Polymorphic Variants of Genes Encoding Angiogenesis-Related Factors in Infertile Women with Recurrent Implantation Failure
Abstract
1. Introduction
2. Results
2.1. Baseline Characteristics of Study Subjects and Control Groups
2.2. Association Studies
2.3. Stratification Analysis
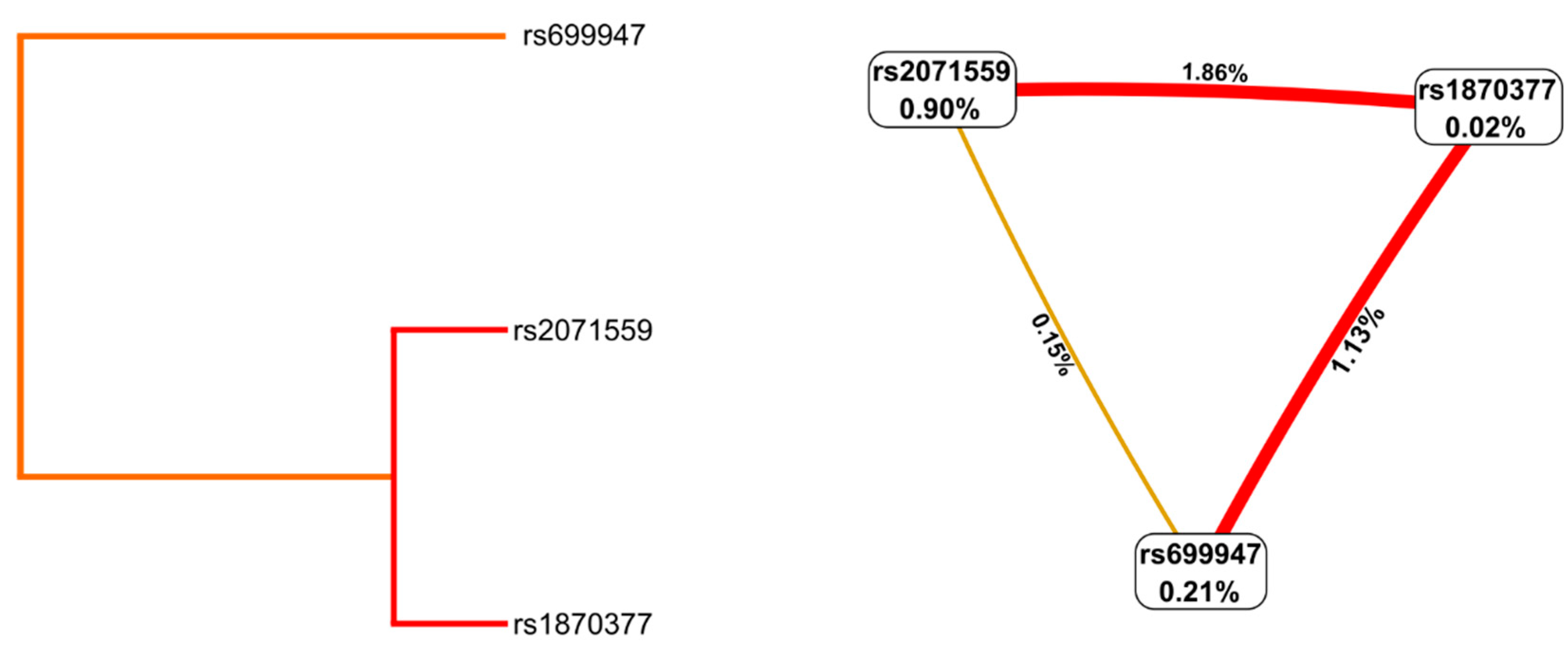
2.4. Haplotype and Gene–Gene Interaction Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Sample Collection for Genetic Testing and DNA Extraction
4.3. DNA Amplification and Genotyping
4.4. Anti-Müllerian Hormone Analyses
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Busnelli, A.; Reschini, M.; Cardellicchio, L.; Vegetti, W.; Somigliana, E.; Vercellini, P. How common is real repeated implantation failure? An indirect estimate of the prevalence. Reprod. Biomed. Online 2020, 40, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Mrozikiewicz, A.E.; Ożarowski, M.; Jędrzejczak, P. Biomolecular markers of recurrent implantation failure—A review. Int. J. Mol. Sci. 2021, 22, 10082. [Google Scholar] [CrossRef] [PubMed]
- Busnelli, A.; Somigliana, E.; Cirillo, F.; Baggiani, A.; Levi-Setti, P.E. Efficacy of therapies and interventions for repeated embryo implantation failure: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 1747. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Zhang, X.; Liang, Y.; Lin, S.; Qian, W.; Fan, S. Alterations in vaginal microbiota and associated metabolome in women with recurrent implantation failure. mBio 2020, 11, e03242-19. [Google Scholar] [CrossRef]
