Circular RNA- and microRNA-Mediated Post-Transcriptional Regulation of Preadipocyte Differentiation in Adipogenesis: From Expression Profiling to Signaling Pathway
Abstract
1. Introduction
1.1. Adipogenesis Is a Crucial Cellular Process
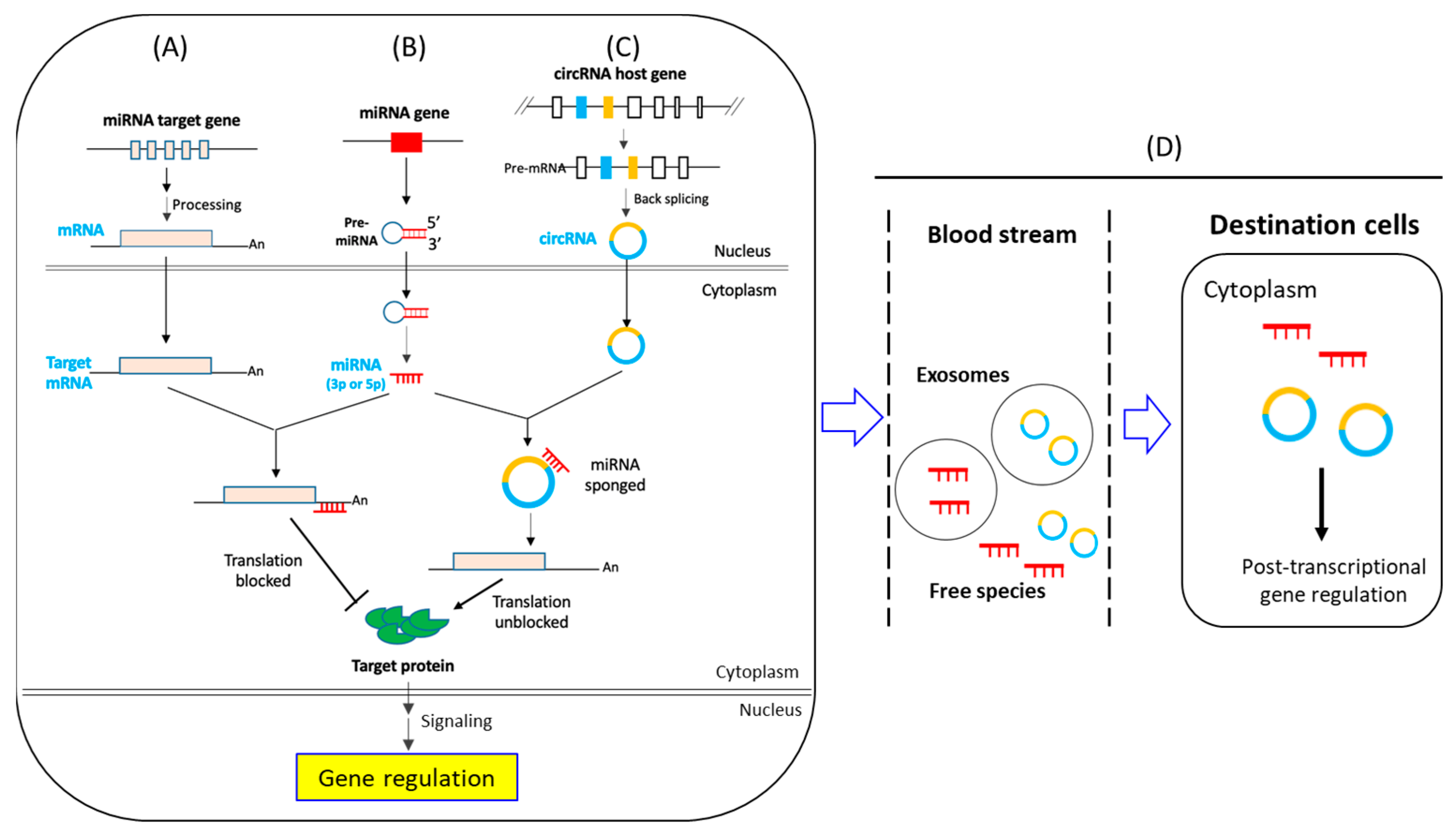
1.2. CircRNA- and miRNA-Mediated Post-Transcriptional Regulation of Gene Expression: An Overview
2. Whole-Transcriptome Profiling of circRNAs in Preadipocyte Differentiation in Adipogenesis: Connecting the Human and Animal Species
2.1. Profiling of Adipogenesis-Associated circRNAs in Different Species: Methodology and Dataset Availability
2.2. Only a Small Number of circRNAs Are Involved in Preadipocyte Differentiation: Findings from Profiling Studies
2.3. Preadipocyte Differentiation-Associated circRNAs in the Adipose Tissue: Extrapolating Animal Data to the Human
3. Cross-Species Conservation of the circRNA–miRNA and miRNA–mRNA Interacting Sequences in Adipogenesis Differentiation
3.1. miRNA Conservation
3.2. Conservation in circRNA–miRNA Interactions
3.3. Conservation in miRNA–mRNA Interactions
4. Selected circRNA- and miRNA-Mediated Post-Transcriptional Regulation of Signaling and Biochemical Pathways in Preadipocyte Differentiation
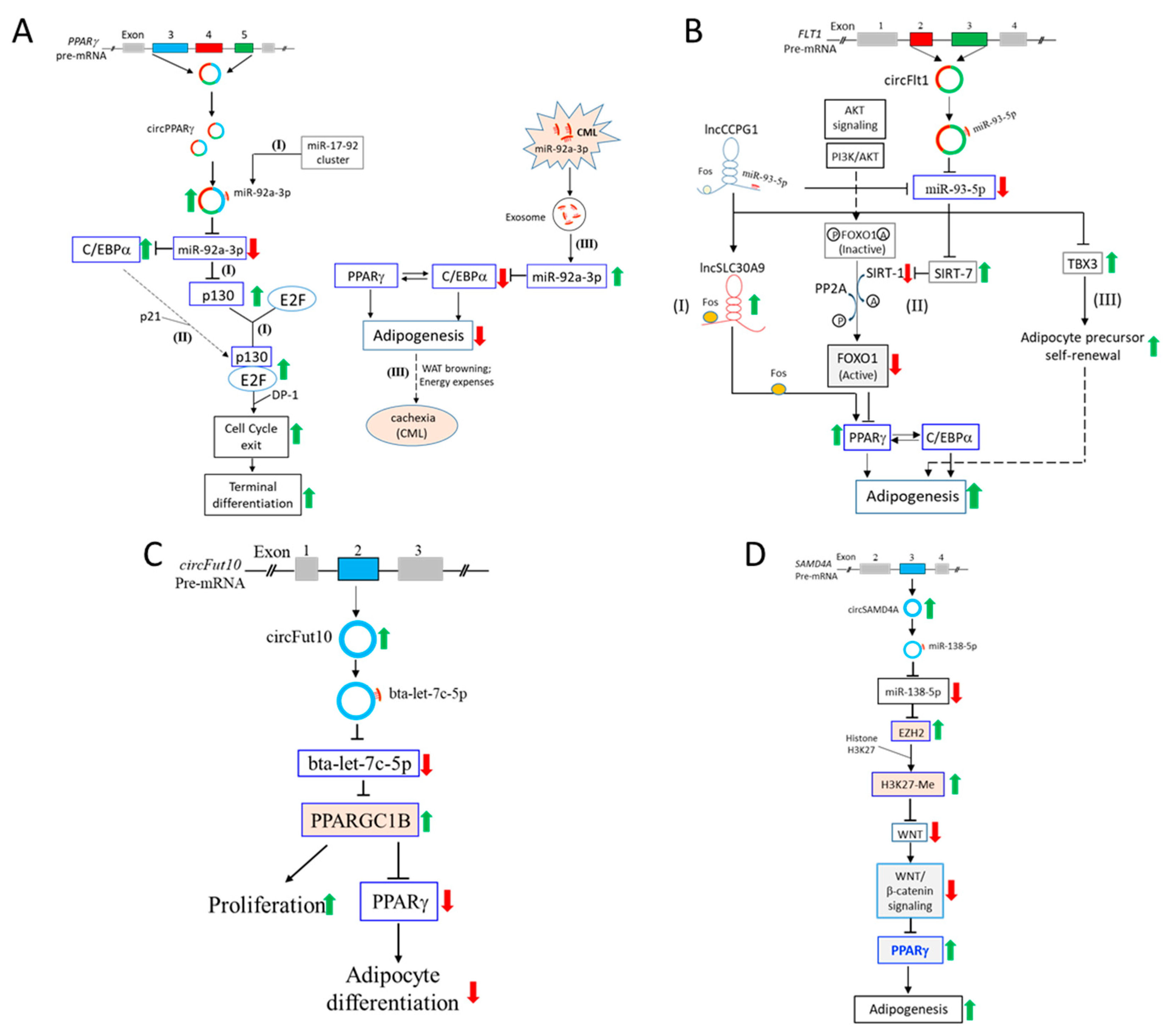
4.1. CircPPARγ Sponges miR-92a-3p to Regulate C/EBPα and p130/Rb2
4.2. CircFLT1 Sponges miR-93-5p to Regulate SIRT-7 and TBX3
4.3. The circFUT10-let-7c-5p-PCG1β Regulatory Pathway
4.4. The circSAMD4A-miR-138-5p-EZH2 Pathway
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zuk, P.A.; Zhu, M.; Ashjian, P.; De Ugarte, D.A.; Huang, J.I.; Mizuno, H.; Alfonso, Z.C.; Fraser, J.K.; Benhaim, P.; Hedrick, M.H. Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell. 2002, 13, 4279–4295. [Google Scholar] [CrossRef]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef]
- Ricoult, S.J.; Manning, B.D. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013, 14, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Karpe, F.; Dickmann, J.R.; Frayn, K.N. Fatty acids, obesity, and insulin resistance: Time for a reevaluation. Diabetes 2011, 60, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Kwok, K.H.; Lam, K.S.; Xu, A. Heterogeneity of white adipose tissue: Molecular basis and clinical implications. Exp. Mol. Med. 2016, 48, e215. [Google Scholar] [CrossRef]
- Hajer, G.R.; van Haeften, T.W.; Visseren, F.L. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Heart J. 2008, 29, 2959–2971. [Google Scholar] [CrossRef]
- Neeland, I.J.; Ayers, C.R.; Rohatgi, A.K.; Turer, A.T.; Berry, J.D.; Das, S.R.; Vega, G.L.; Khera, A.; McGuire, D.K.; Grundy, S.M.; et al. Associations of visceral and abdominal subcutaneous adipose tissue with markers of cardiac and metabolic risk in obese adults. Obesity 2013, 21, E439–E447. [Google Scholar] [CrossRef]
- Daas, S.I.; Rizeq, B.R.; Nasrallah, G.K. Adipose tissue dysfunction in cancer cachexia. J. Cell. Physiol. 2018, 234, 13–22. [Google Scholar] [CrossRef]
- Nunn, E.R.; Shinde, A.B.; Zaganjor, E. Weighing in on Adipogenesis. Front Physiol. 2022, 13, 821278. [Google Scholar] [CrossRef]
