PCR Assay for Rapid Taxonomic Differentiation of Virulent Staphylococcus aureus and Klebsiella pneumoniae Bacteriophages
Abstract
1. Introduction
2. Results
2.1. Sample Collection and Phylogenetic Analysis
2.2. Identification of Signature Genes Suitable for PCR Typing
2.3. PCR-Based Typing Scheme for Rapid Phage Classification
3. Discussion
4. Materials and Methods
4.1. Genome Sampling and Annotation
4.2. Phylogenetic Analysis
4.3. Pangenome Analysis and Homologues Identification
4.4. Primer Design
4.5. PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Review on Antimicrobial Resistance. In Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; HM Government: London, UK, 2014.
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P.; et al. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. 2021, 21, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.L.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef] [PubMed]
- Petrovic Fabijan, A.; Lin, R.C.Y.; Ho, J.; Maddocks, S.; Ben Zakour, N.L.; Iredell, J.R.; Khalid, A.; Venturini, C.; Chard, R.; Morales, S.; et al. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 2020, 5, 465–472. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Loh, B.; Leptihn, S. A Call For a Multidisciplinary Future of Phage Therapy to Combat Multi-drug Resistant Bacterial Infections. Infect. Microbes Dis. 2020, 2, 1–2. [Google Scholar] [CrossRef]
- Cui, Z.; Guo, X.; Feng, T.; Li, L. Exploring the whole standard operating procedure for phage therapy in clinical practice. J. Transl. Med. 2019, 17, 373. [Google Scholar] [CrossRef]
- Harper, D.R. Criteria for Selecting Suitable Infectious Diseases for Phage Therapy. Viruses 2018, 10, 177. [Google Scholar] [CrossRef]
- Hockenberry, A.J.; Wilke, C.O. BACPHLIP: Predicting bacteriophage lifestyle from conserved protein domains. PeerJ 2021, 9, e11396. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Varsani, A.; Tong, Y.; Simmonds, P.; Sabanadzovic, S.; Rubino, L.; Roux, S.; Munoz, A.R.; Lood, C.; Lefkowitz, E.J.; et al. Perspective on taxonomic classification of uncultivated viruses. Curr. Opin. Virol. 2021, 51, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Bin Jang, H.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Grunwald, A.; Dahan, M.; Giesbertz, A.; Nilsson, A.; Nyberg, L.K.; Weinhold, E.; Ambjornsson, T.; Westerlund, F.; Ebenstein, Y. Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 2015, 43, e117. [Google Scholar] [CrossRef] [PubMed]
- Stverakova, D.; Sedo, O.; Benesik, M.; Zdrahal, Z.; Doskar, J.; Pantucek, R. Rapid Identification of Intact Staphylococcal Bacteriophages Using Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry. Viruses 2018, 10, 176. [Google Scholar] [CrossRef]
- Born, Y.; Knecht, L.E.; Eigenmann, M.; Bolliger, M.; Klumpp, J.; Fieseler, L. A major-capsid-protein-based multiplex PCR assay for rapid identification of selected virulent bacteriophage types. Arch. Virol. 2019, 164, 819–830. [Google Scholar] [CrossRef]
- Moisan, M.; Moineau, S. Multilocus sequence typing scheme for the characterization of 936- like phages infecting Lactococcus lactis. Appl. Environ. Microbiol. 2012, 78, 4646–4653. [Google Scholar] [CrossRef]
- Doria, F.; Napoli, C.; Costantini, A.; Berta, G.; Saiz, J.C.; Garcia-Moruno, E. Development of a new method for detection and identification of Oenococcus oeni bacteriophages based on endolysin gene sequence and randomly amplified polymorphic DNA. Appl. Environ. Microbiol. 2013, 79, 4799–4805. [Google Scholar] [CrossRef]