- Bahia, W.; Zitouni, H.; Kanabekova, P.; Bauyrzhanova, Z.; Shaimardanova, M.; Finan, R.R.; Aimagambetova, G.; Almawi, W.Y. Human forkhead box protein 3 gene variants associated with altered susceptibility to idiopathic recurrent pregnancy loss: A retrospective case-control study. Am. J. Reprod. Immunol. 2022, 88, e13551. [Google Scholar] [CrossRef]
- Abdukassimova, M.; Kanabekova, P.; Bauyrzhanova, Z.; Ukybassova, T.; Kaldygulova, L.; Imankulova, B.; Aimagambetova, G.; Almawi, W.Y. Association of Human forkhead box protein 3 (FOXP3) gene polymorphisms with idiopathic recurrent pregnancy loss among Kazakhstani women. Gene 2021, 801, 145835. [Google Scholar] [CrossRef]
- Aimagambetova, G.; Hajjej, A.; Malalla, Z.H.; Finan, R.R.; Sarray, S.; Almawi, W.Y. Maternal HLA-DR, HLA-DQ, and HLA-DP loci are linked with altered risk of recurrent pregnancy loss in Lebanese women: A case-control study. Am. J. Reprod. Immunol. 2019, 82, e13173. [Google Scholar] [CrossRef]
- Shaulov, T.; Sierra, S.; Sylvestre, C. Recurrent implantation failure in IVF: A Canadian Fertility and Andrology Society Clinical Practice Guideline. Reprod. Biomed. Online 2020, 41, 819–833. [Google Scholar] [CrossRef]
- Galán, A.; Herrer, R.; Remohí, J.; Pellicer, A.; Simón, C. Embryonic regulation of endometrial epithelial apoptosis during human implantation. Hum. Reprod. 2000, 15 (Suppl. S6), 74–80. [Google Scholar]
- Idelevich, A.; Vilella, F. Mother and embryo cross-communication. Genes 2020, 11, 376. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, S. Placental Blood Circulation. In Vascular Biology of the Placenta; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2010. Available online: https://www.ncbi.nlm.nih.gov/books/NBK53254/ (accessed on 22 February 2022).