- Cohen, P.; Kajimura, S. The cellular and functional complexity of thermogenic fat. Nat. Rev. Mol. Biol. Cell. 2021, 22, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Verdejo, R.; Marlatt, K.L.; Ravussin, E.; Galgani, J.E. Contribution of brown adipose tissue to human energy metabolism. Mol. Aspects Med. 2019, 68, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, J.; Cereijo, R.; Gavalda-Navarro, A.; Peyrou, M.; Giralt, M.; Villarroya, F. New insights into the secretory functions of brown adipose tissue. J. Endocrinol. 2019, 243, R19–R27. [Google Scholar] [CrossRef] [PubMed]
- Bartelt, A.; Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014, 10, 24–36. [Google Scholar] [CrossRef]
- Wang, W.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef]
- Huang, C.J.; Choo, K.B.; Chen, C.F. The MicroRNA-signaling-peroxisome proliferator-activated receptor gamma connection in the modulation of adipogenesis: Bioinformatics projection on chicken. Poult. Sci. 2022, 101, 101950. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A complex interplay of multiple molecular determinants and pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef]
- Mirza, A.Z.; Althagafi, I.I.; Shamshad, H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019, 166, 502–513. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef]
- Festuccia, W.T.; Oztezcan, S.; Laplante, M.; Berthiaume, M.; Michel, C.; Dohgu, S.; Denis, R.G.; Brito, M.N.; Brito, N.A.; Miller, D.S.; et al. Peroxisome proliferator-activated receptor-gamma-mediated positive energy balance in the rat is associated with reduced sympathetic drive to adipose tissues and thyroid status. Endocrinology 2008, 149, 2121–2130. [Google Scholar] [CrossRef] [PubMed]
- Grozovsky, R.; Ribich, S.; Rosene, M.L.; Mulcahey, M.A.; Huang, S.A.; Patti, M.E.; Bianco, A.C.; Kim, B.W. Type 2 deiodinase expression is induced by peroxisomal proliferator-activated receptor-gamma agonists in skeletal myocytes. Endocrinology 2009, 150, 1976–1983. [Google Scholar] [CrossRef]
- Lu, C.; Cheng, S.Y. Thyroid hormone receptors regulate adipogenesis and carcinogenesis via crosstalk signaling with peroxisome proliferator-activated receptors. J. Mol. Endocrinol. 2010, 44, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Claire D’Andre, H.; Paul, W.; Shen, X.; Jia, X.; Zhang, R.; Sun, L.; Zhang, X. Identification and characterization of genes that control fat deposition in chickens. J. Anim. Sci. Biotechnol. 2013, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Ala, U.; Piro, R.M.; Grassi, E.; Damasco, C.; Silengo, L.; Oti, M.; Provero, P.; Di Cunto, F. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLoS Comput. Biol. 2008, 4, e1000043. [Google Scholar] [CrossRef]
- Huang, H.; Winter, E.E.; Wang, H.; Weinstock, K.G.; Xing, H.; Goodstadt, L.; Stenson, P.D.; Cooper, D.N.; Smith, D.; Alba, M.M.; et al. Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 2004, 5, R47. [Google Scholar] [CrossRef]
- Yu, G.; Yang, Z.; Peng, T.; Lv, Y. Circular RNAs: Rising stars in lipid metabolism and lipid disorders. J. Cell. Physiol. 2021, 236, 4797–4806. [Google Scholar] [CrossRef]
- Li, C.; Ni, Y.Q.; Xu, H.; Xiang, Q.Y.; Zhao, Y.; Zhan, J.K.; He, J.Y.; Li, S.; Liu, Y.S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target Ther. 2021, 6, 383. [Google Scholar]
