Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize
Abstract
1. Introduction
2. Results
2.1. ZmNAC20 Is Upregulated by Drought Stress and ABA
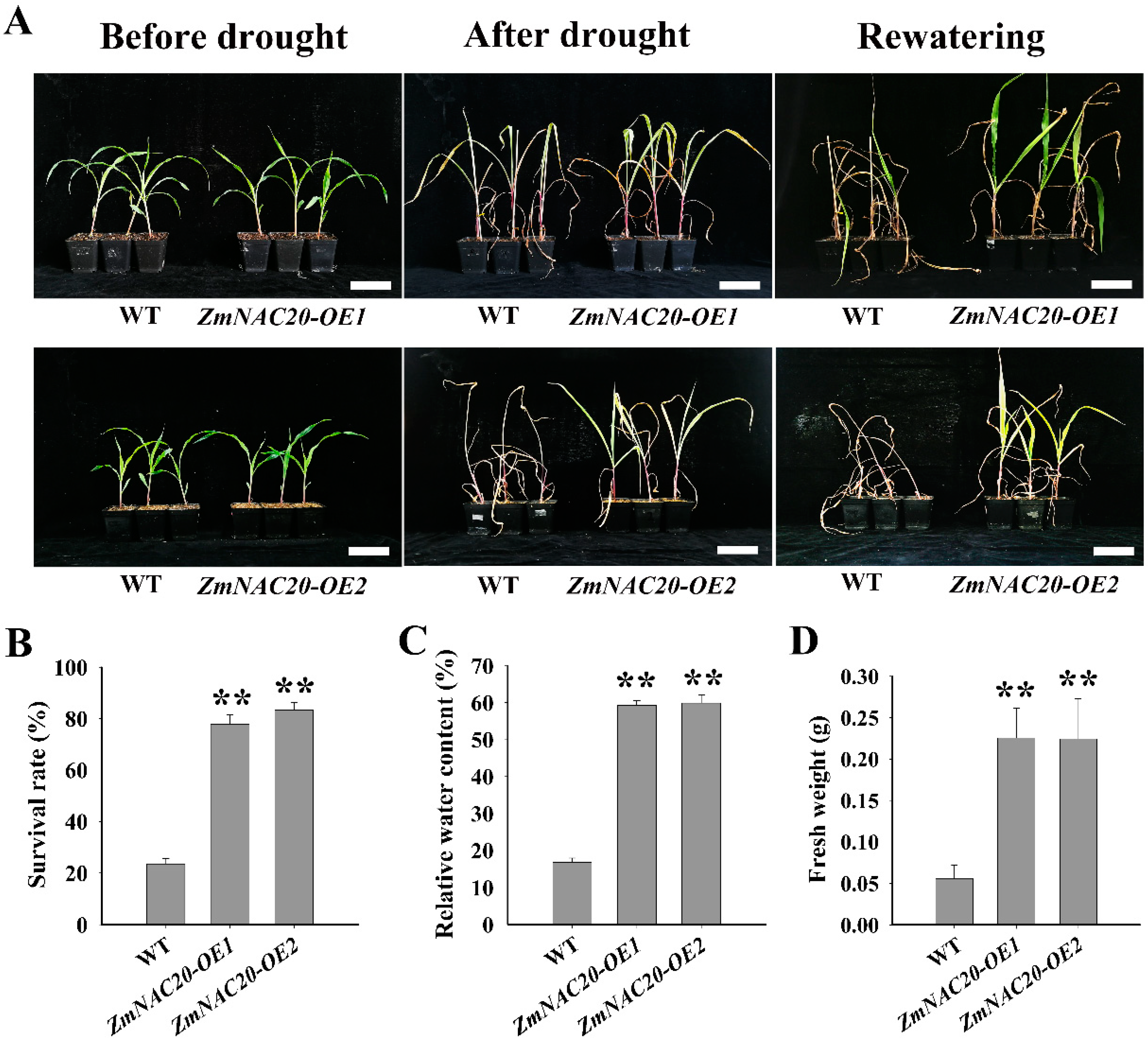
2.2. Overexpression of ZmNAC20 Enhances Drought Resistance in Maize
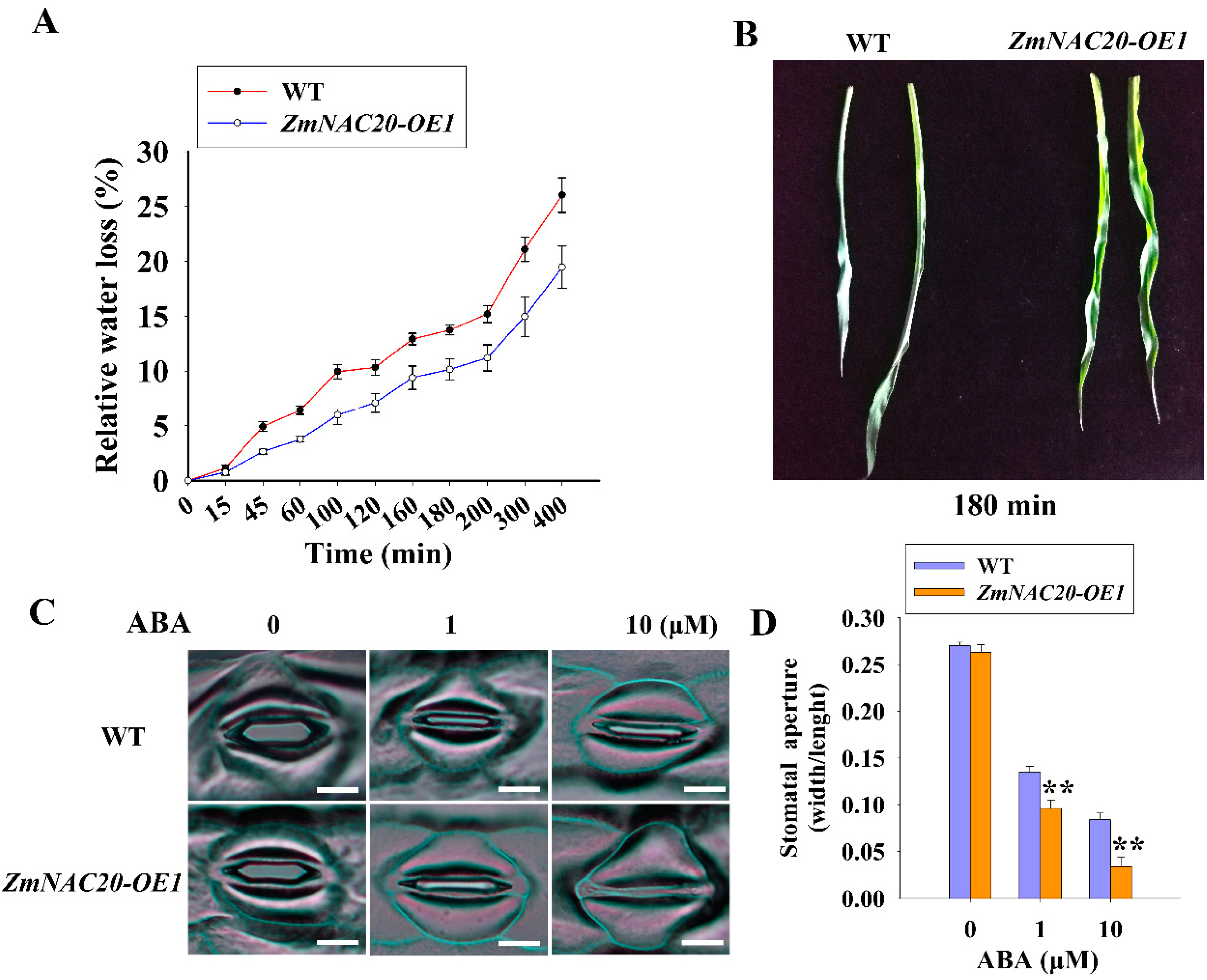
2.3. Overexpression of ZmNAC20 Promotes Stomatal Closure
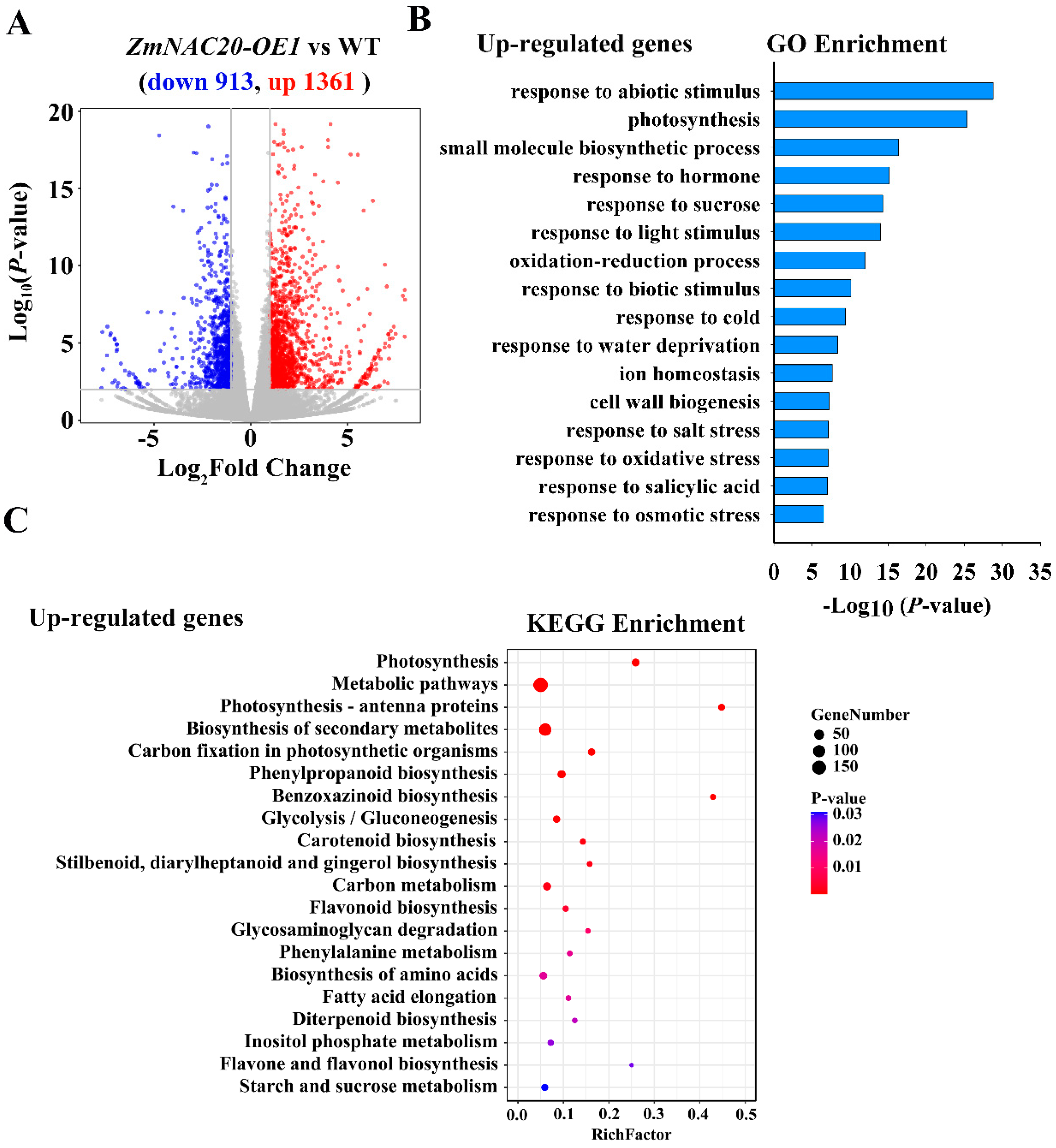
2.4. ZmNAC20 Regulates Multiple Biological Pathways under Drought Stress
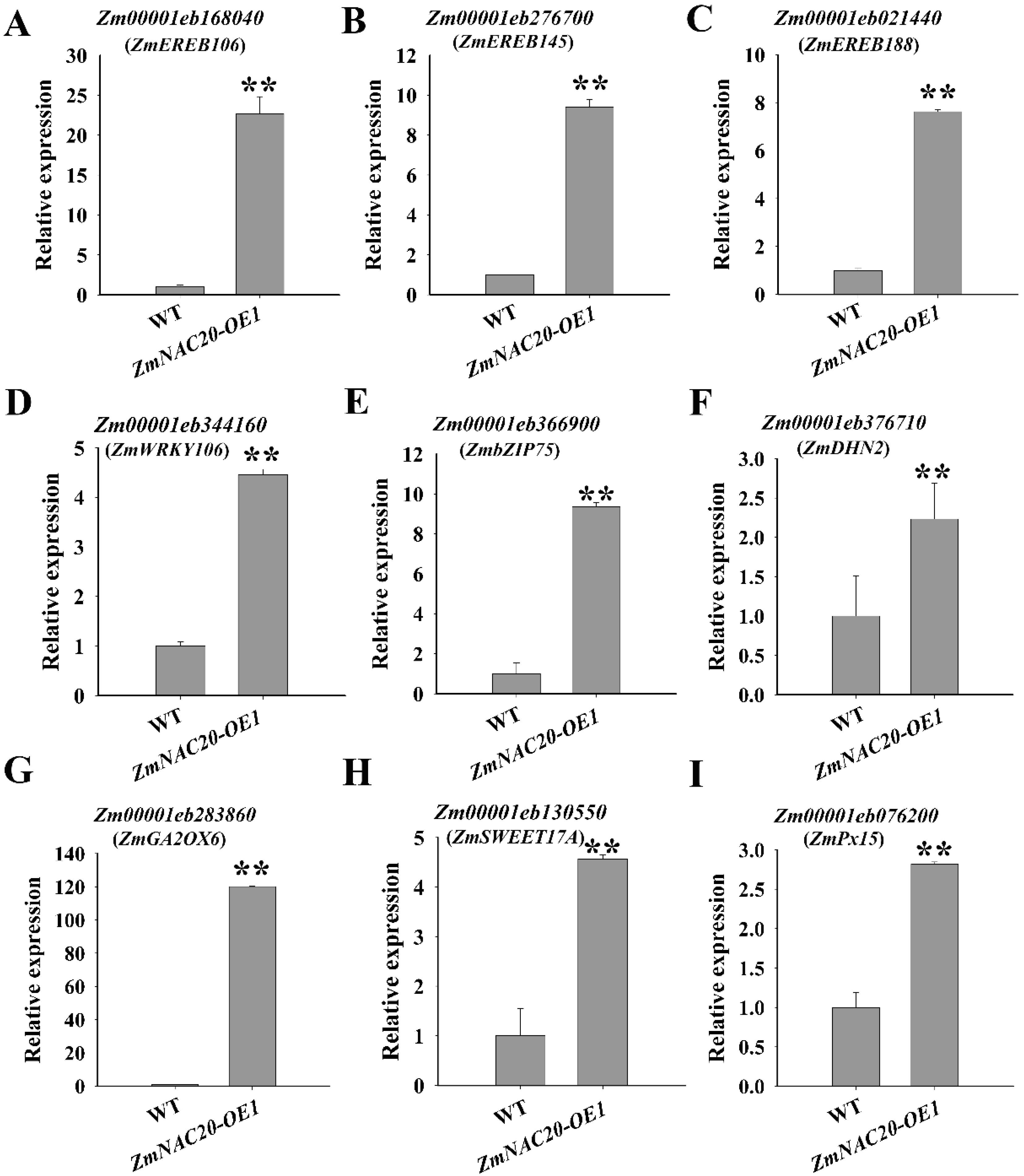
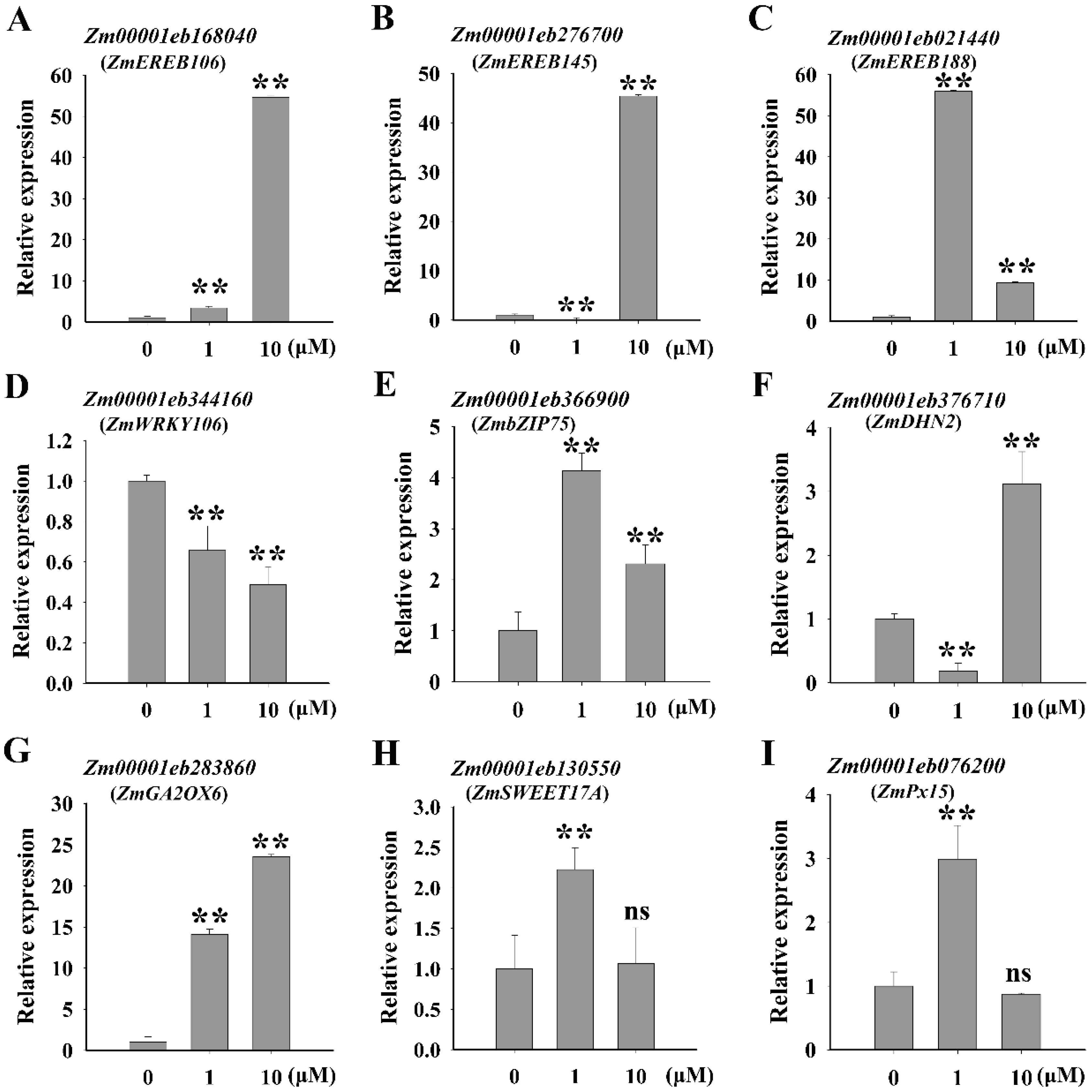
2.5. ZmNAC20 Regulates the Expression of Stress-Responsive Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Drought Treatment
4.3. Relative Water Content Assay
4.4. Detached Leaves Water Loss Assay
4.5. Determination of Biomass
4.6. Stomatal Aperture Assay
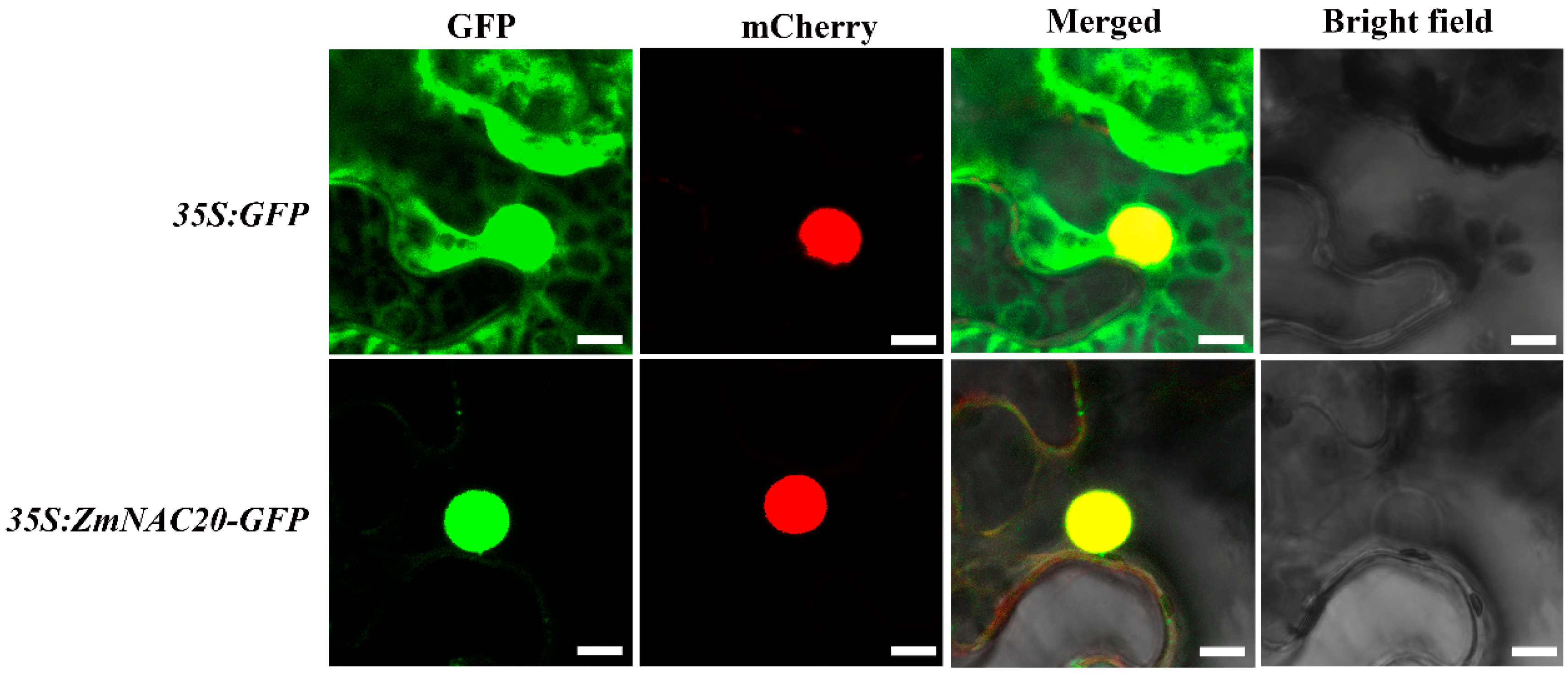
4.7. Subcellular Localization Assay
4.8. qRT-PCR Assay
4.9. RNA-seq and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ault, T.R. On the essentials of drought in a changing climate. Science 2020, 368, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, C.; Li, L.; Reynolds, M.; Mao, X.; Jing, R. Exploitation of drought tolerance-related genes for crop improvement. Int. J. Mol. Sci. 2021, 22, 10265. