Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges
Abstract
1. Introduction
2. Overview of Epigenetics and Epigenome Editing
3. Description of EpiEffector Molecules
4. Description of Epigenome-Editing Methods
5. Challenges in Epigenome-Editing Technologies
5.1. Target Specificity in Epigenome Editing
5.2. Avoidance of Undesirable Genomic Mutations Caused by Epigenome Editing
5.3. Importance of Nuclear Structure in Epigenome Editing
5.4. Selection of Cell Types to Be Subjected to Epigenome Editing
5.5. Method of Administration
6. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef]
- Sharma, R.; Anguela, X.M.; Doyon, Y.; Wechsler, T.; DeKelver, R.C.; Sproul, S.; Paschon, D.E.; Miller, J.C.; Davidson, R.J.; Shivak, D.; et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 2015, 126, 1777–1784. [Google Scholar] [CrossRef]
- Maeder, M.L.; Gersbach, C.A. Genome-editing Technologies for Gene and Cell Therapy. Mol. Ther. 2016, 24, 430–446. [Google Scholar] [CrossRef]
- Ashmore-Harris, C.; Fruhwirth, G.O. The clinical potential of gene editing as a tool to engineer cell-based therapeutics. Clin. Transl. Med. 2020, 9, 15. [Google Scholar] [CrossRef]
- Yeh, C.D.; Richardson, C.D.; Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019, 21, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Goell, J.H.; Hilton, I.B. CRISPR/Cas-Based Epigenome Editing: Advances, Applications, and Clinical Utility. Trends Biotechnol. 2021, 39, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Chaturvedi, A.; Chitkara, D.; Singh, S. CRISPR/Cas mediated epigenome editing for cancer therapy. Semin. Cancer Biol. 2022, 83, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Garriga-Canut, M.; Agustin-Pavon, C.; Herrmann, F.; Sanchez, A.; Dierssen, M.; Fillat, C.; Isalan, M. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. USA 2012, 109, E3136–E3145. [Google Scholar] [CrossRef]
- Zeitler, B.; Froelich, S.; Marlen, K.; Shivak, D.A.; Yu, Q.; Li, D.; Pearl, J.R.; Miller, J.C.; Zhang, L.; Paschon, D.E.; et al. Allele-selective transcriptional repression of mutant HTT for the treatment of Huntington’s disease. Nat. Med. 2019, 25, 1131–1142. [Google Scholar] [CrossRef]
- Jensen, T.L.; Gøtzsche, C.R.; Woldbye, D.P.D. Current and Future Prospects for Gene Therapy for Rare Genetic Diseases Affecting the Brain and Spinal Cord. Front. Mol. Neurosci. 2021, 14, 695937. [Google Scholar] [CrossRef]
- Fine-tuning epigenome editors. Nat. Biotechnol. 2022, 40, 281. [CrossRef]
- Kaiser, J. A gentler way to tweak genes: Epigenome editing. Science 2022, 376, 1034–1035. [Google Scholar] [CrossRef]
- Kumar, S. Genome editing to epigenome editing: Towards unravelling the enigmas in developmental biology. Trends Dev. Biol. 2019, 12, 31–38. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010, 11, 285–296. [Google Scholar] [CrossRef]
- Maeshima, K.; Iida, S.; Tamura, S. Physical Nature of Chromatin in the Nucleus. Cold Spring Harb. Perspect. Biol. 2021, 13, a040675. [Google Scholar] [CrossRef] [PubMed]
- Lennartsson, A.; Ekwall, K. Histone modification patterns and epigenetic codes. Biochim Biophys Acta 2009, 1790, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015, 7, a025064. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.R. A model for all genomes: The role of transcription factories. J. Mol. Biol. 2010, 395, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Musselman, C.A.; Lalonde, M.E.; Cote, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef]
- Kim, J.; Daniel, J.; Espejo, A.; Lake, A.; Krishna, M.; Xia, L.; Zhang, Y.; Bedford, M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006, 7, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; de Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef]
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef] [PubMed]
- de Groote, M.L.; Verschure, P.J.; Rots, M.G. Epigenetic Editing: Targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012, 40, 10596–10613. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Neph, S.; Vierstra, J.; Stergachis, A.B.; Reynolds, A.P.; Haugen, E.; Vernot, B.; Thurman, R.E.; John, S.; Sandstrom, R.; Johnson, A.K.; et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 2012, 489, 83–90. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Kungulovski, G.; Jeltsch, A. Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet. 2016, 32, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Hatano, Y.; Taniguchi, R.; Kobayashi, N.; Yamagata, K. Editing DNA Methylation in Mammalian Embryos. Int. J. Mol. Sci. 2020, 21, 637. [Google Scholar] [CrossRef] [PubMed]