- Zhang, P.; Wu, S.; He, Y.; Li, X.; Zhu, Y.; Lin, X.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. LncRNA-Mediated Adipogenesis in Different Adipocytes. Int. J. Mol. Sci. 2022, 23, 7488. [Google Scholar] [CrossRef]
- Rey, F.; Urrata, V.; Gilardini, L.; Bertoli, S.; Calcaterra, V.; Zuccotti, G.V.; Cancello, R.; Carelli, S. Role of long non-coding RNAs in adipogenesis: State of the art and implications in obesity and obesity-associated diseases. Obes. Rev. 2021, 22, e13203. [Google Scholar] [CrossRef]
- Squillaro, T.; Peluso, G.; Galderisi, U.; Di Bernardo, G. Long non-coding RNAs in regulation of adipogenesis and adipose tissue function. eLife 2020, 9, e59053. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Revi. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Gharanei, S.; Shabir, K.; Brown, J.E.; Weickert, M.O.; Barber, T.M.; Kyrou, I.; Randeva, H.S. Regulatory microRNAs in brown, brite and white adipose tissue. Cells 2020, 9, 2489. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A. microRNAs in human adipose tissue physiology and dysfunction. Cells 2021, 10, 3342. [Google Scholar] [CrossRef] [PubMed]
- Bortolin-Cavaille, M.L.; Dance, M.; Weber, M.; Cavaille, J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009, 37, 3464–3473. [Google Scholar] [CrossRef]
- Nguyen, P.N.; Huang, C.J.; Sugii, S.; Cheong, S.K.; Choo, K.B. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J. Biomed. Sci. 2017, 24, 20. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef]
- Franca, G.S.; Vibranovski, M.D.; Galante, P.A. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 2016, 7, 11438. [Google Scholar] [CrossRef]
- Boivin, V.; Deschamps-Francoeur, G.; Scott, M.S. Protein coding genes as hosts for noncoding RNA expression. Semin. Cell Dev. Biol. 2018, 75, 3–12. [Google Scholar] [CrossRef]
- Choo, K.B.; Soon, Y.L.; Nguyen, P.N.; Hiew, M.S.; Huang, C.J. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 2014, 21, 95. [Google Scholar] [CrossRef]
- Huang, C.J.; Nguyen, P.N.; Choo, K.B.; Sugii, S.; Wee, K.; Cheong, S.K.; Kamarul, T. Frequent co-expression of miRNA-5p and -3p species and cross-targeting in induced pluripotent stem cells. Int. J. Med. Sci. 2014, 11, 824–833. [Google Scholar] [CrossRef]
- Chen, M.; Yan, C.; Zhao, X. Research progress on circular rna in glioma. Front. Oncol. 2021, 11, 705059. [Google Scholar] [CrossRef]
- Wilusz, J.E. A 360 degrees view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Li, Y.; Mao, R.; Yang, H.; Zhang, Y.; Zhang, Y.; Guo, P.; Zhan, D.; Zhang, T. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 2020, 10, 4705–4719. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Li, H.; Yang, J.; Shen, X.; Song, C.; Yang, Z.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; et al. circRNA Profiling Reveals an Abundant circFUT10 that Promotes Adipocyte Proliferation and Inhibits Adipocyte Differentiation via Sponging let-7. Mol. Ther. Nucleic Acids 2020, 20, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Arcinas, C.; Tan, W.; Fang, W.; Desai, T.P.; Teh, D.C.S.; Degirmenci, U.; Xu, D.; Foo, R.; Sun, L. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat. Metab. 2019, 1, 688–703. [Google Scholar] [CrossRef]
