Recent Developments in the Inhibition of Bacterial Adhesion as Promising Anti-Virulence Strategy
Abstract
1. Introduction
2. Adhesion Proteins as Promising Targets for Preventing Antibiotic Resistance
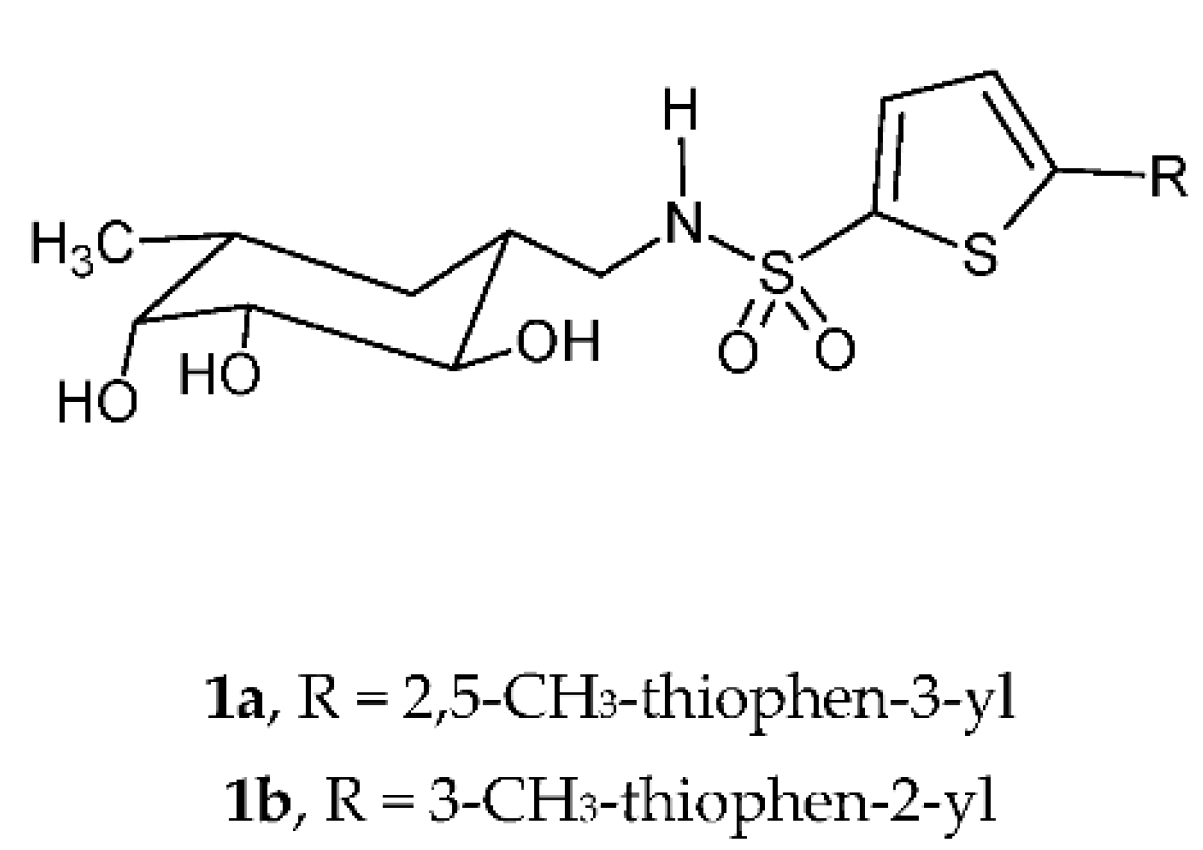
3. Recent Developments of Anti-Adhesion Agents against Relevant Pathogens
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Sarwar, A.; Butt, M.A.; Hafeez, S.; Danish, M.Z. Rapid emergence of antibacterial resistance by bacterial isolates from patients of gynecological infections in Punjab, Pakistan. J. Infect. Public Health 2020, 13, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic Strategies to Counteract Antibiotic Resistance in MRSA Biofilm-Associated Infections. Chemmedchem 2021, 16, 65–80. [Google Scholar] [CrossRef]
- Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Diana, P. Novel strategies in the war against antibiotic resistance. Futur. Med. Chem. 2021, 13, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial Virulence Factors: Secreted for Survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Viljoen, A. Binding Strength of Gram-Positive Bacterial Adhesins. Front. Microbiol. 2020, 11, 1457. [Google Scholar] [CrossRef]
- Parrino, B.; Diana, P.; Cirrincione, G.; Cascioferro, S. Bacterial Biofilm Inhibition in the Development of Effective Anti-Virulence Strategy. Open Med. Chem. J. 2018, 12, 84–87. [Google Scholar] [CrossRef]
- Solanki, V.; Tiwari, M.; Tiwari, V. Host-bacteria interaction and adhesin study for development of therapeutics. Int. J. Biol. Macromol. 2018, 112, 54–64. [Google Scholar] [CrossRef]
- Allen, R.C.; Popat, R.; Diggle, S.P.; Brown, S.P. Targeting virulence: Can we make evolution-proof drugs? Nat. Rev. Microbiol. 2014, 12, 300–308. [Google Scholar] [CrossRef]
- Mühlenkamp, M.; Oberhettinger, P.; Leo, J.C.; Linke, D.; Schütz, M.S. Yersinia adhesin A (YadA)—Beauty & beast. Int. J. Med. Microbiol. 2015, 305, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, N.A.; Ton-That, H. Bacterial Pili and Fimbriae. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2020; pp. 1–13. ISBN 978-0-470-01590-2. [Google Scholar]
- Cascioferro, S.; Cusimano, M.G.; Schillaci, D. Antiadhesion agents against Gram-positive pathogens. Futur. Microbiol. 2014, 9, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V. Pilins in gram-positive bacteria: A structural perspective. IUBMB Life 2015, 67, 533–543. [Google Scholar] [CrossRef]
- Totsika, M.; Vagenas, D.; Paxman, J.; Wang, G.; Dhouib, R.; Sharma, P.; Martin, J.L.; Scanlon, M.; Heras, B. Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens. Antioxid. Redox Signal. 2018, 29, 653–666. [Google Scholar] [CrossRef]
- Inaba, K.; Ito, K. Structure and mechanisms of the DsbB–DsbA disulfide bond generation machine. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2008, 1783, 520–529. [Google Scholar] [CrossRef]
