhang, J.; Connolly, K.; Grzegorzewski, K.J.; Barber, M.C.; Wang, W.; Wathen, K.; Hodge, V.; Fisher, C.L.; et al. A Novel Cytokine Receptor-Ligand Pair. J. Biol. Chem. 2000, 275, 19167–19176. [Google Scholar] [CrossRef] [PubMed]
- Akins, E.L.; Morrissette, M.C. Gross Ovarian Changes during Estrous Cycle of Swine. Am. J. Vet. Res. 1968, 29, 1953–1957. [Google Scholar]
- Kunicka, Z.; Mierzejewski, K.; Kurzyńska, A.; Stryiński, R.; Mateos, J.; Carrera, M.; Bogacka, I. Analysis of Changes in the Proteomic Profile of Porcine Corpus Luteum during Different Stages of the Estrous Cycle: Effects of PPAR Gamma Ligands. Reprod. Fertil. Dev. 2022, 34, 776–788. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Mierzejewski, K.; Paukszto, Ł.; Kurzyńska, A.; Kunicka, Z.; Jastrzębski, J.P.; Bogacka, I. Transcriptome Analysis of Porcine Endometrium after LPS-Induced Inflammation: Effects of the PPAR-Gamma Ligands in Vitro. Biol. Reprod. 2021, 104, 130–143. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Maehly, A.C. [136] Assay of catalases and peroxidases. In Methods in Enzymology; Elsevier Inc.: Amsterdam, The Netherlands, 1955; Volume 2, pp. 764–775. [Google Scholar] [CrossRef]
- Aebi, H. [13] Catalase in Vitro. In Methods in Enzymology; Elsevier Inc.: Amsterdam, The Netherlands, 1984; Volume 104, pp. 121–126. [Google Scholar] [CrossRef]
- Podczasy, J.J.; Wei, R. Reduction of Iodonitrotetrazolium Violet by Superoxide Radicals. Biochem Biophys Res. Commun. 1988, 150, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Rice-Evans, C.; Miller, N.J. [241 Total antioxidant status in plasma and body fluids. In Methods in Enzymology; Elsevier Inc.: Amsterdam, The Netherlands, 1994; Volume 234, pp. 279–293. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mierzejewski, K.; Kurzyńska, A.; Gerwel, Z.; Golubska, M.; Stryiński, R.; Bogacka, I. PPARβ/δ Ligands Regulate Oxidative Status and Inflammatory Response in Inflamed Corpus Luteum—An In Vitro Study. Int. J. Mol. Sci. 2023, 24, 4993. https://doi.org/10.3390/ijms24054993
Mierzejewski K, Kurzyńska A, Gerwel Z, Golubska M, Stryiński R, Bogacka I. PPARβ/δ Ligands Regulate Oxidative Status and Inflammatory Response in Inflamed Corpus Luteum—An In Vitro Study. International Journal of Molecular Sciences. 2023; 24(5):4993. https://doi.org/10.3390/ijms24054993
Chicago/Turabian StyleMierzejewski, Karol, Aleksandra Kurzyńska, Zuzanna Gerwel, Monika Golubska, Robert Stryiński, and Iwona Bogacka. 2023. "PPARβ/δ Ligands Regulate Oxidative Status and Inflammatory Response in Inflamed Corpus Luteum—An In Vitro Study" International Journal of Molecular Sciences 24, no. 5: 4993. https://doi.org/10.3390/ijms24054993
APA StyleMierzejewski, K., Kurzyńska, A., Gerwel, Z., Golubska, M., Stryiński, R., & Bogacka, I. (2023). PPARβ/δ Ligands Regulate Oxidative Status and Inflammatory Response in Inflamed Corpus Luteum—An In Vitro Study. International Journal of Molecular Sciences, 24(5), 4993. https://doi.org/10.3390/ijms24054993