- Sun, W.; Sun, X.; Chu, W.; Yu, S.; Dong, F.; Xu, G. CircRNA expression profiles in human visceral preadipocytes and adipocytes. Mol. Med. Rep. 2020, 21, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.P.; Han, Q.; Sheng, M.X.; Du, C.Y.; Wang, Y.L.; Cheng, X.F.; Xu, H.X.; Li, C.C.; Xu, Y.J. Identification of circular RNA expression profiles in white adipocytes and their roles in adipogenesis. Front. Physiol. 2021, 12, 728208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sheng, M.; Du, C.; Chao, Z.; Xu, H.; Cheng, X.; Li, C.; Xu, Y. Assessment of CircRNA Expression profiles and potential functions in brown adipogenesis. Front. Genet. 2021, 12, 769690. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Pei, J.; Chu, M.; Ding, X.; Wu, X.; Liang, C.; Yan, P. CircRNA Expression profile during yak adipocyte differentiation and screen potential circRNAs for adipocyte differentiation. Genes 2020, 11, 414. [Google Scholar] [CrossRef]
- Liu, X.; Liu, K.; Shan, B.; Wei, S.; Li, D.; Han, H.; Wei, W.; Chen, J.; Liu, H.; Zhang, L. A genome-wide landscape of mRNAs, lncRNAs, and circRNAs during subcutaneous adipogenesis in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 76. [Google Scholar] [CrossRef]
- Li, A.; Huang, W.; Zhang, X.; Xie, L.; Miao, X. Identification and Characterization of circRNAs of two pig breeds as a new biomarker in metabolism-related diseases. Cell Physiol. Biochem. 2018, 47, 2458–2470. [Google Scholar] [CrossRef] [PubMed]
- Song, X.H.; He, N.; Xing, Y.T.; Jin, X.Q.; Li, Y.W.; Liu, S.S.; Gao, Z.Y.; Guo, C.; Wang, J.J.; Huang, Y.Y.; et al. A novel age-related circular RNA circ-ATXN2 inhibits proliferation, promotes cell death and adipogenesis in rat adipose tissue-derived stromal cells. Front. Genet. 2021, 12, 761926. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, S.; Yue, B.; Zhang, S.; Jiang, E.; Chen, H.; Lan, X. circRNA profiling reveals circPPARgamma modulates adipogenic differentiation via sponging miR-92a-3p. J. Agric. Food Chem. 2022, 70, 6698–6708. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.C.; Wang, J.; Kong, J.; Qi, Y.; Quigg, R.J.; Li, X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. USA 2008, 105, 2889–2894. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, N.A.; Wilde, M.; Iakova, P.; Albrecht, J.H.; Darlington, G.J. E2F/p107 and E2F/p130 complexes are regulated by C/EBPalpha in 3T3-L1 adipocytes. Nucleic Acids Res. 1999, 27, 3621–3630. [Google Scholar] [CrossRef]
- Wan, Z.; Chen, X.; Gao, X.; Dong, Y.; Zhao, Y.; Wei, M.; Fan, W.; Yang, G.; Liu, L. Chronic myeloid leukemia-derived exosomes attenuate adipogenesis of adipose derived mesenchymal stem cells via transporting miR-92a-3p. J. Cell. Physiol. 2019, 234, 21274–21283. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, S.; Jiang, E.; Wang, X.; Wang, Z.; Chen, H.; Lan, X. circFLT1 and lncCCPG1 Sponges miR-93 to Regulate the Proliferation and Differentiation of Adipocytes by Promoting lncSLC30A9 Expression. Mol. Ther. Nucleic Acids 2020, 22, 484–499. [Google Scholar] [CrossRef]