- Waryah, C.B.; Moses, C.; Arooj, M.; Blancafort, P. Zinc Fingers, TALEs, and CRISPR Systems: A Comparison of Tools for Epigenome Editing. Methods Mol. Biol. 2018, 1767, 19–63. [Google Scholar] [PubMed]
- Pei, W.D.; Zhang, Y.; Yin, T.L.; Yu, Y. Epigenome editing by CRISPR/Cas9 in clinical settings: Possibilities and challenges. Brief. Funct. Genom. 2020, 19, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Amabile, A.; Migliara, A.; Capasso, P.; Biffi, M.; Cittaro, D.; Naldini, L.; Lombardo, A. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell 2016, 167, 219–232.e14. [Google Scholar] [CrossRef]
- Chavez, A.; Tuttle, M.; Pruitt, B.W.; Ewen-Campen, B.; Chari, R.; Ter-Ovanesyan, D.; Haque, S.J.; Cecchi, R.J.; Kowal, E.J.K.; Buchthal, J.; et al. Comparison of Cas9 activators in multiple species. Nat. Methods 2016, 13, 563–567. [Google Scholar] [CrossRef]
- Chen, T.; Gao, D.; Zhang, R.; Zeng, G.; Yan, H.; Lim, E.; Liang, F.S. Chemically Controlled Epigenome Editing through an Inducible dCas9 System. J. Am. Chem. Soc. 2017, 139, 11337–11340. [Google Scholar] [CrossRef]
- Okada, M.; Kanamori, M.; Someya, K.; Nakatsukasa, H.; Yoshimura, A. Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenet. Chromatin 2017, 10, 24. [Google Scholar] [CrossRef]
- Shrimp, J.H.; Grose, C.; Widmeyer, S.R.T.; Thorpe, A.L.; Jadhav, A.; Meier, J.L. Chemical Control of a CRISPR-Cas9 Acetyltransferase. ACS Chem. Biol. 2018, 13, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Shan, L.; Han, L.; Ma, S.; Zhang, Y.; Hao, B.; Lin, Y.; Rong, Z. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell 2018, 9, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Mammadov, R.; Czikora, A.; Unlu, H.; Tufan, T.; Fischer, N.L.; Arslan, S.; Bekiranov, S.; Kanemaki, M.; Adli, M. Temporal and Spatial Epigenome Editing Allows Precise Gene Regulation in Mammalian Cells. J. Mol. Biol. 2019, 431, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, Y.; Cao, H.; Zhang, Y.; Gu, Z.; Liu, X.; Yu, A.; Kaphle, P.; Dickerson, K.E.; Ni, M.; et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 2020, 11, 485. [Google Scholar] [CrossRef]
- Bohnsack, J.P.; Zhang, H.; Wandling, G.M.; He, D.; Kyzar, E.J.; Lasek, A.W.; Pandey, S.C. Targeted epigenomic editing ameliorates adult anxiety and excessive drinking after adolescent alcohol exposure. Sci. Adv. 2022, 8, eabn2748. [Google Scholar] [CrossRef]
- Cheng, A.W.; Jillette, N.; Lee, P.; Plaskon, D.; Fujiwara, Y.; Wang, W.; Taghbalout, A.; Wang, H. Casilio: A versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016, 26, 254–257. [Google Scholar] [CrossRef]
- Chiarella, A.M.; Butler, K.V.; Gryder, B.E.; Lu, D.; Wang, T.A.; Yu, X.; Pomella, S.; Khan, J.; Jin, J.; Hathaway, N.A. Dose-dependent activation of gene expression is achieved using CRISPR and small molecules that recruit endogenous chromatin machinery. Nat. Biotechnol. 2020, 38, 50–55. [Google Scholar] [CrossRef]
- Liao, H.K.; Hatanaka, F.; Araoka, T.; Reddy, P.; Wu, M.Z.; Sui, Y.; Yamauchi, T.; Sakurai, M.; O’Keefe, D.D.; Nunez-Delicado, E.; et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 2017, 171, 1495–1507.e15. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Zhao, Y.T.; Lamonica, J.M.; Zhou, Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun. 2017, 8, 15315. [Google Scholar] [CrossRef]
- Kim, J.M.; Kim, K.; Schmidt, T.; Punj, V.; Tucker, H.; Rice, J.C.; Ulmer, T.S.; An, W. Cooperation between SMYD3 and PC4 drives a distinct transcriptional program in cancer cells. Nucleic Acids Res. 2015, 43, 8868–8883. [Google Scholar] [CrossRef]
- Cano-Rodriguez, D.; Gjaltema, R.A.; Jilderda, L.J.; Jellema, P.; Dokter-Fokkens, J.; Ruiters, M.H.; Rots, M.G. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 2016, 7, 12284. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.M.G.; Kirkland, J.G.; Chory, E.J.; Husmann, D.; Calarco, J.P.; Crabtree, G.R. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat. Commun. 2017, 8, 560. [Google Scholar] [CrossRef] [PubMed]
- Kearns, N.A.; Pham, H.; Tabak, B.; Genga, R.M.; Silverstein, N.J.; Garber, M.; Maehr, R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods 2015, 12, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 12, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- O’Geen, H.; Ren, C.; Nicolet, C.M.; Perez, A.A.; Halmai, J.; Le, V.M.; Mackay, J.P.; Farnham, P.J.; Segal, D.J. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res. 2017, 45, 9901–9916. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, R.; Du, Z.; Bai, L.; Li, L.; Cui, J.; Li, W.; Hoffman, A.R.; Hu, J.F. Epigenetic Targeting of Granulin in Hepatoma Cells by Synthetic CRISPR dCas9 Epi-suppressors. Mol. Ther. Nucleic Acids 2018, 11, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; Kwon, J.B.; Nelson, C.E.; Rouse, D.C.; Gemberling, M.P.; Oliver, M.L.; Gersbach, C.A. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat. Commun. 2018, 9, 1674. [Google Scholar] [CrossRef]