- Kahankova, J.; Pantucek, R.; Goerke, C.; Ruzickova, V.; Holochova, P.; Doskar, J. Multilocus PCR typing strategy for differentiation of Staphylococcus aureus siphoviruses reflecting their modular genome structure. Environ. Microbiol. 2010, 12, 2527–2538. [Google Scholar] [CrossRef]
- Ko, D.S.; Seong, W.J.; Kim, D.; Kim, E.K.; Kim, N.H.; Lee, C.Y.; Kim, J.H.; Kwon, H.J. Molecular prophage typing of Staphylococcus aureus isolates from bovine mastitis. J. Vet. Sci. 2018, 19, 771–781. [Google Scholar] [CrossRef]
- Sanchini, A.; Del Grosso, M.; Villa, L.; Ammendolia, M.G.; Superti, F.; Monaco, M.; Pantosti, A. Typing of Panton-Valentine leukocidin-encoding phages carried by methicillin-susceptible and methicillin-resistant Staphylococcus aureus from Italy. Clin. Microbiol. Infect. 2014, 20, O840–O846. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Smith, M.C.M.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef]
- Keen, E.C.; Adhya, S.L.; Wormser, G.P. Phage Therapy: Current Research and Applications. Clin. Infect. Dis. 2015, 61, 141. [Google Scholar] [CrossRef]
- Grose, J.H.; Casjens, S.R. Understanding the enormous diversity of bacteriophages: The tailed phages that infect the bacterial family Enterobacteriaceae. Virology 2014, 468–470, 421–443. [Google Scholar] [CrossRef] [PubMed]
- Philipson, C.W.; Voegtly, L.J.; Lueder, M.R.; Long, K.A.; Rice, G.K.; Frey, K.G.; Biswas, B.; Cer, R.Z.; Hamilton, T.; Bishop-Lilly, K.A. Characterizing Phage Genomes for Therapeutic Applications. Viruses 2018, 10, 188. [Google Scholar] [CrossRef]
- Russell, D.A. Sequencing, Assembling, and Finishing Complete Bacteriophage Genomes. Methods Mol. Biol. 2018, 1681, 109–125. [Google Scholar] [PubMed]
- Deghorain, M.; Van Melderen, L. The staphylococci phages family: An overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef]
- Henry, M.; Bobay, L.-M.; Chevallereau, A.; Saussereau, E.; Ceyssens, P.-J.; Debarbieux, L. The Search for Therapeutic Bacteriophages Uncovers One New Subfamily and Two New Genera of Pseudomonas-Infecting Myoviridae. PLoS ONE 2015, 10, e0117163. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Ackermann, H.-W.; Anany, H.; Blasdel, B.; Connerton, I.F.; Goulding, D.; Griffiths, M.W.; Hooton, S.P.; Kutter, E.M.; Kropinski, A.M.; et al. A suggested new bacteriophage genus: “Viunalikevirus”. Arch. Virol. 2012, 157, 2035–2046. [Google Scholar] [CrossRef]
- Smith, K.C.; Castro-Nallar, E.; Fisher, J.N.; Breakwell, D.P.; Grose, J.H.; Burnett, S.H. Phage cluster relationships identified through single gene analysis. BMC Genom. 2013, 14, 410. [Google Scholar] [CrossRef]
- Barylski, J.; Enault, F.; Dutilh, B.E.; Schuller, M.B.P.; Edwards, R.A.; Gillis, A.; Klumpp, J.; Knezevic, P.; Krupovic, M.; Kuhn, J.H.; et al. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Phages. Syst. Biol. 2020, 69, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Grazziotin, A.L.; Koonin, E.V.; Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017, 45, D491–D498. [Google Scholar] [CrossRef] [PubMed]
- Terzian, P.; Olo Ndela, E.; Galiez, C.; Lossouarn, J.; Perez Bucio, R.E.; Mom, R.; Toussaint, A.; Petit, M.A.; Enault, F. PHROG: Families of prokaryotic virus proteins clustered using remote homology. NAR Genomics Bioinforma. 2021, 3, lqab067. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinforma. 2020, 69, e96. [Google Scholar] [CrossRef]
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Larsson, J. Qualpalr: Automatic Generation of Qualitative Color Palettes. R Package Version 0.4.3. Available online: https://CRAN.R-project.org/package=qualpalr (accessed on 20 July 2022).
- Muller, K. Here: A Simpler Way to Find Your Files. R Package Version 1.0.1. Available online: https://CRAN.R-project.org/package=here (accessed on 20 July 2022).