- Cioffi, M.; Vallespinos-Serrano, M.; Trabulo, S.M.; Fernandez-Marcos, P.J.; Firment, A.N.; Vazquez, B.N.; Vieira, C.R.; Mulero, F.; Camara, J.A.; Cronin, U.P.; et al. MiR-93 Controls Adiposity via Inhibition of Sirt7 and Tbx3. Cell Rep. 2015, 12, 1594–1605. [Google Scholar] [CrossRef] [PubMed]
- Tanzer, A.; Stadler, P.F. Evolution of microRNAs. Methods Mol. Biol. 2006, 342, 335–350. [Google Scholar] [PubMed]
- Guo, L.; Zhao, Y.; Zhang, H.; Yang, S.; Chen, F. Integrated evolutionary analysis of human miRNA gene clusters and families implicates evolutionary relationships. Gene 2014, 534, 24–32. [Google Scholar] [CrossRef]
- Prince, A.M.; May, J.S.; Burton, G.R.; Lyle, R.E.; McGehee, R.E., Jr. Proteasomal degradation of retinoblastoma-related p130 during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2002, 290, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.; Kim, C.Y. Natural Products and Obesity: A Focus on the Regulation of Mitotic Clonal Expansion during Adipogenesis. Molecules 2019, 24, 1157. [Google Scholar] [CrossRef]
- Cobrinik, D.; Whyte, P.; Peeper, D.S.; Jacks, T.; Weinberg, R.A. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993, 7, 2392–2404. [Google Scholar] [CrossRef]
- Bell, L.A.; Ryan, K.M. Life and death decisions by E2F-1. Cell Death Differ. 2004, 11, 137–142. [Google Scholar] [CrossRef]
- Capasso, S.; Alessio, N.; Di Bernardo, G.; Cipollaro, M.; Melone, M.A.; Peluso, G.; Giordano, A.; Galderisi, U. Silencing of RB1 and RB2/P130 during adipogenesis of bone marrow stromal cells results in dysregulated differentiation. Cell Cycle 2014, 13, 482–490. [Google Scholar] [CrossRef]
- Linhart, H.G.; Ishimura-Oka, K.; DeMayo, F.; Kibe, T.; Repka, D.; Poindexter, B.; Bick, R.J.; Darlington, G.J. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 2001, 98, 12532–12537. [Google Scholar] [CrossRef]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Tontonoz, P.; Spiegelman, B.M. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc. Natl. Acad. Sci. USA 1995, 92, 9856–9860. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmant, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef]
- Wang, X.; Yang, P.; Liu, J.; Wu, H.; Yu, W.; Zhang, T.; Fu, H.; Liu, Y.; Hai, C. RARgamma-C-Fos-PPARgamma2 signaling rather than ROS generation is critical for all-trans retinoic acid-inhibited adipocyte differentiation. Biochimie 2014, 106, 121–130. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; He, M.; Liu, Y.; Paredes, S.; Villanova, L.; Brown, K.; Qiu, X.; Nabavi, N.; Mohrin, M.; Wojnoonski, K.; et al. SIRT7 represses Myc activity to suppress ER stres and prevent fatty liver disease. Cell Rep. 2013, 5, 654–665. [Google Scholar] [CrossRef]
- Fang, J.; Ianni, A.; Smolka, C.; Vakhrusheva, O.; Nolte, H.; Kruger, M.; Wietelmann, A.; Simonet, N.G.; Adrian-Segarra, J.M.; Vaquero, A.; et al. Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc. Natl. Acad. Sci. USA 2017, 114, E8352–E8361. [Google Scholar] [CrossRef]
- Yan, L.; Lavin, V.A.; Moser, L.R.; Cui, Q.; Kanies, C.; Yang, E. PP2A regulates the pro-apoptotic activity of FOXO1. J. Biol. Chem. 2008, 283, 7411–7420. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Imamura, T.; Sonoda, N.; Sears, D.D.; Patsouris, D.; Kim, J.J.; Olefsky, J.M. FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J. Biol. Chem. 2009, 284, 12188–12197. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, Y.; Tian, M.; Huang, Q. Molecular mechanisms of FOXO1 in adipocyte differentiation. J. Mol. Endocrinol. 2019, 62, R239–R253. [Google Scholar] [CrossRef]