- O’Geen, H.; Bates, S.L.; Carter, S.S.; Nisson, K.A.; Halmai, J.; Fink, K.D.; Rhie, S.K.; Farnham, P.J.; Segal, D.J. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenet. Chromatin 2019, 12, 26. [Google Scholar] [CrossRef]
- Backstrom, J.R.; Sheng, J.; Wang, M.C.; Bernardo-Colon, A.; Rex, T.S. Optimization of S. aureus dCas9 and CRISPRi Elements for a Single Adeno-Associated Virus that Targets an Endogenous Gene. Mol. Ther. Methods Clin. Dev. 2020, 19, 139–148. [Google Scholar] [CrossRef]
- Nunez, J.K.; Chen, J.; Pommier, G.C.; Cogan, J.Z.; Replogle, J.M.; Adriaens, C.; Ramadoss, G.N.; Shi, Q.; Hung, K.L.; Samelson, A.J.; et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 2021, 184, 2503–2519.e17. [Google Scholar] [CrossRef]
- Oberkofler, V.; Baurle, I. Inducible epigenome editing probes for the role of histone H3K4 methylation in Arabidopsis heat stress memory. Plant. Physiol. 2022, 189, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.S.; Takeda, H.; Nakamura, R. Targeted in vivo epigenome editing of H3K27me3. Epigenet. Chromatin 2019, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA Methylation in the Mammalian Genome. Cell 2016, 167, 233–247.e17. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.I.; Celik, H.; Rois, L.E.; Fishberger, G.; Fowler, T.; Rees, R.; Kramer, A.; Martens, A.; Edwards, J.R.; Challen, G.A. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol. Open 2016, 5, 866–874. [Google Scholar] [CrossRef]
- Huang, Y.H.; Su, J.; Lei, Y.; Brunetti, L.; Gundry, M.C.; Zhang, X.; Jeong, M.; Li, W.; Goodell, M.A. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 2017, 18, 176. [Google Scholar] [CrossRef]
- Galonska, C.; Charlton, J.; Mattei, A.L.; Donaghey, J.; Clement, K.; Gu, H.; Mohammad, A.W.; Stamenova, E.K.; Cacchiarelli, D.; Klages, S.; et al. Genome-wide tracking of dCas9-methyltransferase footprints. Nat. Commun. 2018, 9, 597. [Google Scholar] [CrossRef]
- Pflueger, C.; Tan, D.; Swain, T.; Nguyen, T.; Pflueger, J.; Nefzger, C.; Polo, J.M.; Ford, E.; Lister, R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018, 28, 1193–1206. [Google Scholar] [CrossRef]
- Lin, L.; Liu, Y.; Xu, F.; Huang, J.; Daugaard, T.F.; Petersen, T.S.; Hansen, B.; Ye, L.; Zhou, Q.; Fang, F.; et al. Genome-wide determination of on-target and off-target characteristics for RNA-guided DNA methylation by dCas9 methyltransferases. Gigascience 2018, 7, giy011. [Google Scholar] [CrossRef]
- Kantor, B.; Tagliafierro, L.; Gu, J.; Zamora, M.E.; Ilich, E.; Grenier, C.; Huang, Z.Y.; Murphy, S.; Chiba-Falek, O. Downregulation of SNCA Expression by Targeted Editing of DNA Methylation: A Potential Strategy for Precision Therapy in PD. Mol. Ther. 2018, 26, 2638–2649. [Google Scholar] [CrossRef]
- Marx, N.; Grunwald-Gruber, C.; Bydlinski, N.; Dhiman, H.; Ngoc Nguyen, L.; Klanert, G.; Borth, N. CRISPR-Based Targeted Epigenetic Editing Enables Gene Expression Modulation of the Silenced Beta-Galactoside Alpha-2,6-Sialyltransferase 1 in CHO Cells. Biotechnol. J. 2018, 13, e1700217. [Google Scholar] [CrossRef]
- Mkannez, G.; Gagne-Ouellet, V.; Jalloul Nsaibia, M.; Boulanger, M.C.; Rosa, M.; Argaud, D.; Hadji, F.; Gaudreault, N.; Rheaume, G.; Bouchard, L.; et al. DNA methylation of a PLPP3 MIR transposon-based enhancer promotes an osteogenic programme in calcific aortic valve disease. Cardiovasc. Res. 2018, 114, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Ortega, J.A.; Quinlan, K.A.; Santos, D.P.; Gu, H.; Martin, E.J.; Galonska, C.; Pop, R.; Maidl, S.; Di Pardo, A.; et al. Dissecting the Functional Consequences of De Novo DNA Methylation Dynamics in Human Motor Neuron Differentiation and Physiology. Cell Stem Cell 2018, 22, 559–574.e9. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lang, J.; Zhang, Q.; Yang, C.R.; Zhao, Z.A.; Zhang, Y.; Du, Y.; Sun, Y. DNA methylation analysis and editing in single mammalian oocytes. Proc. Natl. Acad. Sci. USA 2019, 116, 9883–9892. [Google Scholar] [CrossRef] [PubMed]
- Saunderson, E.A.; Stepper, P.; Gomm, J.J.; Hoa, L.; Morgan, A.; Allen, M.D.; Jones, J.L.; Gribben, J.G.; Jurkowski, T.P.; Ficz, G. Hit-and-run epigenetic editing prevents senescence entry in primary breast cells from healthy donors. Nat. Commun. 2017, 8, 1450. [Google Scholar] [CrossRef]