- Geva, E.; Ginzinger, D.G.; Zaloudek, C.J.; Moore, D.H.; Byrne, A.; Jaffe, R.B. Human placental vascular development: Vasculogenic and angiogenic (branching and nonbranching) transformation is regulated by vascular endothelial growth factor-A, angiopoietin-1, and angiopoietin-2. J. Clin. Endocrinol. Metab. 2002, 87, 4213–4224. [Google Scholar] [CrossRef]
- Chen, D.B.; Zheng, J. Regulation of placental angiogenesis. Microcirculation 2014, 21, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Zygmunt, M.; Herr, F.; Münstedt, K.; Lang, U.; Liang, O.D. Angiogenesis and vasculogenesis in pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 2003, 110 (Suppl. S1), S10–S18. [Google Scholar] [CrossRef] [PubMed]
- Alitalo, K.; Carmeliet, P. Molecular mechanisms of lymphangiogenesis in health and disease. Cancer Cell 2002, 1, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Shaik, F.; Cuthbert, G.A.; Homer-Vanniasinkam, S.; Muench, S.P.; Ponnambalam, S.; Harrison, M.A. Structural basis for vascular endothelial growth factor receptor activation and implications for disease therapy. Biomolecules 2020, 10, 1673. [Google Scholar] [CrossRef] [PubMed]
- Senger, D.R.; Galli, S.J.; Dvorak, A.M.; Perruzzi, C.A.; Harvey, V.S.; Dvorak, H.F. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science 1983, 219, 983–985. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.I.; Zachary, I. The vascular endothelial growth factor (VEGF) family: Angiogenic factors in health and disease. Genome Biol. 2005, 6, 209. [Google Scholar] [CrossRef]
- Olsson, A.K.; Dimberg, A.; Kreuger, J.; Claesson-Welsh, L. VEGF receptor signalling—In control of vascular function. Nat. Rev. Mol. Cell Biol. 2006, 7, 359–371. [Google Scholar] [CrossRef]
- Guo, X.; Yi, H.; Li, T.C.; Wang, Y.; Wang, H.; Chen, X. Role of vascular endothelial growth factor (VEGF) in human embryo implantation: Clinical implications. Biomolecules 2021, 11, 253. [Google Scholar] [CrossRef]
- Pandita, S.; Maurya, D.; Ramachandran, V.; Verma, J.; Kohli, S.; Saxena, R.; Verma, I.C. Vascular endothelial growth factor (VEGF) gene promoter polymorphisms and disease progression in north indian cohort with autosomal dominant polycystic kidney disease. Int. J. Mol. Cell. Med. 2017, 6, 164–173. [Google Scholar] [CrossRef]
- Turienzo, A.; Lledó, B.; Ortiz, J.A.; Morales, R.; Sanz, J.; Llácer, J.; Bernabeu, R. Prevalence of candidate single nucleotide polymorphisms on p53, IL-11, IL-10, VEGF and APOE in patients with repeated implantation failure (RIF) and pregnancy loss (RPL). Hum. Fertil. 2020, 23, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Vagnini, L.D.; Nascimento, A.M.; Canas, M.C.; Renzi, A.; Oliveira-Pelegrin, G.R.; Petersen, C.G.; Mauri, A.L.; Oliveira, J.B.; Baruffi, R.L.; Cavagna, M.; et al. The relationship between vascular endothelial growth factor 1154G/A polymorphism and recurrent implantation failure. Med. Princ. Pract. 2015, 24, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.W.; Kim, J.O.; Rah, H.; Kim, J.H.; Kim, Y.R.; Lee, Y.; Lee, W.S.; Kim, N.K. Genetic variants of vascular endothelial growth factor are associated with recurrent implantation failure in Korean women. Reprod. Biomed. Online 2016, 32, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.; Jeyendran, R.S.; Coulam, C.B. Vascular endothelial growth factor gene polymorphism and implantation failure. Reprod. Biomed. Online 2008, 16, 720–723. [Google Scholar] [CrossRef]