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Mukarram, M.; Choudhary, S.; Kurjak, D.; Petek, A.; Khan, M.M.A. Drought: Sensing, signalling, effects and tolerance in higher plants. Physiol. Plant 2021, 172, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Leng, P.; Zhao, J. Transcription factors as molecular switches to regulate drought adaptation in maize. Theor. Appl. Genet. 2020, 133, 1455–1465. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Hrmova, M.; Hussain, S.S. Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Karim, M.R.; Harikrishna, J.A.; Shimizu, T.; Sasaya, T.; Omura, T.; Haque, M.A.; Hasan, S.M.; et al. NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus, Rice black-streaked dwarf virus, Rice grassy stunt virus, Rice ragged stunt virus, and Rice transitory yellowing virus. Front. Plant Sci. 2015, 6, 676. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Mao, H.; Yu, L.; Han, R.; Li, Z.; Liu, H. ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2016, 105, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Zhang, D.; Cao, L.; Zhang, W.; Zheng, H.; Liu, Z.; Han, S.; Dong, Y.; Zhu, F.; Liu, H.; et al. Functions and regulatory framework of ZmNST3 in maize under lodging and drought stress. Plant Cell Environ. 2020, 43, 2272–2286. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Xu, J.; Wang, Q.; Li, G.; Tang, X.; Liu, T.; Feng, X.; Wu, F.; Li, M.; Xie, W.; et al. A natural antisense transcript acts as a negative regulator for the maize drought stress response gene ZmNAC48. J. Exp. Bot. 2021, 72, 2790–2806. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Bian, X.; Wei, T.; Han, T.; Yan, J.; Zhang, A. The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J. Exp. Bot. 2021, 72, 1399–1410. [Google Scholar] [CrossRef]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Voitsik, A.M.; Muench, S.; Deising, H.B.; Voll, L.M. Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant Biol. 2013, 13, 85. [Google Scholar] [CrossRef]
- Liu, W.; Zhao, B.G.; Chao, Q.; Wang, B.; Zhang, Q.; Zhang, C.; Li, S.; Jin, F.; Yang, D.; Li, X. Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis. Plant Physiol. Biochem. 2019, 145, 174–183. [Google Scholar] [CrossRef]
- Dong, Z.; Xu, Z.; Xu, L.; Galli, M.; Gallavotti, A.; Dooner, H.K.; Chuck, G. Necrotic upper tips1 mimics heat and drought stress and encodes a protoxylem-specific transcription factor in maize. Proc. Natl. Acad. Sci. USA 2020, 117, 20908–20919. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cheng, M.; Chen, Y.; Liu, B.; Wang, X.; Li, G.; Zhou, Y.; Luo, P.; Xi, Z.; Yong, H.; et al. Natural variations in the non-coding region of ZmNAC080308 contributes maintaining grain yield under drought stress in maize. BMC Plant Biol. 2021, 21, 305. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Chen, Y.; Rong, K.; Lu, Y.; Wang, N.; Xu, Z.; Pang, B.; Zhou, D.; Weng, J.; Li, M.; et al. ZmSNAC13, a maize NAC transcription factor conferring enhanced resistance to multiple abiotic stresses in transgenic Arabidopsis. Plant Physiol. Biochem. 2022, 170, 160–170. [Google Scholar] [CrossRef]
- Han, T.; Yan, J.; Xiang, Y.; Zhang, A. Phosphorylation of ZmNAC84 at Ser-113 enhances the drought tolerance by directly modulating ZmSOD2 expression in maize. Biochem. Biophys. Res. Commun. 2021, 567, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The maize ABA receptors ZmPYL8, 9, and 12 facilitate plant drought resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef]
- Lu, F.; Li, W.; Peng, Y.; Cao, Y.; Qu, J.; Sun, F.; Yang, Q.; Lu, Y.; Zhang, X.; Zheng, L.; et al. ZmPP2C26 alternative splicing variants negatively regulate drought tolerance in maize. Front. Plant Sci. 2022, 13, 851531. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Gao, S.; Qin, F.; Dai, M. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol. Plant 2017, 10, 456–469. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Sun, F.; Yu, H.; Qu, J.; Cao, Y.; Ding, L.; Feng, W.; Khalid, M.H.B.; Li, W.; Fu, F. Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 996. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Lu, X.; Wang, G.; Zhang, Q.; Zhang, X.; Fan, Z.; Cao, Y.; Wei, L.; Wang, T.; Wang, Z. Maize ZmbZIP33 is involved in drought resistance and recovery ability through an abscisic acid-dependent signaling pathway. Front. Plant Sci. 2021, 12, 629903. [Google Scholar] [CrossRef]