- Stepper, P.; Kungulovski, G.; Jurkowska, R.Z.; Chandra, T.; Krueger, F.; Reinhardt, R.; Reik, W.; Jeltsch, A.; Jurkowski, T.P. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res. 2017, 45, 1703–1713. [Google Scholar] [CrossRef]
- Hofacker, D.; Broche, J.; Laistner, L.; Adam, S.; Bashtrykov, P.; Jeltsch, A. Engineering of Effector Domains for Targeted DNA Methylation with Reduced Off-Target Effects. Int. J. Mol. Sci. 2020, 21, 502. [Google Scholar] [CrossRef]
- Yamazaki, T.; Hatano, Y.; Handa, T.; Kato, S.; Hoida, K.; Yamamura, R.; Fukuyama, T.; Uematsu, T.; Kobayashi, N.; Kimura, H.; et al. Targeted DNA methylation in pericentromeres with genome editing-based artificial DNA methyltransferase. PLoS ONE 2017, 12, e0177764. [Google Scholar] [CrossRef]
- Xiong, T.; Meister, G.E.; Workman, R.E.; Kato, N.C.; Spellberg, M.J.; Turker, F.; Timp, W.; Ostermeier, M.; Novina, C.D. Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci. Rep. 2017, 7, 6732. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, X.; Su, J.; Jeong, M.; Gundry, M.C.; Huang, Y.H.; Zhou, Y.; Li, W.; Goodell, M.A. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017, 8, 16026. [Google Scholar] [CrossRef]
- Ghoshal, B.; Picard, C.L.; Vong, B.; Feng, S.; Jacobsen, S.E. CRISPR-based targeting of DNA methylation in Arabidopsis thaliana by a bacterial CG-specific DNA methyltransferase. Proc. Natl. Acad. Sci. USA 2021, 118, e2125016118. [Google Scholar] [CrossRef]
- Morita, S.; Noguchi, H.; Horii, T.; Nakabayashi, K.; Kimura, M.; Okamura, K.; Sakai, A.; Nakashima, H.; Hata, K.; Nakashima, K.; et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 2016, 34, 1060–1065. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992.e6. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Morita, S.; Hino, S.; Kimura, M.; Hino, Y.; Kogo, H.; Nakao, M.; Hatada, I. Successful generation of epigenetic disease model mice by targeted demethylation of the epigenome. Genome Biol. 2020, 21, 77. [Google Scholar] [CrossRef]
- Hanzawa, N.; Hashimoto, K.; Yuan, X.; Kawahori, K.; Tsujimoto, K.; Hamaguchi, M.; Tanaka, T.; Nagaoka, Y.; Nishina, H.; Morita, S.; et al. Targeted DNA demethylation of the Fgf21 promoter by CRISPR/dCas9-mediated epigenome editing. Sci. Rep. 2020, 10, 5181. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tan, X.; Tampe, B.; Wilhelmi, T.; Hulshoff, M.S.; Saito, S.; Moser, T.; Kalluri, R.; Hasenfuss, G.; Zeisberg, E.M.; et al. High-fidelity CRISPR/Cas9- based gene-specific hydroxymethylation rescues gene expression and attenuates renal fibrosis. Nat. Commun. 2018, 9, 3509. [Google Scholar] [CrossRef]
- Baumann, V.; Wiesbeck, M.; Breunig, C.T.; Braun, J.M.; Koferle, A.; Ninkovic, J.; Gotz, M.; Stricker, S.H. Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat. Commun. 2019, 10, 2119. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W., 3rd; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant. Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.F.; Voytas, D.F.; Zhang, Y.; Qi, Y. Robust Transcriptional Activation in Plants Using Multiplexed CRISPR-Act2.0 and mTALE-Act Systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef]
- Matharu, N.; Rattanasopha, S.; Tamura, S.; Maliskova, L.; Wang, Y.; Bernard, A.; Hardin, A.; Eckalbar, W.L.; Vaisse, C.; Ahituv, N. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 2019, 363, eaau0629. [Google Scholar] [CrossRef]
- Kemaladewi, D.U.; Bassi, P.S.; Erwood, S.; Al-Basha, D.; Gawlik, K.I.; Lindsay, K.; Hyatt, E.; Kember, R.; Place, K.M.; Marks, R.M.; et al. A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature 2019, 572, 125–130. [Google Scholar] [CrossRef]
- Peter, C.J.; Saito, A.; Hasegawa, Y.; Tanaka, Y.; Nagpal, M.; Perez, G.; Alway, E.; Espeso-Gil, S.; Fayyad, T.; Ratner, C.; et al. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat. Commun. 2019, 10, 4112. [Google Scholar] [CrossRef]
- Colasante, G.; Lignani, G.; Brusco, S.; Di Berardino, C.; Carpenter, J.; Giannelli, S.; Valassina, N.; Bido, S.; Ricci, R.; Castoldi, V.; et al. dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol. Ther. 2020, 28, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Bohm, S.; Splith, V.; Riedmayr, L.M.; Rotzer, R.D.; Gasparoni, G.; Nordstrom, K.J.V.; Wagner, J.E.; Hinrichsmeyer, K.S.; Walter, J.; Wahl-Schott, C.; et al. A gene therapy for inherited blindness using dCas9-VPR-mediated transcriptional activation. Sci. Adv. 2020, 6, eaba5614. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Wei, H.J.; Lin, L.; George, H.; Wang, T.; Lee, I.H.; Zhao, H.Y.; Wang, Y.; Kan, Y.; Shrock, E.; et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017, 357, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Brocken, D.J.W.; Tark-Dame, M.; Dame, R.T. dCas9: A Versatile Tool for Epigenome Editing. Curr. Issues Mol. Biol. 2018, 26, 15–32. [Google Scholar] [CrossRef]