- Bayliss, S.C.; Thorpe, H.A.; Coyle, N.M.; Sheppard, S.K.; Feil, E.J. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience 2019, 8, giz119. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual 2012 Edition (Final); Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
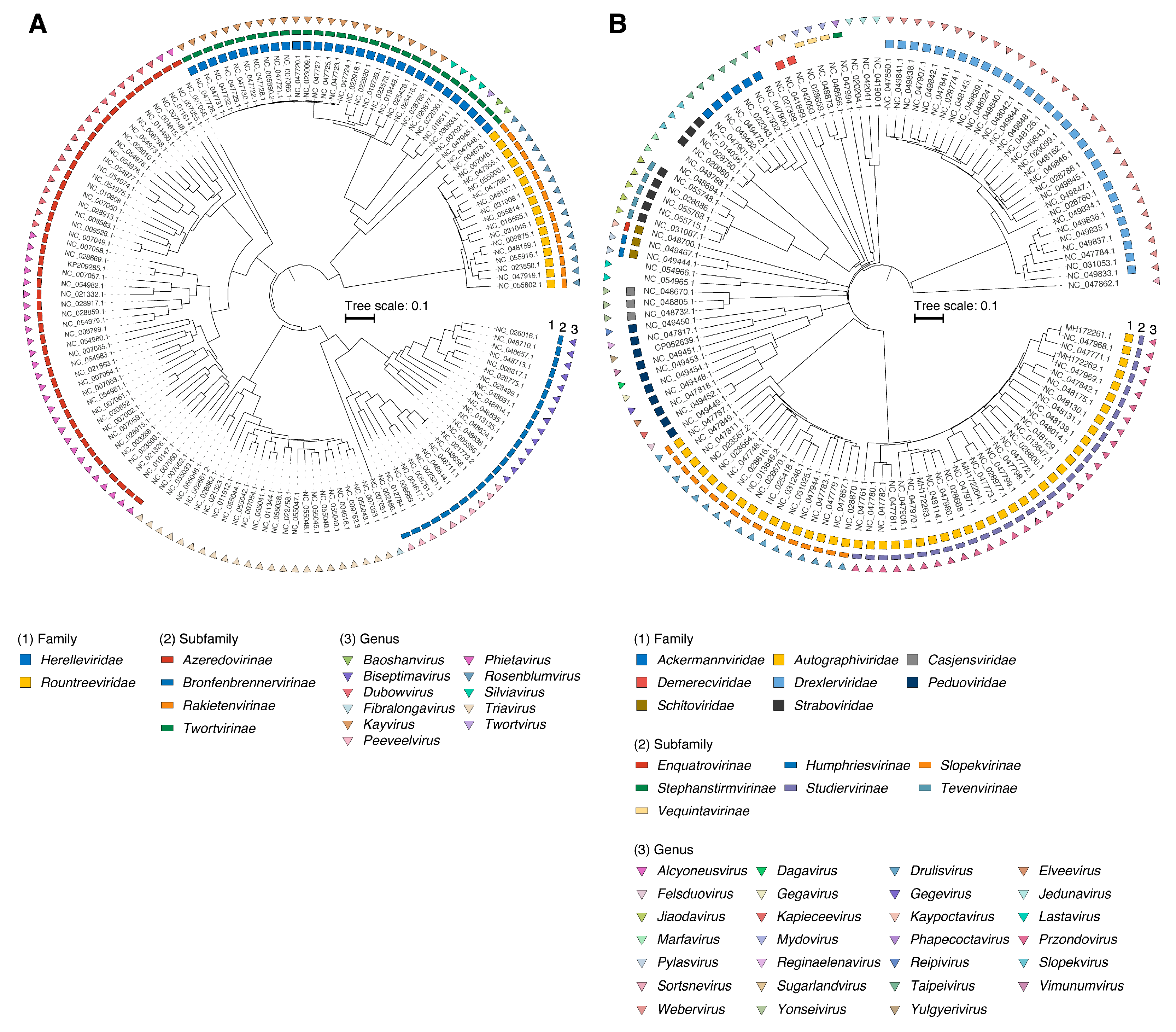

| Family | Subfamily | Genus | NCBI (incl. ICTV) | Average Genome Length, Kbp | Orthologous Gene Families (N) |
|---|---|---|---|---|---|
| Staphylococcus aureus phages | |||||
| Herelleviridae | Twortvirinae | Baoshanvirus * | 2 (2) | 146.1 | 14 |
| Kayvirus * | 54 (24) | 142.0 | 0 | ||
| Silviavirus * | 9 (4) | 135.9 | 3 | ||
| Twortvirus * | 1 (1) | 130.7 | |||
| Unclassified * | 5 | ||||
| Unclassified | Unclassified * | 1 | |||
| Rountreeviridae | Rakietenvirinae | Rosenblumvirus * | 29 (16) | 17.7 | 9 |
| Azeredovirinae | Dubowvirus | 32 (18) | 43.1 | 4 | |
| Phietavirus | 39 (29) | 44.4 | 2 | ||
| Bronfenbrennervirinae | Biseptimavirus | 20 (14) | 42.6 | 0 | |
| Peeveelvirus | 22 (10) | 42.0 | 0 | ||
| Fibralongavirus | 1 (1) | 41.3 | 0 | ||
| Triavirus | 42 (23) | 45.8 | 3 | ||
| Unclassified | Unclassified | Unclassified | 12 | ||
| Klebsiella pneumoniae phages | |||||
| Ackermannviridae | Taipeivirus * | 7 (6) | 158.5 | 63 | |
| Autographiviridae | Slopekvirinae | Drulisvirus * | 48 (17) | 44.1 | 13 |
| Studiervirinae | Przondovirus * | 79 (31) | 40.6 | 10 | |
| Unclassified * | 12 | ||||
| Unclassified | Unclassified | 4 | |||
| Casjensviridae | Yonseivirus * | 9 (3) | 58.8 | 2 | |
| Demerecviridae | Sugarlandvirus * | 11 (2) | 112.6 | 55 | |
| Drexlerviridae | Webervirus * | 75 (30) | 49.6 | 7 | |
| Peduoviridae | Reipivirus | 1 (1) | 38.3 | ||
| Dagavirus | 1 (1) | 39.0 | |||
| Elveevirus | 1 (1) | 33.5 | |||
| Felsduovirus * | 1 (1) | 18.2 | |||
| Gegavirus | 1 (1) | 33.8 | |||
| Gegevirus | 1 (1) | 39.6 | |||
| Kapieceevirus * | 1 (1) | 32.3 | |||
| Reginaelenavirus | 1 (1) | 35.1 | |||