- Baston-Buest, D.M.; Porn, A.C.; Schanz, A.; Kruessel, J.S.; Janni, W.; Hess, A.P. Expression of the vascular endothelial growth factor receptor neuropilin-1 at the human embryo-maternal interface. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 154, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Demir, R. Expression of VEGF receptors VEFGR-1 and VEGFR-2, angiopoietin receptors Tie-1 and Tie-2 in chorionic villi tree during early pregnancy. Folia Histochem. Cytobiol. 2009, 47, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.J.; Maynard, S.E.; Qian, C.; Lim, K.H.; England, L.J.; Yu, K.F.; Schisterman, E.F.; Thadhani, R.; Sachs, B.P.; Epstein, F.H.; et al. Circulating angiogenic factors and the risk of preeclampsia. N. Engl. J. Med. 2004, 350, 672–683. [Google Scholar] [CrossRef]
- Amin-Beidokhti, M.; Gholami, M.; Abedin-Do, A.; Pirjani, R.; Sadeghi, H.; Karamoddin, F.; Yassaee, V.R.; Mirfakhraie, R. An intron variant in the FLT1 gene increases the risk of preeclampsia in Iranian women. Clin. Exp. Hypertens. 2019, 41, 697–701. [Google Scholar] [CrossRef]
- Amosco, M.D.; Villar, V.A.; Naniong, J.M.; David-Bustamante, L.M.; Jose, P.A.; Palmes-Saloma, C.P. VEGF-A and VEGFR1 SNPs associate with preeclampsia in a Philippine population. Clin. Exp. Hypertens. 2016, 38, 578–585. [Google Scholar] [CrossRef]
- Powers, R.W.; Jeyabalan, A.; Clifton, R.G.; Van Dorsten, P.; Hauth, J.C.; Klebanoff, M.A.; Lindheimer, M.D.; Sibai, B.; Landon, M.; Miodovnik, M. Soluble fms-Like tyrosine kinase 1 (sFlt1), endoglin and placental growth factor (PlGF) in preeclampsia among high risk pregnancies. PLoS ONE 2010, 5, e13263. [Google Scholar] [CrossRef]
- Wujcicka, W.I.; Kacerovsky, M.; Krekora, M.; Kaczmarek, P.; Grzesiak, M. Single nucleotide polymorphisms from CSF2, FLT1, TFPI and TLR9 genes are associated with prelabor rupture of membranes. Genes 2021, 12, 1725. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bove, A.M.; Simone, G.; Ma, B. Molecular bases of VEGFR-2-mediated physiological function and pathological role. Front. Cell Dev. Biol. 2020, 8, 599281. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Tan, C.; Ding, J.; Wang, J.; Ma’ayan, A.; Gouon-Evans, V. Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications. Stem Cell Res. 2018, 30, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ge, H.; Peng, L.; Wang, B. A meta-analysis of the relationship between VEGFR2 polymorphisms and atherosclerotic cardiovascular diseases. Clin. Cardiol. 2019, 42, 860–865. [Google Scholar] [CrossRef]
- Miramontes-González, J.P.; Usategui-Martín, R.; Pérez, I.L.; Alonso, R.; Muñiz-Grijalvo, O.; Díaz-Díaz, J.L.; Zambón, D.; Jiménez, F.F.; Martín-Vallejo, J.; Rodríguez Gude, A.E.; et al. VEGFR2 and OPG genes modify the risk of subclinical coronary atherosclerosis in patients with familial hypercholesterolemia. Atherosclerosis 2019, 285, 17–22. [Google Scholar] [CrossRef]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef]
- Cox, C.M.; Poole, T.J. Angioblast differentiation is influenced by the local environment: FGF-2 induces angioblasts and patterns vessel formation in the quail embryo. Dev. Dyn. 2000, 218, 371–382. [Google Scholar] [CrossRef]
- Poole, T.J.; Finkelstein, E.B.; Cox, C.M. The role of FGF and VEGF in angioblast induction and migration during vascular development. Dev. Dyn. 2001, 220, 1–17. [Google Scholar] [CrossRef]
- Beránek, M.; Tschöplová, S.; Kanková, K.; Kuhrová, V.; Vácha, J. Genetic variation in the promoter region of the basic fibroblast growth factor gene. Hum. Immunol. 2003, 64, 374–377. [Google Scholar] [CrossRef]
- SNP Database (dbSNP) of the National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/snp/ (accessed on 22 February 2022).