- Feng, X.; Xiong, J.; Zhang, W.; Guan, H.; Zheng, D.; Xiong, H.; Jia, L.; Hu, Y.; Zhou, H.; Wen, Y.; et al. ZmLBD5, a class-II LBD gene, negatively regulates drought tolerance by impairing abscisic acid synthesis. Plant J. 2022, 112, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef]
- Li, X.D.; Gao, Y.Q.; Wu, W.H.; Chen, L.M.; Wang, Y. Two calcium-dependent protein kinases enhance maize drought tolerance by activating anion channel ZmSLAC1 in guard cells. Plant Biotechnol. J. 2022, 20, 143–157. [Google Scholar] [CrossRef]
- Zhu, D.; Chang, Y.; Pei, T.; Zhang, X.; Liu, L.; Li, Y.; Zhuang, J.; Yang, H.; Qin, F.; Song, C.; et al. MAPK-like protein 1 positively regulates maize seedling drought sensitivity by suppressing ABA biosynthesis. Plant J. 2020, 102, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yuan, Z.; Zhang, P.; Liu, Z.; Wang, T.; Wei, L. Genome-wide analysis of NAC transcription factor family in maize under drought stress and rewatering. Physiol. Mol. Biol. Plants 2020, 26, 705–717. [Google Scholar] [CrossRef]
- Agurla, S.; Gahir, S.; Munemasa, S.; Murata, Y.; Raghavendra, A.S. Mechanism of stomatal closure in plants exposed to drought and cold stress. Adv. Exp. Med. Biol. 2018, 1081, 215–232. [Google Scholar]