- Tsai, P.J.; Lai, Y.H.; Manne, R.K.; Tsai, Y.S.; Sarbassov, D.; Lin, H.K. Akt: A key transducer in cancer. J. Biomed. Sci. 2022, 29, 76. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Cho, H.H.; Kim, H.K.; Bae, Y.C.; Baik, H.S.; Jung, J.S. Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells. Mol. Cell Biochem. 2007, 296, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Yuan, P.; Yang, H.; Zhang, J.; Soh, B.S.; Li, P.; Lim, S.L.; Cao, S.; Tay, J.; Orlov, Y.L.; et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 2010, 463, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Pedersen, T.A.; Hagenbeek, D.; Moulos, P.; Siersbaek, R.; Megens, E.; Denissov, S.; Borgesen, M.; Francoijs, K.J.; Mandrup, S.; et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008, 22, 2953–2967. [Google Scholar] [CrossRef] [PubMed]
- Hamza, M.S.; Pott, S.; Vega, V.B.; Thomsen, J.S.; Kandhadayar, G.S.; Ng, P.W.; Chiu, K.P.; Pettersson, S.; Wei, C.L.; Ruan, Y.; et al. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PloS ONE 2009, 4, e4907. [Google Scholar] [CrossRef]
- Wirtenberger, M.; Tchatchou, S.; Hemminki, K.; Schmutzhard, J.; Sutter, C.; Schmutzler, R.K.; Meindl, A.; Wappenschmidt, B.; Kiechle, M.; Arnold, N.; et al. Associations of genetic variants in the estrogen receptor coactivators PPARGC1A, PPARGC1B and EP300 with familial breast cancer. Carcinogenesis 2006, 27, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Petr, M.; Stastny, P.; Zajac, A.; Tufano, J.J.; Maciejewska-Skrendo, A. The role of peroxisome proliferator-activated receptors and their transcriptional coactivators gene variations in human trainability: A systematic review. Int. J. Mol. Sci. 2018, 19, 1472. [Google Scholar] [CrossRef]
- Ji, H.; Lu, R.H.; Chang, Z.G.; Su, S.S.; Yang, G.S. PGC-1beta modulates the expression of genes involved in mitochondrial function and adipogenesis during preadipocyte differentiation. Reprod. Domest. Anim. 2012, 47, 419–427. [Google Scholar] [CrossRef]
- Song, W.; Zhong, C.; Yuan, Y.; Zhu, Q.; Wang, Y.; Yin, H.; Li, D.; Zhang, Z.; Shu, G.; Yang, C.; et al. Peroxisome proliferator-activated receptor-coactivator 1-beta (PGC-1beta) modulates the expression of genes involved in adipogenesis during preadipocyte differentiation in chicken. Gene 2020, 741, 144516. [Google Scholar] [CrossRef]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef]
- Yang, Z.; Bian, C.; Zhou, H.; Huang, S.; Wang, S.; Liao, L.; Zhao, R.C. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011, 20, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008, 647, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Batool, A.; Jin, C.; Liu, Y.X. Role of EZH2 in cell lineage determination and relative signaling pathways. Front. Biosci. 2019, 24, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Tonini, T.; D’Andrilli, G.; Fucito, A.; Gaspa, L.; Bagella, L. Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements. J. Cell. Physiol. 2008, 214, 295–300. [Google Scholar] [CrossRef]