| Vimunumvirus * | 1 (1) | 34.1 | |||
| Yulgyerivirus | 1 (1) | 32.3 | |||
| Unclassified | Unclassified | 61 | |||
| Schitoviridae | Enquatrovirinae | Kaypoctavirus * | 1 (1) | 73.7 | |
| Humphriesvirinae | Pylasvirus * | 2 (2) | 70.5 | 12 | |
| Straboviridae | Tevenvirinae | Jiaodavirus * | 15 (5) | 116.8 | 23 |
| Slopekvirus * | 9 (3) | 175.4 | 39 | ||
| Vequintavirinae | Mydovirus * | 6 (3) | 143.7 | 41 | |
| Stephanstirmvirinae | Phapecoctavirus * | 1 (1) | 150.9 | ||
| Alcyoneusvirus * | 3 (1) | 347.0 | 207 | ||
| Jedunavirus * | 12 (3) | 47.5 | 12 | ||
| Lastavirus | 3 (2) | 62.1 | 18 | ||
| Marfavirus * | 10 (2) | 170.1 | 16 | ||
| Sortsnevirus * | 1 (1) | 42.5 | |||
| Unclassified | Unclassified | Unclassified | 101 | ||
| Target | Taxon | Primer Name | Nucleotide Sequence (5′—3′) | Product Size (bp) |
|---|---|---|---|---|
| S. aureus bacteriophage typing | ||||
| resolvase | Herelleviridae family | Herell_f | GAATTAACTTCTTGGTGGGG | 307 |
| Herell_r | ATACTTTTTCATCATAMGGTAA | |||
| major capsid protein | Rountreeviridae family | Rountr_f | TCAATTTCCAAACATTAGCAG | 780 |
| Rountr_f | GGATTTACATCTTGGTCAGTA | |||
| K. pneumoniae bacteriophage typing | ||||
| internal virion protein D | Przondovirus genus | Przond_f | CGTACAACCAAGGKGAAGG | 463 |
| Przond_r | TCCGTGAACACATCRTACCC | |||
| putative tail tube associated baseplate protein | Taipeivirus genus | Taipei_f | AGTTCTGAACACCAAAGGC | 326 |
| Taipei_r | CCAACTCAGAGCCGTTCC | |||
| capsid protein | Drulisvirus genus | Drulis_f | CGCTCCGTAACGATAAGATG | 728 |
| Drulis_r | ACGCAGACCGATGTTGTAC | |||
| putative major head subunit precursor | Webervirus genus | Weber_f | CCTATGATGACGACTCAAAC | 422 |
| Weber_r | ATTGCCAGCCATCTTATCAG | |||
| alpha-glucosyl-transferase | Jiaodavirus genus | Jiaoda_f | TGAACATCAAAGCAATTCGTG | 502 |
| Jiaoda_r | AACCACAGAATGCCAGAATC | |||
| putative replicative DNA helicase | Sugarlandvirus genus | Sugarl_f | GATCTACCAAGCTGTCCAG | 307 |