- Koperwas, M.; Głowacka, M. Problem niepłodności wśród kobiet i mężczyzn—Epidemiologia, czynniki ryzyka i świadomość społeczna. Asp. Zdrowia Chor. 2017, 2, 31–49. [Google Scholar]
- Kumar, K.V.A.; Kavitha, S.; Sreekanth, K.S. Regulatory proteins in placental angiogenesis. Biomedicine 2021, 41, 694–700. [Google Scholar] [CrossRef]
- Shim, S.H.; Kim, J.O.; Jeon, Y.J.; An, H.J.; Lee, H.A.; Kim, J.H.; Ahn, E.H.; Lee, W.S.; Kim, N.K. Association between vascular endothelial growth factor promoter polymorphisms and the risk of recurrent implantation failure. Exp. Ther. Med. 2018, 15, 2109–2119. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Hu, L.; Xie, H.; Ma, W.; Quan, S. Polymorphisms of vascular endothelial growth factor and recurrent implantation failure: A systematic review and meta-analysis. Arch. Gynecol. Obstet. 2021, 304, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Boudjenah, R.; Molina-Gomes, D.; Wainer, R.; de Mazancourt, P.; Selva, J.; Vialard, F. The vascular endothelial growth factor (VEGF) +405 G/C polymorphism and its relationship with recurrent implantation failure in women in an IVF programme with ICSI. J. Assist. Reprod. Genet. 2012, 29, 1415–1420. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, S.K.; Morrison, A.C.; Andrela, C.M.; Elovitz, M.A. Allelic variations in angiogenic pathway genes are associated with preeclampsia. Am. J. Obstet. Gynecol. 2010, 202, 445.e1–445.e11. [Google Scholar] [CrossRef]
- Keskin, U.; Ulubay, M.; Dede, M.; Ozgurtas, T.; Koçyiğit, Y.K.; Aydin, F.N.; Ergün, A. The relationship between the VEGF/sVEGFR-1 ratio and threatened abortion. Arch. Gynecol. Obstet. 2015, 291, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Wei, Z.; Li, O.; Huang, R.; Qin, J.; Chen, H.; Fan, X.; Chen, Z.J. An increase in vascular endothelial growth factor (VEGF) and VEGF soluble receptor-1 (sFlt-1) are associated with early recurrent spontaneous abortion. PLoS ONE 2013, 8, e75759. [Google Scholar] [CrossRef]
- Bansal, R.; Ford, B.; Bhaskaran, S.; Thum, M.; Bansal, A. Elevated levels of serum vascular endothelial growth factor-A are not related to NK cell parameters in recurrent IVF failure. J. Reprod. Infertil. 2017, 18, 280–287. [Google Scholar]
- Rah, H.; Jeon, Y.J.; Lee, B.E.; Choi, D.H.; Yoon, T.K.; Lee, W.S.; Kim, N.K. Association of kinase insert domain-containing receptor (KDR) gene polymorphisms with idiopathic recurrent spontaneous abortion in Korean women. Fertil. Steril. 2013, 99, 753–760. [Google Scholar] [CrossRef]
- Freiin von Hövel, F.; Kefalakes, E.; Grothe, C. What can we learn from FGF-2 isoform-specific mouse mutants? Differential insights into FGF-2 physiology in vivo. Int. J. Mol. Sci. 2020, 22, 390. [Google Scholar] [CrossRef]
- Lin, H.J.; Wan, L.; Tsai, Y.; Liu, S.C.; Chen, W.C.; Tsai, S.W.; Tsai, F.J. Sclera-related gene polymorphisms in high myopia. Mol. Vis. 2009, 15, 1655–1663. [Google Scholar] [PubMed]
- Priščáková, P.; Minárik, G.; Repiská, V. Candidate gene studies of diabetic retinopathy in human. Mol. Biol. Rep. 2016, 43, 1327–1345. [Google Scholar] [CrossRef] [PubMed]