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA–guided activation of endogenous human genes. Nat. Methods 2013, 10, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef]
- Wright, A.V.; Sternberg, S.H.; Taylor, D.W.; Staahl, B.T.; Bardales, J.A.; Kornfeld, J.E.; Doudna, J.A. Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. USA 2015, 112, 2984–2989. [Google Scholar] [CrossRef]
- Nihongaki, Y.; Kawano, F.; Nakajima, T.; Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015, 33, 755–760. [Google Scholar] [CrossRef]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef]
- De Groot, A.S.; Scott, D.W. Immunogenicity of protein therapeutics. Trends Immunol. 2007, 28, 482–490. [Google Scholar] [CrossRef]
- Baker, M.P.; Reynolds, H.M.; Lumicisi, B.; Bryson, C.J. Immunogenicity of protein therapeutics: The key causes, consequences and challenges. Self Nonself 2010, 1, 314–322. [Google Scholar] [CrossRef]
- Crudele, J.M.; Chamberlain, J.S. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 2018, 9, 314–322. [Google Scholar] [CrossRef]
- Kaminskas, E.; Farrell, A.T.; Wang, Y.C.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist 2005, 10, 176–182. [Google Scholar] [CrossRef]
- Grant, S.; Easley, C.; Kirkpatrick, P. Vorinostat. Nat. Rev. Drug Discov. 2007, 6, 21–22. [Google Scholar] [CrossRef]
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Altucci, L.; Rots, M.G. Epigenetic drugs: From chemistry via biology to medicine and back. Clin. Epigenetics 2016, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, S.; van der Gaag, B.; Cortese, F.A.B. RNAi mechanisms in Huntington’s disease therapy: siRNA versus shRNA. Transl. Neurodegener. 2017, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Caron, N.S.; Southwell, A.L.; Brouwers, C.C.; Cengio, L.D.; Xie, Y.; Black, H.F.; Anderson, L.M.; Ko, S.; Zhu, X.; van Deventer, S.J.; et al. Potent and sustained huntingtin lowering via AAV5 encoding miRNA preserves striatal volume and cognitive function in a humanized mouse model of Huntington disease. Nucleic Acids Res. 2020, 48, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Osterwalder, M.; Barozzi, I.; Tissieres, V.; Fukuda-Yuzawa, Y.; Mannion, B.J.; Afzal, S.Y.; Lee, E.A.; Zhu, Y.; Plajzer-Frick, I.; Pickle, C.S.; et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 2018, 554, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Wu, Q.; Chew, J.L.; Vega, V.B.; Zhang, W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef]
- Do, D.V.; Ueda, J.; Messerschmidt, D.M.; Lorthongpanich, C.; Zhou, Y.; Feng, B.; Guo, G.; Lin, P.J.; Hossain, M.Z.; Zhang, W.; et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013, 27, 1378–1390. [Google Scholar] [CrossRef]
- Morisaka, H.; Yoshimi, K.; Okuzaki, Y.; Gee, P.; Kunihiro, Y.; Sonpho, E.; Xu, H.; Sasakawa, N.; Naito, Y.; Nakada, S.; et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat. Commun. 2019, 10, 5302. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed]
- Dolan, A.E.; Hou, Z.; Xiao, Y.; Gramelspacher, M.J.; Heo, J.; Howden, S.E.; Freddolino, P.L.; Ke, A.; Zhang, Y. Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Mol. Cell 2019, 74, 936–950.e5. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R.; Schor, I.E.; Alló, M.; Dujardin, G.; Petrillo, E.; Muñoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef]
- Laughery, M.F.; Mayes, H.C.; Pedroza, I.K.; Wyrick, J.J. R-loop formation by dCas9 is mutagenic in Saccharomyces cerevisiae. Nucleic Acids Res. 2019, 47, 2389–2401. [Google Scholar] [CrossRef] [PubMed]
- Doi, G.; Okada, S.; Yasukawa, T.; Sugiyama, Y.; Bala, S.; Miyazaki, S.; Kang, D.; Ito, T. Catalytically inactive Cas9 impairs DNA replication fork progression to induce focal genomic instability. Nucleic Acids Res. 2021, 49, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Meshorer, E.; Yellajoshula, D.; George, E.; Scambler, P.J.; Brown, D.T.; Misteli, T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 2006, 10, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Maehara, K.; Mashiko, D.; Ichinose, T.; Yao, T.; Hori, M.; Sato, Y.; Kimura, H.; Ohkawa, Y.; Yamagata, K. Heterochromatin dynamics during the differentiation process revealed by the DNA methylation reporter mouse, MethylRO. Stem Cell Rep. 2014, 2, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Boskovic, A.; Eid, A.; Pontabry, J.; Ishiuchi, T.; Spiegelhalter, C.; Raghu Ram, E.V.; Meshorer, E.; Torres-Padilla, M.E. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014, 28, 1042–1047. [Google Scholar] [CrossRef]