- Cascioferro, S.; Totsika, M.; Schillaci, D. Sortase A: An ideal target for anti-virulence drug development. Microb. Pathog. 2014, 77, 105–112. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Richter, A.M.; Hengge, R.; Stahlhut, S.G.; Chattopadhyay, S.; Kisiela, D.I.; Hvidtfeldt, K.; Clegg, S.; Struve, C.; Sokurenko, E.V.; et al. Cellulose as an Architectural Element in Spatially Structured Escherichia coli Biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef] [PubMed]
- Niba, E.T.E.; Naka, Y.; Nagase, M.; Mori, H.; Kitakawa, M. A Genome-wide Approach to Identify the Genes Involved in Biofilm Formation in E. coli. DNA Res. 2007, 14, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, R.R.; Everett, M.L.; Wahl, S.D.; Lee, Y.-H.; Orndorff, P.E.; Parker, W. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: Role of type 1 pili. Mol. Immunol. 2006, 43, 378–387. [Google Scholar] [CrossRef]
- Sharma, V.; von Ossowski, I.; Krishnan, V. Exploiting pilus-mediated bacteria-host interactions for health benefits. Mol. Asp. Med. 2021, 81, 100998. [Google Scholar] [CrossRef]
- Pinkner, J.S.; Remaut, H.; Buelens, F.; Miller, E.; Åberg, V.; Pemberton, N.; Hedenström, M.; Larsson, A.; Seed, P.; Waksman, G.; et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 17897–17902. [Google Scholar] [CrossRef]
- Greene, S.E.; Pinkner, J.; Chorell, E.; Dodson, K.W.; Shaffer, C.L.; Conover, M.S.; Livny, J.; Hadjifrangiskou, M.; Almqvist, F.; Hultgren, S.J. Pilicide ec240 Disrupts Virulence Circuits in Uropathogenic Escherichia coli. Mbio 2014, 5, e02038-14. [Google Scholar] [CrossRef] [PubMed]
- Sivick, K.E.; Mobley, H.L.T. Waging War against Uropathogenic Escherichia coli: Winning Back the Urinary Tract. Infect. Immun. 2010, 78, 568–585. [Google Scholar] [CrossRef]
- Schembri, M.A.; Christiansen, G.; Klemm, P. FimH-mediated autoaggregation of Escherichia coli. Mol. Microbiol. 2001, 41, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Dautin, N. Folding Control in the Path of Type 5 Secretion. Toxins 2021, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V Secretion Systems: An Overview of Passenger Domain Functions. Front. Microbiol. 2019, 10, 1163. [Google Scholar] [CrossRef] [PubMed]
- Paxman, J.J.; Lo, A.W.; Sullivan, M.J.; Panjikar, S.; Kuiper, M.; Whitten, A.E.; Wang, G.; Luan, C.-H.; Moriel, D.G.; Tan, L.; et al. Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules. Nat. Commun. 2019, 10, 1967. [Google Scholar] [CrossRef] [PubMed]
- Sarshar, M.; Behzadi, P.; Ambrosi, C.; Zagaglia, C.; Palamara, A.T.; Scribano, D. FimH and Anti-Adhesive Therapeutics: A Disarming Strategy Against Uropathogens. Antibiotics 2020, 9, 397. [Google Scholar] [CrossRef]