- Liu, W.; Zhao, B.-G.; Chao, Q.; Wang, B.; Zhang, Q.; Zhang, C.; Li, S.; Jin, F.; Yang, D.; Li, X. The maize AP2/EREBP transcription factor ZmEREB160 enhances drought tolerance in Arabidopsis. Trop. Plant Biol. 2020, 13, 251–261. [Google Scholar] [CrossRef]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Li, M.; Zhao, D.; Yang, J.F.; Fu, J.D.; Xu, Z.S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Yang, J.F.; Li, M.; Xu, Z.S.; Fu, J.D. The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Riyazuddin, R.; Nisha, N.; Singh, K.; Verma, R.; Gupta, R. Involvement of dehydrin proteins in mitigating the negative effects of drought stress in plants. Plant Cell Rep. 2022, 41, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Zhang, R.; Qu, Y.; Miao, Z.; Zhang, Y.; Shen, X.; Wang, T.; Dong, J. Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 2012, 195, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Feng, H.; Deng, J.; Zhang, R.; Wen, J.; Dong, J.; Wang, T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 2020, 16, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Cheriker, H.; Kilambi, H.V.; Illouz Eliaz, N.; Blum, S.; Amsellem, Z.; Tarkowská, D.; Aharoni, A.; Eshed, Y.; Weiss, D. Inhibition of gibberellin accumulation by water deficiency promotes fast and long-term ‘drought avoidance’ responses in tomato. New Phytol. 2021, 232, 1985–1998. [Google Scholar] [CrossRef]