- Shah, S.; Rashid, M.; Verma, T.; Gupta, S. Chromatin, histones, and histone modifications in health and disease. In Genome Plasticity in Health and Disease; Academic Press: Cambridge, MA, USA, 2020; pp. 109–135. [Google Scholar]
- Mattout, A.; Meshorer, E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010, 22, 334–341. [Google Scholar] [CrossRef]
- Eshghi, I.; Eaton, J.A.; Zidovska, A. Interphase Chromatin Undergoes a Local Sol-Gel Transition upon Cell Differentiation. Phys. Rev. Lett. 2021, 126, 228101. [Google Scholar] [CrossRef] [PubMed]
- Horlbeck, M.A.; Witkowsky, L.B.; Guglielmi, B.; Replogle, J.M.; Gilbert, L.A.; Villalta, J.E.; Torigoe, S.E.; Tjian, R.; Weissman, J.S. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife 2016, 5, e12677. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, M.R. The new mouse genetics: Altering the genome by gene targeting. Trends Genet. 1989, 5, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Horii, T.; Kimura, M.; Hatada, I. Synergistic Upregulation of Target Genes by TET1 and VP64 in the dCas9-SunTag Platform. Int. J. Mol. Sci. 2020, 21, 1574. [Google Scholar] [CrossRef] [PubMed]
- Lostalé-Seijo, I.; Montenegro, J. Synthetic materials at the forefront of gene delivery. Nat. Rev. Chem. 2018, 2, 258–277. [Google Scholar] [CrossRef]
- Degors, I.M.S.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019, 52, 1750–1760. [Google Scholar] [CrossRef]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef]
- Qin, X.; Yu, C.; Wei, J.; Li, L.; Zhang, C.; Wu, Q.; Liu, J.; Yao, S.Q.; Huang, W. Rational Design of Nanocarriers for Intracellular Protein Delivery. Adv. Mater. 2019, 31, 1902791. [Google Scholar] [CrossRef]
- Zhao, Z.; Ukidve, A.; Kim, J.; Mitragotri, S. Targeting Strategies for Tissue-Specific Drug Delivery. Cell 2020, 181, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Lagerborg, K.A.; Stanton, A.; King, E.M.; Ye, S.; Tellez, L.; Krunnfusz, A.; Tavakoli, S.; Widrick, J.J.; Messemer, K.A.; et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 2021, 184, 4919–4938.e22. [Google Scholar] [CrossRef] [PubMed]
- Boisselier, E.; Astruc, D. Gold nanoparticles in nanomedicine: Preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 2009, 38, 1759–1782. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Gomez, J.A.; Beitnere, U.; Segal, D.J. Live-Animal Epigenome Editing: Convergence of Novel Techniques. Trends Genet. 2019, 35, 527–541. [Google Scholar] [CrossRef]
- Amina, S.J.; Guo, B. A Review on the Synthesis and Functionalization of Gold Nanoparticles as a Drug Delivery Vehicle. Int. J. Nanomed. 2020, 15, 9823–9857. [Google Scholar] [CrossRef]
- Surosky, R.T.; Urabe, M.; Godwin, S.G.; McQuiston, S.A.; Kurtzman, G.J.; Ozawa, K.; Natsoulis, G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 1997, 71, 7951–7959. [Google Scholar] [CrossRef]
- van Haasteren, J.; Li, J.; Scheideler, O.J.; Murthy, N.; Schaffer, D.V. The delivery challenge: Fulfilling the promise of therapeutic genome editing. Nat. Biotechnol. 2020, 38, 845–855. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, H.; Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010, 18, 80–86. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Ibraheim, R.; Song, C.Q.; Mir, A.; Amrani, N.; Xue, W.; Sontheimer, E.J. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol. 2018, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasPhi from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chemparathy, A.; Zeng, L.; Kempton, H.R.; Shang, S.; Nakamura, M.; Qi, L.S. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 2021, 81, 4333–4345.e4. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Bell, P.; McMenamin, D.; He, Z.; White, J.; Yu, H.; Xu, C.; Morizono, H.; Musunuru, K.; et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016, 34, 334–338. [Google Scholar] [CrossRef]
- McClements, M.E.; MacLaren, R.E. Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med. 2017, 90, 611–623. [Google Scholar] [PubMed]
- Marrone, L.; Marchi, P.M.; Azzouz, M. Circumventing the packaging limit of AAV-mediated gene replacement therapy for neurological disorders. Expert Opin. Biol. Ther. 2022, 22, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.J.; Kuhner, K.; Kuhn, R.; Werfel, S.; Engelhardt, S.; Wurst, W.; Ortiz, O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015, 43, 6450–6458. [Google Scholar] [CrossRef]