- Wróbel, T.; Butrym, A.; Łacina, P.; Rybka, J.; Gębura, K.; Mazur, G.; Bogunia-Kubik, K. bFGF polymorphism is associated with disease progression and response to chemotherapy in multiple myeloma patients. Anticancer. Res. 2017, 37, 1799–1803. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, T.; Wu, T.; Hetmanski, J.B.; Ruczinski, I.; Schwender, H.; Liang, K.Y.; Murray, T.; Fallin, M.D.; Redett, R.J.; et al. The FGF and FGFR gene family and risk of cleft lip with or without cleft palate. Cleft Palate Craniofacial J. 2013, 50, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Osadnik, T.; Strzelczyk, J.K.; Reguła, R.; Bujak, K.; Fronczek, M.; Gonera, M.; Gawlita, M.; Wasilewski, J.; Lekston, A.; Kurek, A.; et al. The relationships between polymorphisms in genes encoding the growth factors TGF-β1, PDGFB, EGF, bFGF and VEGF-A and the restenosis process in patients with stable coronary artery disease treated with bare metal stent. PLoS ONE 2016, 11, e0150500. [Google Scholar] [CrossRef]
- Weizman, N.F.; Defer, M.K.; Montbriand, J.; Pasquale, J.M.; Silver, A.; Librach, C.L. Does body mass index impact assisted reproductive technology treatment outcomes in gestational carriers. Reprod. Biol. Endocrinol. 2020, 18, 35. [Google Scholar] [CrossRef]
- Paik, J.H.; Kollipara, R.; Chu, G.; Ji, H.; Xiao, Y.; Ding, Z.; Miao, L.; Tothova, Z.; Horner, J.W.; Carrasco, D.R.; et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 2007, 128, 309–323. [Google Scholar] [CrossRef]
- Potente, M.; Urbich, C.; Sasaki, K.; Hofmann, W.K.; Heeschen, C.; Aicher, A.; Kollipara, R.; DePinho, R.A.; Zeiher, A.M.; Dimmeler, S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Investig. 2005, 115, 2382–2392. [Google Scholar] [CrossRef]
- Li-Villarreal, N.; Wong, R.L.Y.; Garcia, M.D.; Udan, R.S.; Poché, R.A.; Rasmussen, T.L.; Rhyner, A.M.; Wythe, J.D.; Dickinson, M.E. FOXO1 represses sprouty 2 and sprouty 4 expression to promote arterial specification and vascular remodeling in the mouse yolk sac. Development 2022, 149, dev200131. [Google Scholar] [CrossRef]
- Li, X.; Gao, Y.; Li, J.; Zhang, K.; Han, J.; Li, W.; Hao, Q.; Zhang, W.; Wang, S.; Zeng, C.; et al. FOXP3 inhibits angiogenesis by downregulating VEGF in breast cancer. Cell Death Dis. 2018, 3, 744. [Google Scholar] [CrossRef]
- Lin, Y.; Qi, J.; Sun, Y. Platelet-rich plasma as a potential new strategy in the endometrium treatment in assisted reproductive technology. Front. Endocrinol. 2021, 12, 707584. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lu, H.; Huo, Z.; Ma, Z.; Dang, J.; Dang, W.; Pan, L.; Chen, J.; Zhong, H. MicroRNA-16 inhibits fetomaternal angiogenesis and causes recurrent spontaneous abortion by targeting vascular endothelial growth factor. Sci. Rep. 2016, 6, 35536. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lv, W.; Zhu, Y. Comprehensive Analysis of circRNA-Mediated ceRNA Regulatory Networks in relation to Recurrent Implantation Failure. Evid. Based Complement. Alternat. Med. 2022, 2022, 8314838. [Google Scholar] [CrossRef] [PubMed]