- Foroogh, N.; Rezvan, M.; Ahmad, K.; Mahmood, S. Structural and functional characterization of the FimH adhesin of uropathogenic Escherichia coli and its novel applications. Microb. Pathog. 2021, 161, 105288. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, Y.; García-Heredia, A.; Merino-Mascorro, A.; García, S.; Solís-Soto, L.; Heredia, N. Natural and synthetic antimicrobials reduce adherence of enteroaggregative and enterohemorrhagic Escherichia coli to epithelial cells. PLoS ONE 2021, 16, e0251096. [Google Scholar] [CrossRef] [PubMed]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842. [Google Scholar] [CrossRef]
- Sommer, R.; Rox, K.; Wagner, S.; Hauck, D.; Henrikus, S.S.; Newsad, S.; Arnold, T.; Ryckmans, T.; Brönstrup, M.; Imberty, A.; et al. Anti-biofilm Agents against Pseudomonas aeruginosa: A Structure–Activity Relationship Study of C-Glycosidic LecB Inhibitors. J. Med. Chem. 2019, 62, 9201–9216. [Google Scholar] [CrossRef]
- Ha, U.-H.; Wang, Y.; Jin, S. DsbA of Pseudomonas aeruginosa Is Essential for Multiple Virulence Factors. Infect. Immun. 2003, 71, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.M.; Parrino, B.; Carbone, D.; Schillaci, D.; Giovannetti, E.; Cirrincione, G.; Diana, P. Thiazoles, Their Benzofused Systems, and Thiazolidinone Derivatives: Versatile and Promising Tools to Combat Antibiotic Resistance. J. Med. Chem. 2020, 63, 7923–7956. [Google Scholar] [CrossRef]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Schillaci, D.; Cusimano, M.G.; Cirrincione, G.; Diana, P. Thiazole Analogues of the Marine Alkaloid Nortopsentin as Inhibitors of Bacterial Biofilm Formation. Molecules 2020, 26, 81. [Google Scholar] [CrossRef] [PubMed]
- Parrino, B.; Carbone, D.; Cascioferro, S.; Pecoraro, C.; Giovannetti, E.; Deng, D.; Di Sarno, V.; Musella, S.; Auriemma, G.; Cusimano, M.G.; et al. 1,2,4-Oxadiazole topsentin analogs as staphylococcal biofilm inhibitors targeting the bacterial transpeptidase sortase A. Eur. J. Med. Chem. 2021, 209, 112892. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.; Parrino, B.; Petri, G.L.; Cusimano, M.G.; Schillaci, D.; Di Sarno, V.; Musella, S.; Giovannetti, E.; Cirrincione, G.; Diana, P. 2,6-Disubstituted imidazo[2,1-b][1,3,4]thiadiazole derivatives as potent staphylococcal biofilm inhibitors. Eur. J. Med. Chem. 2019, 167, 200–210. [Google Scholar] [CrossRef]
- Ragab, A.; Abusaif, M.S.; Gohar, N.A.; Aboul-Magd, D.S.; Fayed, E.A.; Ammar, Y.A. Development of new spiro[1,3]dithiine-4,11′-indeno[1,2-b]quinoxaline derivatives as S. aureus Sortase A inhibitors and radiosterilization with molecular modeling simulation. Bioorg. Chem. 2023, 131, 106307. [Google Scholar] [CrossRef]