- Fine, E.J.; Appleton, C.M.; White, D.E.; Brown, M.T.; Deshmukh, H.; Kemp, M.L.; Bao, G. Trans-spliced Cas9 allows cleavage of HBB and CCR5 genes in human cells using compact expression cassettes. Sci. Rep. 2015, 5, 10777. [Google Scholar] [CrossRef]
- Chew, W.L.; Tabebordbar, M.; Cheng, J.K.; Mali, P.; Wu, E.Y.; Ng, A.H.; Zhu, K.; Wagers, A.J.; Church, G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, C.; Wang, Y.; Koivisto, O.; Zhou, J.; Shu, Y.; Zhang, H. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv. Drug Deliv. Rev. 2021, 176, 113891. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Saw, P.E.; Cui, G.H.; Xu, X. Nanoparticles-Mediated CRISPR/Cas Gene Editing Delivery System. ChemMedChem 2022, 17, e202100777. [Google Scholar] [CrossRef] [PubMed]
- Pogribna, M.; Hammons, G. Epigenetic Effects of Nanomaterials and Nanoparticles. J. Nanobiotechnology 2021, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L. Ex vivo gene transfer and correction for cell-based therapies. Nat. Rev. Genet. 2011, 12, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gomez-Gonzalez, B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008, 9, 204–217. [Google Scholar] [CrossRef]
- Zeps, N.; Lysaght, T.; Chadwick, R.; Erler, A.; Foo, R.; Giordano, S.; San Lai, P.; Schaefer, G.O.; Xafis, V.; Chew, W.L.; et al. Ethics and regulatory considerations for the clinical translation of somatic cell human epigenetic editing. Stem Cell Rep. 2021, 16, 1652–1655. [Google Scholar] [CrossRef]
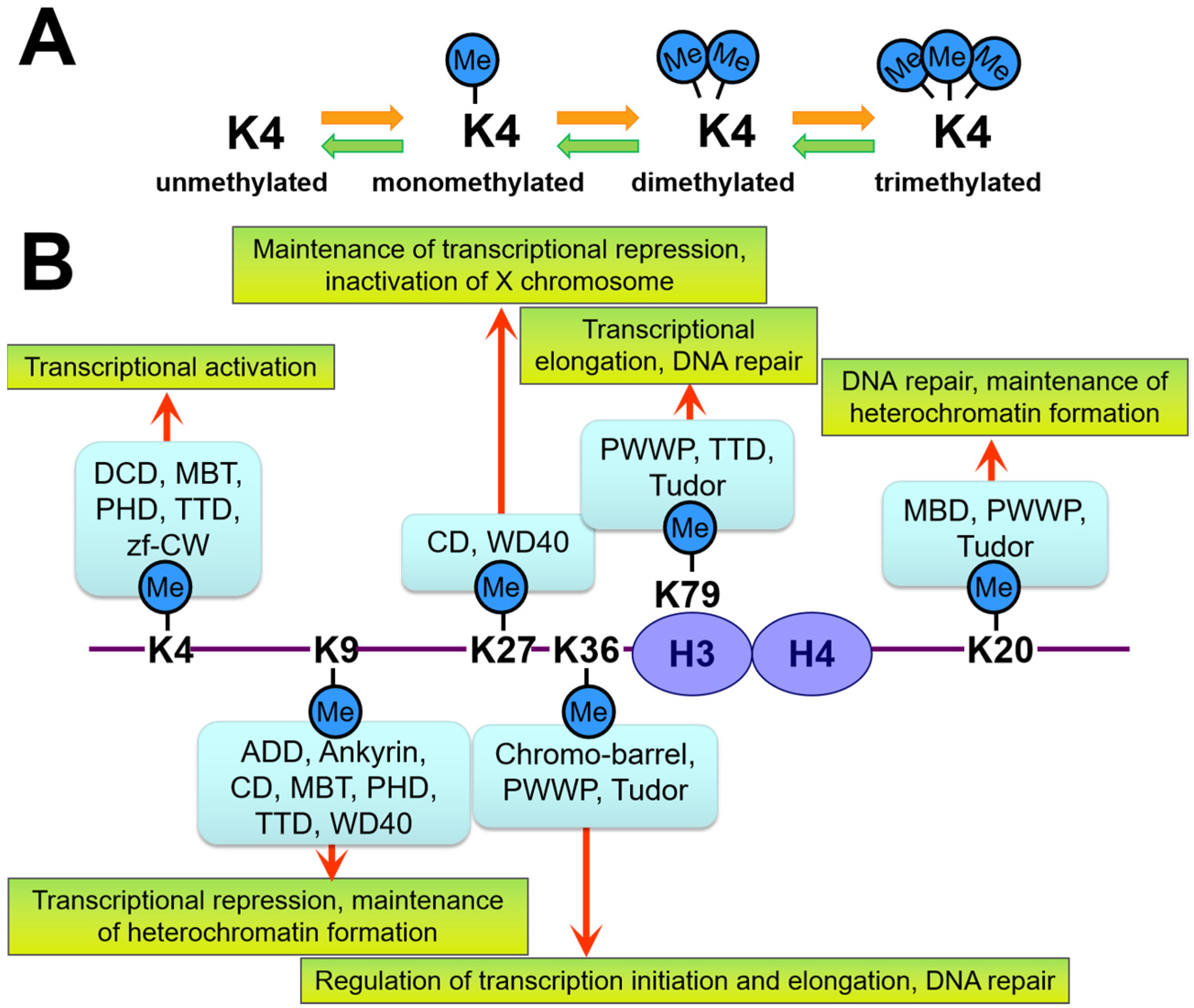
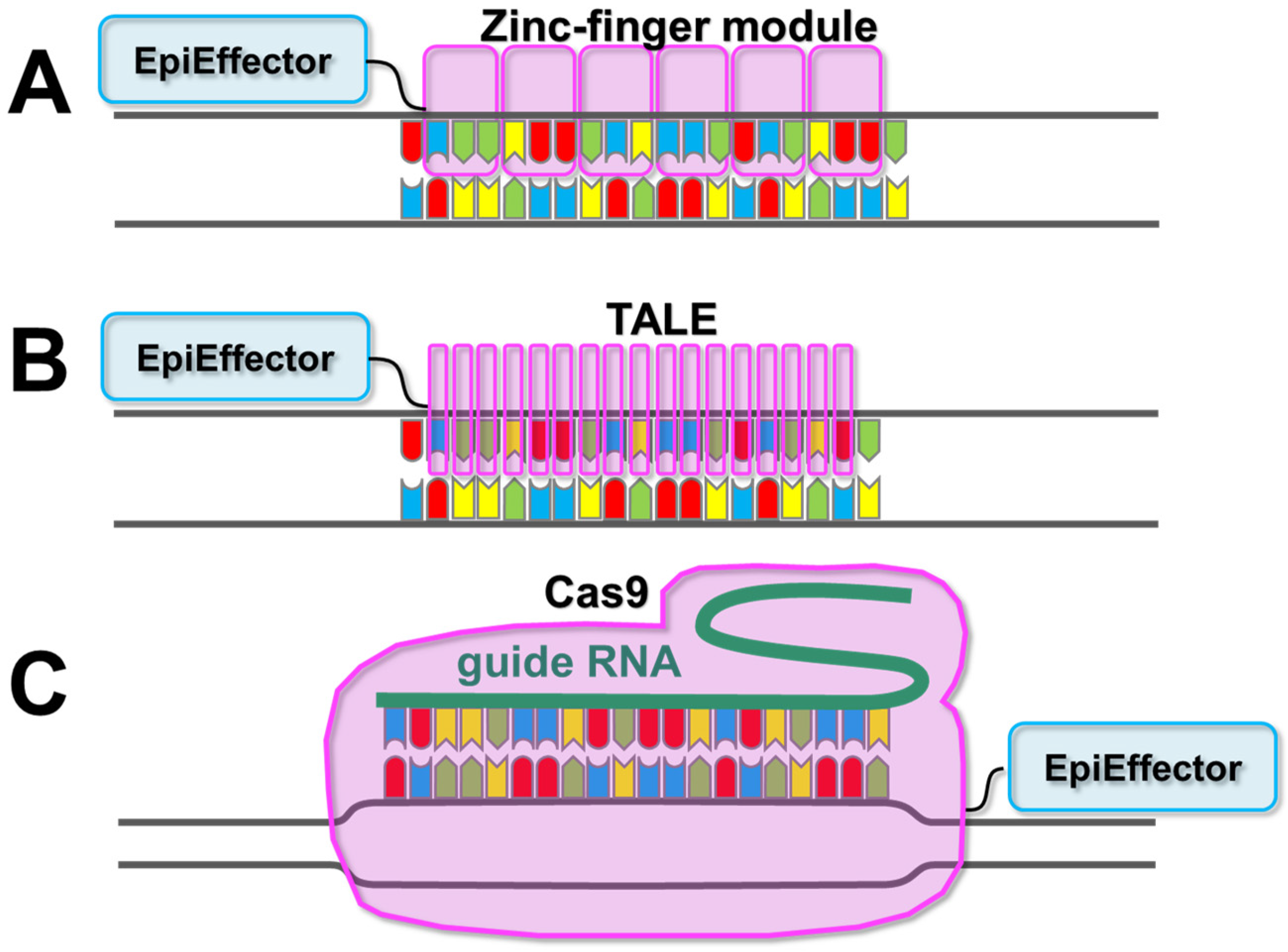

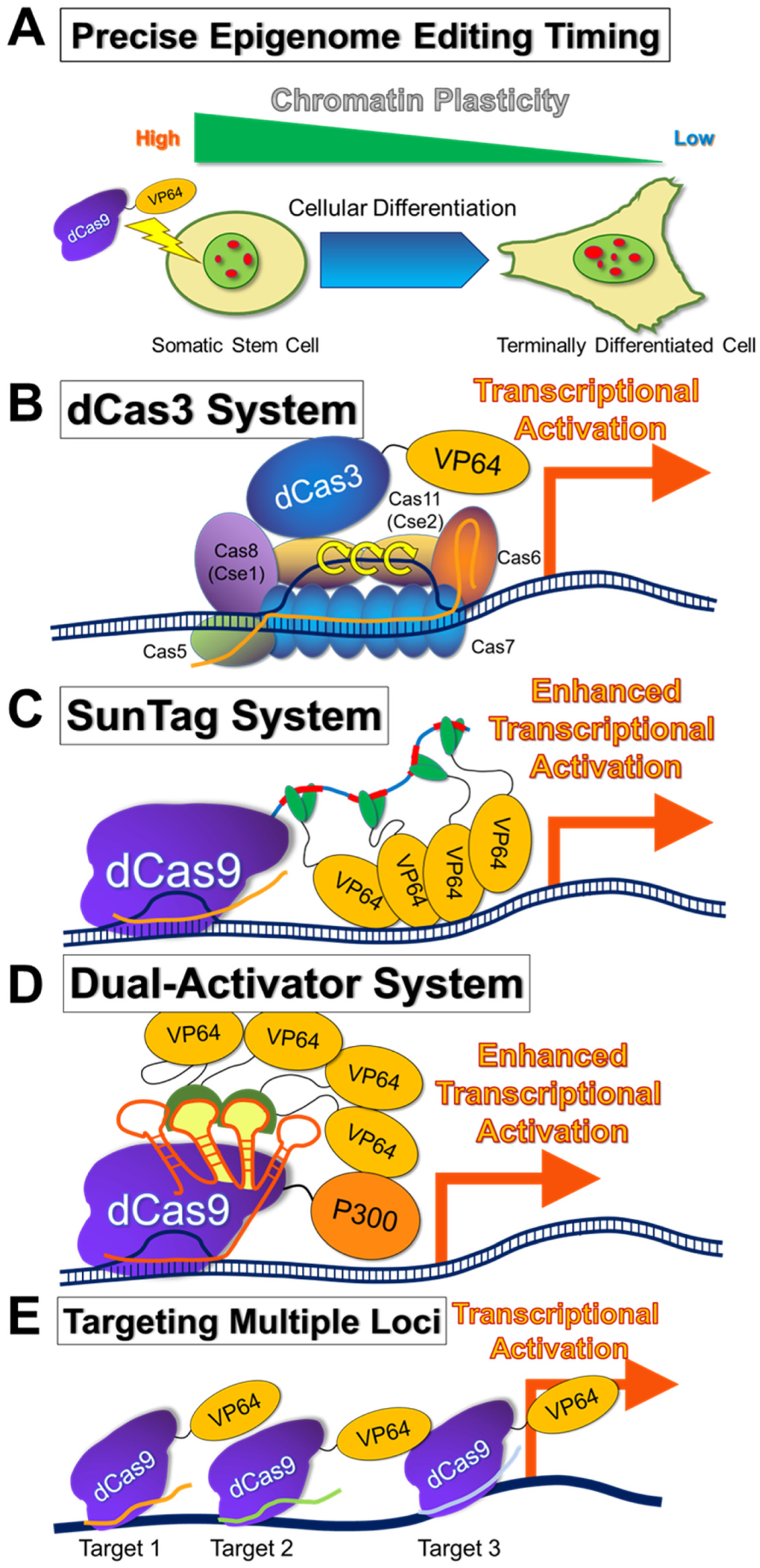
| Application | EpiEffector | Induced Epigenetic Changes |
|---|---|---|
| H3K27 acetylation (Gene activation) | P300 [46,47,48,49,50,51,52,53,54,55] cAMP-response element binding protein (CREB)-binding protein (CBP) [56] P300 and/or CBP [57] VP64 + P300 [54] MS2-P65-HSF1 (MPH) [58] | Increase in H3K27 acetylation, H3 acetylation Enhanced expression of target genes Enhanced expression of target genes Increase in H3K4 trimethylation and H3K27 acetylation |
| H3K27 deacetylation | Histone deacetylase 3 (HDAC3) [59] | Decrease in H3K27 acetylation |
| H3K4 methylation (Gene activation) | SET and MYND domain-containing protein 3 (SMYD3) [60] PR domain zinc finger protein 9 (PRDM9) [61] Disruptor of telomeric silencing 1-like (DOT1L) [61] Ubiquitin-conjugating enzyme E2 A (UBE2A) [61] BRG1/BRM associated factor (BAF) (SS18 subunit) [62] | Increase in H3K4 methylation Increase in H3K4 trimethylation Increase in H3K79 trimethylation Loss of H3K27 trimethylation and increase in H3K4 trimethylation |
| H3K9 and H3K27 methylation (Gene repression) | Lysine-specific demethylase 1 (LSD1) [54,63] Krüppel-associated box (KRAB) [14,15,54,55,56,63,64,65,66,67,68,69,70] JUMONJI (JMJ) [71] G9A (also known as Euchromatic histone-lysine N-methyltransferase 2 (EHMT2)) [65] Suppressor of Variegation 3–9 Homolog 1 (SUV39H1) [65] Enhancer of zeste homolog 2 (EZH2) [65,66,68,72] Friend of GATA protein 1 (FOG1) [65,68] LSD1 + KRAB [54] heterochromatin protein 1 (HP1) [62] | Decrease in H3K9 dimethylation and H3K27 acetylation Decrease in H3K27 acetylation and increase in H3K27 trimethylation Decrease in H3K4 trimethylation Increase in H3K9 trimethylation Increase in H3K9 trimethylation Increase in H3K27 trimethylation Decrease in H3K27 acetylation and increase in H3K27 trimethylation Decrease in H3K4 mono- and dimethylation Increase in H3K9 trimethylation |
| DNA methylation (Gene repression) | DNMT3A [65,66,73,74,75,76,77,78,79,80,81,82,83] DNMT3A + DNMT3L [70,84,85,86] KRAB/EZH2/FOG1 + DNMT3A [65] KRAB + DNMT3A (+ DNMT3L) [47,70] M. SssI MQ1 [87,88,89,90] DNMT1 [78] DNMT3B [78] | Increase in DNA methylation Increase in DNA methylation |
| DNA demethylation (Gene activation) | TET1 [47,70,73,83,91,92,93,94] TET3 [95] CRISPR activation (CRISPRa) + TET1 [96] | Decrease in DNA methylation Increase in 5-hydroxymethylcytosine Decrease in DNA methylation |
| Platform | EpiEffectors | Species | Target Diseases | Effects | Carrier, Gene Delivery Methods | Reference | |
|---|---|---|---|---|---|---|---|
| 1 | Zinc-finger protein | Kox-1 KRAB domain | Mouse | Huntington’s disease | Repression of mutant htt gene | Stereotaxic injection | [14] |
| 2 | dCas9 | VP64 or three copies of transcriptional repressor domain SRDX | Arabidopsis | Not applicable | Activation or repression of target genes (activation: AtPAP1, miR319; repression: AtCSTF64, miR159A, miR159B) | Transgenic plant | [98] |