- Gullo, G.; Carlomagno, G.; Unfer, V.; D’Anna, R. Myo-inositol: From induction of ovulation to menopausal disorder management. Minerva Ginecol. 2015, 67, 485–486. [Google Scholar] [PubMed]
- Bezerra, E.M.S.; Lagana, A.S.; Bilotta, G.; Gullo, G.; Aragona, C.; Unfer, V. D-chiro-inositol Induces Ovulation in Non-Polycystic Ovary Syndrome (PCOS), Non-Insulin-Resistant Young Women, Likely by Modulating Aromatase Expression: A Report of 2 Cases. Am. J. Case Rep. 2021, 22, e932722. [Google Scholar] [CrossRef]
- Prapas, Y.; Petousis, S.; Panagiotidis, Y.; Gullo, G.; Kasapi, L.; Papadeothodorou, A.; Prapas, N. Injection of embryo culture supernatant to the endometrial cavity does not affect outcomes in IVF/ICSI or oocyte donation cycles: A randomized clinical trial. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 162, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Goudakou, M.; Kalogeraki, A.; Matalliotakis, I.; Panagiotidis, Y.; Gullo, G.; Prapas, Y. Cryptic sperm defects may be the cause for total fertilization failure in oocyte donor cycles. Reprod. Biomed. Online 2012, 24, 148–152. [Google Scholar] [CrossRef]
- Medenica, S.; Zivanovic, D.; Batkoska, L.; Marinelli, S.; Basile, G.; Perino, A.; Cucinella, G.; Gullo, G.; Zaami, S. The Future Is Coming: Artificial Intelligence in the Treatment of Infertility Could Improve Assisted Reproduction Outcomes-The Value of Regulatory Frameworks. Diagnostics 2022, 12, 2979. [Google Scholar] [CrossRef]
- Medenica, S.; Abazovic, D.; Ljubić, A.; Vukovic, J.; Begovic, A.; Cucinella, G.; Zaami, S.; Gullo, G. The Role of Cell and Gene Therapies in the Treatment of Infertility in Patients with Thyroid Autoimmunity. Int. J. Endocrinol. 2022, 2022, 4842316. [Google Scholar] [CrossRef]
- Burgio, S.; Polizzi, C.; Buzzaccarini, G.; Lagana, A.S.; Gullo, G.; Perricone, G.; Perino, A.; Cucinella, G.; Alesi, M. Psychological variables in medically assisted reproduction: A systematic review. Menopause Rev. 2022, 21, 47–63. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Kang, S.; Wang, N.; Zhou, R.M.; Duan, Y.N.; Sun, D.L.; Qin, J.J.; Zhao, W.; Zhao, L. Association of vascular endothelial growth factor gene polymorphisms with susceptibility to epithelial ovarian cancer. Int. J. Gynecol. Cancer 2010, 20, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, Y.; Zhang, W.; Yu, H.; Lou, K.; Zhang, Y.; Qin, Q.; Zhao, B.; Yang, Y.; Hui, R. Polymorphisms of KDR gene are associated with coronary heart disease. J. Am. Coll. Cardiol. 2007, 50, 760–767. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 22 February 2022).
- Gonzalez, J.R.; Moreno, V.; SNPassoc: SNPs-Based Whole Genome Association Studies. R Package Version 2.0-2. 2020. Available online: https://CRAN.R-project.org/package=SNPassoc (accessed on 22 February 2022).
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2004, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Cherny, S.S.; Sham, P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003, 19, 149–150. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Wang, Y.; Lin, C.; Zhang, D.; Chen, J.; Ouyang, L.; Wu, F.; Zhang, J.; Chen, L. Recent progress on vascular endothelial growth factor receptor inhibitors with dual targeting capabilities for tumor therapy. J. Hematol. Oncol. 2022, 15, 89. [Google Scholar] [CrossRef]