- Valifard, M.; Le Hir, R.; Müller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Fatima, U.; Anjali, A.; Senthil-Kumar, M. AtSWEET11 and AtSWEET12: The twin traders of sucrose. Trends Plant Sci. 2022, 27, 958–960. [Google Scholar] [CrossRef]
- Qamer, Z.; Chaudhary, M.T.; Du, X.; Hinze, L.; Azhar, M.T. Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. J. Cotton Res. 2021, 4, 9. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription Factors Interact with ABA through Gene Expression and Signaling Pathways to Mitigate Drought and Salinity Stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Wei, Z.; Liu, F. ABA-mediated modulation of elevated CO2 on stomatal response to drought. Curr. Opin. Plant Biol. 2020, 56, 174–180. [Google Scholar] [CrossRef]
- Thakur, M.; Anand, A. Hydrogen sulfide: An emerging signaling molecule regulating drought stress response in plants. Physiol. Plant 2021, 172, 1227–1243. [Google Scholar] [CrossRef] [PubMed]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.; Finley, T.; Chappell, H.; Veena, V.; Wang, K. An improved agrobacterium-mediated transformation and genome-editing method for maize inbred B104 using a ternary vector system and immature embryos. Front. Plant Sci. 2022, 13, 860971. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Milhiet, T.; Couvreur, V.; Nelissen, H.; Meziane, A.; Parent, B.; Aesaert, S.; Van Lijsebettens, M.; Inzé, D.; Tardieu, F.; et al. Modification of the expression of the aquaporin ZmPIP2;5 affects water relations and plant growth. Plant Physiol. 2020, 182, 2154–2165. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Song, S.; Liu, M.; Mu, Y.; Li, Y.; Xuan, Y.; Niu, L.; Zhang, H.; Wang, W. Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize. Int. J. Mol. Sci. 2023, 24, 4712. https://doi.org/10.3390/ijms24054712
Liu H, Song S, Liu M, Mu Y, Li Y, Xuan Y, Niu L, Zhang H, Wang W. Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize. International Journal of Molecular Sciences. 2023; 24(5):4712. https://doi.org/10.3390/ijms24054712
Chicago/Turabian StyleLiu, Hui, Songbo Song, Mengyao Liu, Yangwei Mu, Ying Li, Yuxin Xuan, Liangjie Niu, Hui Zhang, and Wei Wang. 2023. "Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize" International Journal of Molecular Sciences 24, no. 5: 4712. https://doi.org/10.3390/ijms24054712
APA StyleLiu, H., Song, S., Liu, M., Mu, Y., Li, Y., Xuan, Y., Niu, L., Zhang, H., & Wang, W. (2023). Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize. International Journal of Molecular Sciences, 24(5), 4712. https://doi.org/10.3390/ijms24054712