| Sugarl_r | AGTCGTTGTTACTCGTTCC | |||
| putative baseplate hub | Slopekvirus genus | Slopek_f | TCAAAGAACAATACGAAGAGG | 411 |
| Slopek_r | TTGCCATTGCTTCCAGAGAG | |||
| virion structural protein | Jedunavirus genus | Jeduna_f | ACTTCTATTGTCATGGCTGG | 349 |
| Jeduna_r | CACCTTACAGTTTAGCGTC | |||
| hypothetical protein | Marfavirus genus | Marfa_f | GCACCTGAAGGCATTACCC | 252 |
| Marfa_r | CCCATCAATAGAATAAAGCAC | |||
| putative helicase | Mydovirus genus | Mydo_f | GATCGAAAAGAATGTCTGGG | ast302 |
| Mydo_r | TTGGTCTACGATAATATCACG | |||
| DNA polymerase I | Yonseivirus genus | Yonsei_f | GCACGCCGACCTATCCCG | 482 |
| Yonsei_r | GCCACGGTCATTGATAAGC | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kornienko, M.; Bespiatykh, D.; Malakhova, M.; Gorodnichev, R.; Kuptsov, N.; Shitikov, E. PCR Assay for Rapid Taxonomic Differentiation of Virulent Staphylococcus aureus and Klebsiella pneumoniae Bacteriophages. Int. J. Mol. Sci. 2023, 24, 4483. https://doi.org/10.3390/ijms24054483
Kornienko M, Bespiatykh D, Malakhova M, Gorodnichev R, Kuptsov N, Shitikov E. PCR Assay for Rapid Taxonomic Differentiation of Virulent Staphylococcus aureus and Klebsiella pneumoniae Bacteriophages. International Journal of Molecular Sciences. 2023; 24(5):4483. https://doi.org/10.3390/ijms24054483
Chicago/Turabian StyleKornienko, Maria, Dmitry Bespiatykh, Maja Malakhova, Roman Gorodnichev, Nikita Kuptsov, and Egor Shitikov. 2023. "PCR Assay for Rapid Taxonomic Differentiation of Virulent Staphylococcus aureus and Klebsiella pneumoniae Bacteriophages" International Journal of Molecular Sciences 24, no. 5: 4483. https://doi.org/10.3390/ijms24054483
APA StyleKornienko, M., Bespiatykh, D., Malakhova, M., Gorodnichev, R., Kuptsov, N., & Shitikov, E. (2023). PCR Assay for Rapid Taxonomic Differentiation of Virulent Staphylococcus aureus and Klebsiella pneumoniae Bacteriophages. International Journal of Molecular Sciences, 24(5), 4483. https://doi.org/10.3390/ijms24054483