- Wang, X.; Luan, Y.; Hou, J.; Jiang, T.; Zhao, Y.; Song, W.; Wang, L.; Kong, X.; Guan, J.; Song, D.; et al. The protection effect of rhodionin against methicillin-resistant Staphylococcus aureus-induced pneumonia through sortase A inhibition. World J. Microbiol. Biotechnol. 2022, 39, 18. [Google Scholar] [CrossRef] [PubMed]
- Jaudzems, K.; Kurbatska, V.; Jēkabsons, A.; Bobrovs, R.; Rudevica, Z.; Leonchiks, A.; Kurbatska, V.; Leonciks, A. Targeting Bacterial Sortase A with Covalent Inhibitors: 27 New Starting Points for Structure-Based Hit-to-Lead Optimization. ACS Infect. Dis. 2020, 6, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making Use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [PubMed]

| Proteins | Functions | Role in Pathogenesis |
|---|---|---|
| Protein A (Spa) | Binds Fc domain for immunoglobulins; binds complement protein C3 | Inhibition of innate and adaptive immune responses |
| Fibronectin-binding protein A (FnbpA) | Adhesin for fibrinogen, fibronectin and elastin | Adhesion; colonization; biofilm formation |
| Fibronectin-binding protein homolog (FnbpB) | Adhesin for fibronectin and elastin | Adhesion; colonization; biofilm formation |
| Clumping factor A (ClfA) | Platelet adhesion (fibrin-mediated) | Adhesion; colonization; evasion of innate immune defenses |
| Clumping factor B (ClfB) | Platelet adhesion (fibrin-mediated) | Adhesion; colonization; evasion of innate immune defenses |
| Collagen-binding protein (Cna) | Adhesin for collagen (type I and IV) | Adhesion |
| Serine-aspartate repeat protein C (SdrC) | Adhesin | Adhesion; colonization |
| Serine-aspartate repeat protein D (SdrD) | Adhesin | Adhesion; colonization |
| Serine-aspartate repeat protein E (SdrE) | Adhesin | Adhesion; colonization |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecoraro, C.; Carbone, D.; Parrino, B.; Cascioferro, S.; Diana, P. Recent Developments in the Inhibition of Bacterial Adhesion as Promising Anti-Virulence Strategy. Int. J. Mol. Sci. 2023, 24, 4872. https://doi.org/10.3390/ijms24054872
Pecoraro C, Carbone D, Parrino B, Cascioferro S, Diana P. Recent Developments in the Inhibition of Bacterial Adhesion as Promising Anti-Virulence Strategy. International Journal of Molecular Sciences. 2023; 24(5):4872. https://doi.org/10.3390/ijms24054872
Chicago/Turabian StylePecoraro, Camilla, Daniela Carbone, Barbara Parrino, Stella Cascioferro, and Patrizia Diana. 2023. "Recent Developments in the Inhibition of Bacterial Adhesion as Promising Anti-Virulence Strategy" International Journal of Molecular Sciences 24, no. 5: 4872. https://doi.org/10.3390/ijms24054872
APA StylePecoraro, C., Carbone, D., Parrino, B., Cascioferro, S., & Diana, P. (2023). Recent Developments in the Inhibition of Bacterial Adhesion as Promising Anti-Virulence Strategy. International Journal of Molecular Sciences, 24(5), 4872. https://doi.org/10.3390/ijms24054872