| 3 | dCas9-SunTag | scFV-TET1 | Mouse | Not applicable | Demethylation of Gfap regulatory region. | Electroporation | [91] |
| 4 | dCas9 | DNMT3A or TET1 | Mouse | Not applicable | Demethylation of BDNF promoter or de novo methylation of CTCF motifs | Stereotaxic injection of lentivirus | [73] |
| 5 | dCas9 | An engineered prokaryotic DNA methyltransferase MQ1 | Mouse | Not applicable | DNA methylation of H19 locus | Microinjection of gene expressing plasmid | [89] |
| 6 | TALE | A bacterial CpG methyltransferase MQ1 (SssI) | Mouse | Not applicable | Methylation of major satellite DNA | Microinjection of mRNA into the embryo | [87] |
| 7 | dCas9 + dead sgRNA (dgRNA) | MS2-P65-HSF1 (MPH) | Mouse | Duchenne muscular dystrophy, acute kidney injury, diabetes | Activation of Klotho, Utrophin, Fst, and Pdx1 | Tail vein injection of AAV9 | [58] |
| 8 | Staphylococcus aureus dCas9 | KRAB | Mouse | To lower low-density lipoprotein cholesterol levels | Repression of Pcsk9 expression | AAV, dual-vector AAV8 system | [67] |
| 9 | high-fidelity dCas9 | TET3 catalytic domain | Mouse | Fibrosis | Activation of Rasal1 and Klotho expression | Renal artery/vein injection of lentivirus | [95] |
| 10 | CRISPR-Act2.0 and mTALE-Act | VP64 | Arabidopsis | Not applicable | Activation of multiple (CSTF64, GL1, and RBP-DR1) genes | Transgenic plant | [99] |
| 11 | dCas9 | TET1 | Mouse | Fragile X syndrome | Activation of FMR1 expression | Epigenome-edited neural precursor cells were injected into the brain | [92] |
| 12 | dCas9 | VP64 | Mouse | Obesity | Activation of Mc4r expression | Stereotaxic injection of AAV-DJ | [100] |
| 13 | dCas9 | VP64 | Mouse | Muscular dystrophy | Activation of Lama1 expression | Tail vein injection of AAV9 | [101] |
| 14 | dCas9 | DNMT3A or TET1 | Mouse | Not applicable | Repression or activation of Avy locus | Microinjection | [83] |
| 15 | dCas9 | Oryzias latipes EZH2 | Medaka | Not applicable | H3K27 methylation of Arhgap35, Nanos3, Pfkfb4a, Dcx, Tbx16, and Slc41a2a | Injection of mRNA | [72] |
| 16 | dCas9-SunTag | scFv-C11orf46 | Mouse | Hypoplasia of the corpus callosum | Normalization of Sema6a expression | In utero electroporation | [102] |
| 17 | Zinc-finger protein | KRAB | Mouse | Huntington’s disease | Repression of mutant htt | Stereotaxic injection of AAV2/6 or AAV2/9 | [15] |
| 18 | dCas9 | VP64 | Mouse | Dravet syndrome | Activation of Scn1a expression | Intracerebroventricular injection of AAV9 | [103] |
| 19 | dCas9 | VPR | Mouse | Blindness | Activation of Opn1mw expression | Dual adeno-associated viral vectors | [104] |
| 20 | dCas9-SunTag | TET1 catalytic domain | Mouse | Generation of Silver–Russell syndrome disease model | Demethylation of H19-DMR and repression of Igf2 | Microinjection of mRNA, transgenic mice | [93] |
| 21 | dCas9-SunTag | scFv-TET1 catalytic domain | Mouse | Not applicable | Activation of Fgf21 expression | Hydrodynamic tail vein injection | [94] |
| 22 | enCRISPRi | LSD1 and KRAB | Mouse | Not applicable | Perturbation of enhancers during hematopoiesis | Tetracycline-inducible knock-in mice | [54] |
| 23 | Staphylococcus aureus dCas9 | KRAB | Mouse | To lower low-density lipoprotein cholesterol levels | Repression of Pcsk9 expression | Tail vein injection of AAV8 | [69] |
| 24 | dCas9 | A bacterial CG-specific DNA methyltransferase MQ1 Q147L | Arabidopsis | Not applicable | Repression of FWA expression | Transgenic plant | [90] |
| 25 | dCas9 | P300 or KRAB | Rat | Alcohol use disorder | Activation or repression of Arc expression | Stereotaxic injection of lentivirus | [55] |
| 26 | dCas9 | VP64, JMJ | Arabidopsis | Not applicable | Repression of APX2 expression | Transgenic plant | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, J.; Yamazaki, T.; Funakoshi, H. Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges. Int. J. Mol. Sci. 2023, 24, 4778. https://doi.org/10.3390/ijms24054778
Ueda J, Yamazaki T, Funakoshi H. Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges. International Journal of Molecular Sciences. 2023; 24(5):4778. https://doi.org/10.3390/ijms24054778
Chicago/Turabian StyleUeda, Jun, Taiga Yamazaki, and Hiroshi Funakoshi. 2023. "Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges" International Journal of Molecular Sciences 24, no. 5: 4778. https://doi.org/10.3390/ijms24054778
APA StyleUeda, J., Yamazaki, T., & Funakoshi, H. (2023). Toward the Development of Epigenome Editing-Based Therapeutics: Potentials and Challenges. International Journal of Molecular Sciences, 24(5), 4778. https://doi.org/10.3390/ijms24054778