- Wang, L.; Jin, Q.; Lee, J.E.; Su, I.H.; Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7317–7322. [Google Scholar] [CrossRef]
- Yi, S.A.; Um, S.H.; Lee, J.; Yoo, J.H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Jeon, Y.J.; Park, J.W.; et al. S6K1 Phosphorylation of H2B Mediates EZH2 Trimethylation of H3: A determinant of early adipogenesis. Mol. Cell 2016, 62, 443–452. [Google Scholar] [CrossRef]
- Wan, D.; Liu, C.; Sun, Y.; Wang, W.; Huang, K.; Zheng, L. MacroH2A1.1 cooperates with EZH2 to promote adipogenesis by regulating Wnt signaling. J. Mol. Cell Biol. 2017, 9, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef]
- Yanbin, Z.; Jing, Z. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem. Biophys. Res. Commun. 2019, 516, 102–111. [Google Scholar] [CrossRef]
- Zou, S.; Gao, Y.; Zhang, S. lncRNA HCP5 acts as a ceRNA to regulate EZH2 by sponging miR1385p in cutaneous squamous cell carcinoma. Int. J. Oncol. 2021, 59, 56. [Google Scholar] [CrossRef]
- Deng, Y.; Cheng, L.; Lv, Z.; Zhu, H.; Meng, X. lncRNA SNHG7 promotes cell proliferation in glioma by acting as a competing endogenous RNA and sponging miR-138-5p to regulate EZH2 expression. Oncol. Lett. 2021, 22, 565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Wang, P.; Zhang, X.; Han, S.H.; Huo, F. LncRNA DS cell adhesion molecule antisense RNA1 facilitates oral squamous cell carcinoma progression through the microRNA-138-5p/ Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit Axis. Cell. J. 2022, 24, 222–229. [Google Scholar] [PubMed]
- Qiu, S.; Huang, D.; Yin, D.; Li, F.; Li, X.; Kung, H.F.; Peng, Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim. Biophys. Acta 2013, 1832, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Chen, Z.; Jin, Y.; Wang, Y.; Kolokythas, A.; Dai, Y.; Zhou, X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011, 440, 23–31. [Google Scholar] [CrossRef]
- Si, F.; Sun, J.; Wang, C. MicroRNA-138 suppresses cell proliferation in laryngeal squamous cell carcinoma via inhibiting EZH2 and PI3K/AKT signaling. Exp. Ther. Med. 2021, 22, 1090. [Google Scholar] [CrossRef] [PubMed]


| No. | Species | Adipose Tissue Profiled | Platform a (RNA Prep’n) | Profiling Dataset Availability | Number in circRNA | Reference | |
|---|---|---|---|---|---|---|---|
| Total | DE (Up/Down) on Different’n/Comparison | ||||||
| (A) Preadipocyte Differentiation Studies | |||||||
| A1 | Human | VAT | circRNA microarray (total RNA) | Yes | NA | 4080 (2215/1865) | [50] |
| A2 | Mouse | WAT (stromal cells) | RNA-seq (circRNA-enriched) | Yes | 3771 | 41 (1.09%) (28/13) | [51] |
| A3 | Mouse | BAT (stromal cells) | RNA-seq (circRNA-enriched) | Yes | 3869 | 117 (3.02%) (77/40) | [52] |
| A4 | Yak | SAT | RNA-seq (circRNA-enriched) | Yes | 7203 | 136 (1.89%) (92/44) | [53] |
| A5 | Pig | SAT | RNA-seq | Yes (chrom’l sites only) | 2172 | 297 (13.67%) (Also, lncRNA and mRNA) | [54] |
| (B) Differentiation and Comparative Studies | |||||||
| B1 | Human | VAT | circRNA microarray (circRNA-enriched) | Yes | NA | Obese vs. lean: 244 (143/101) | [47] |
| B2 | Human | VAT and SAT | RNA-seq | Yes | 6925 | NA | [49] |
| B3 | Mouse | Epididymal and inguinal fat (WAT) | RNA-seq | Yes | 2380 | NA | [49] |
| B4 | Pig | SAT | RNA-seq (circRNA-enriched) | No | 29,763 (combined) | Two breeds: 275 (70/205) | [55] |
| B5 | Rat | SAT (stromal cells) | RNA-seq | No | Young: 4860 Old: 4952 | Young vs. old: 67 (1.38%) (33/34) | [56] |
| B6 | Cattle | SAT | RNA-seq | No | Calf: 4337 Adult: 5465 | Calf vs. adult: 307 (7.08%) (156/151) | [48] |
| Dataset No. | Dataset a | No. of DE circRNAs | No. of Overlapping circRNAs in WAT-1 (Percentage) a | Reference | |
|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||
| 1 | Mouse WAT-1 | 28 | 13 | 41 (100%) | [51] |
| 2 | Mouse WAT-2 | 20 | 5 | 25 (61.0%) | [49] |
| 3 | Human WAT | 18 | 5 | 23 (56.1%) | [49] |
| 4 | Yak WAT | 14 | 5 | 19 (46.3%) | [53] |
| 5 | Mouse BAT | 25 | 7 | 32 (78.0%) | [52] |
| CircRNA | Presence in WAT/BAT Dataset a | miRNA (Family) | mRNA (Gene Symbol) | Reference | ||
|---|---|---|---|---|---|---|
| Host Gene | CircRNA (circBase ID) (Species) | Exons | ||||
| PPARγ(Peroxisome Proliferator-Activated Receptor-gamma) | bta_circ_Pparγ (bta_circ_0010660) (Bovine) | 3-5 | h, m, y | miR-92a-3p/(mir-25) |
| [57,58,59,60] |
| FLT1(Fms-Related Receptor Tyrosine Kinase 1 | bta_circ_Flt1 (bta_circ_002673) (Bovine) | 2-3 | y | miR-93-5p (mir-17) (and lncCCPG1 and lncSL30A9) |
| [61,62] |
| FUT10(Fucosyltransferase 10) | bta_circ_Fut10 (NA) (Bovine) | 2 | h, y | let-7c-5p (let-7) |
| [48] |
| SAMD4A(Sterile Alpha Motif Domain Containing 4A) | hsa_circ_SAMD4A (hsa_circ_0004846) (Human) | 3 | h, m, y | miR-138-5p (mir-138) |
| [47] |
| circRNA | miRNA | mRNA | miRNA Seed Sequence Conservation | ||
|---|---|---|---|---|---|
| miRNA in Different Species a | cirRNA–miRNA Interaction b | miRNA–mRNA Interaction a | |||
| circPPARγ | miR-92a-3p |
| All 7/7 | All 7/7 |
|
| circFLT1 | miR-93-5p |
| All 7/7 (NA for pig and chicken) | All 7/7 except mouse: 6/7 |
|
| circFUT10 | let-7c-5p | PGC1β | All 6/6 | All 6/6 except mouse: 4/6 |
|
| circSAMD4A | miR-138-5p | EZH2 | All 7/7 | All 6/6 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-J.; Choo, K.B. Circular RNA- and microRNA-Mediated Post-Transcriptional Regulation of Preadipocyte Differentiation in Adipogenesis: From Expression Profiling to Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 4549. https://doi.org/10.3390/ijms24054549
Huang C-J, Choo KB. Circular RNA- and microRNA-Mediated Post-Transcriptional Regulation of Preadipocyte Differentiation in Adipogenesis: From Expression Profiling to Signaling Pathway. International Journal of Molecular Sciences. 2023; 24(5):4549. https://doi.org/10.3390/ijms24054549
Chicago/Turabian StyleHuang, Chiu-Jung, and Kong Bung Choo. 2023. "Circular RNA- and microRNA-Mediated Post-Transcriptional Regulation of Preadipocyte Differentiation in Adipogenesis: From Expression Profiling to Signaling Pathway" International Journal of Molecular Sciences 24, no. 5: 4549. https://doi.org/10.3390/ijms24054549
APA StyleHuang, C.-J., & Choo, K. B. (2023). Circular RNA- and microRNA-Mediated Post-Transcriptional Regulation of Preadipocyte Differentiation in Adipogenesis: From Expression Profiling to Signaling Pathway. International Journal of Molecular Sciences, 24(5), 4549. https://doi.org/10.3390/ijms24054549