| Characteristic | Cases n = 247 | Controls n = 120 | p |
|---|---|---|---|
| Age (years), mean ± SD | 33.11 ± 3.51 | 32.50 ± 3.60 | 0.123 |
| min-max | 24–39 | 23–40 | |
| BMI (kg/m2), mean ± SD | 23.36 ± 4.17 | 20.71 ± 1.79 | <0.001 |
| Normal or Underweight (BMI ≤ 25), n (%) | 183 (74.1) | 120 (100.0) | <0.001 * |
| Overweight or Obese (BMI > 25), n (%) | 64 (25.9) | 0 (0.0) | |
| AMH (pmol/L), median (IQR) | 21.00 (11.17–30.79) | — | — |
| Indications for ART, n (%) | |||
| Idiopathic infertility | 119 (48.2) | — | — |
| Male factor | 89 (36.0) | — | |
| Female factor (Oviduct + Ovulatory) | 39 (15.8) | — | |
| Number of failed transfers, n (%) | |||
| 1–2 | 175 (70.9) | — | — |
| RIF (≥3) | 72 (29.1) | — | |
| Number of pregnancies, n (%) | 188 (100.0) | 293 (100.0) | <0.001 * |
| 0 | 95 (38.5) | 0 (0.0) | |
| 1 | 120 (48.6) | 0 (0.0) | |
| 2 | 28 (11.3) | 81 (67.5) | |
| ≥3 | 4 (1.6) | 39 (32.5) | |
| Embryo transfers, n (%) | — | — | |
| Fresh Embryo Transfer | 55 (29.3) | ||
| Frozen Embryo Transfer | 133 (70.7) |
| SNP | Group | Major Allele n (%) | Minor Allele n (%) | HWE p Value | OR (95% CI) | Chi2 (df = 2) | Pearson’s p |
|---|---|---|---|---|---|---|---|
| VEGFA rs699947 C > A | Case | 247 (0.50) | 247 (0.50) | 0.375 | 0.860 (0.632–1.172) | 0.909 | 0.340 |
| Controls | 111 (0.46) | 129 (0.54) | 0.856 | ||||
| FLT1 rs722503 T > C | Case | 371 (0.75) | 123 (0.25) | 0.865 | 0.995 (0.696–1.420) | 0.001 | 0.976 |
| Controls | 180 (0.75) | 60 (0.25) | 0.809 | ||||
| KDR rs2071559 A > G | Case | 224 (0.45) | 270 (0.55) | 0.249 | 1.378 (1.011–1.877) | 4.131 | 0.042 |
| Controls | 128 (0.53) | 112 (0.47) | 0.278 | ||||
| KDR rs1870377 T > A | Case | 351 (0.71) | 143 (0.29) | 0.646 | 1.052 (0.747–1.482) | 0.084 | 0.772 |
| Controls | 173 (0.72) | 67 (0.28) | 1.000 | ||||
| FGF2 rs308395 C > G | Case | 427 (0.86) | 67 (0.14) | 0.096 | 1.142 (0.717–1.819) | 0.311 | 0.577 |
| Controls | 211 (0.88) | 29 (0.12) | 0.375 |
| SNP (rs ID)/Model | Genotypes | Controls n (%) | Case n (%) | Crude | Adjusted | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | AIC | AOR (95% CI) | p-Value | AIC | ||||
| VEGFA (rs699947) | |||||||||
| Codominant | CC | 25 (20.8) | 58 (23.5) | 1.00 | 0.585 | 468.8 | 1.00 | 0.696 | 417.9 |
| CA | 61 (50.8) | 131 (53.0) | 0.93 (0.53–1.62) | 0.97 (0.53–1.77) | |||||
| AA | 34 (28.3) | 58 (23.5) | 0.74 (0.39–1.38) | 0.78 (0.39–1.54) | |||||
| Dominant | CC vs. CA-AA | 95 (79.2) | 189 (76.5) | 0.86 (0.50–1.46) | 0.569 | 467.6 | 0.90 (0.51–1.60) | 0.727 | 416.5 |
| Recessive | CC-CA vs. AA | 86 (71.7) | 189 (76.5) | 0.78 (0.47–1.27) | 0.314 | 466.9 | 0.79 (0.47–1.35) | 0.397 | 415.9 |
| Overdominant | AA-CC vs. CA | 59 (49.2) | 116 (47.0) | 1.09 (0.71–1.69) | 0.692 | 467.7 | 1.11 (0.70–1.78) | 0.651 | 416.4 |
| log-Additive | 0,1,2 | 120 (32.7) | 247 (67.3) | 1.17 (0.85–1.61) | 0.328 | 466.9 | 1.14 (0.81–1.60) | 0.454 | 416.1 |
| FLT1 (rs722503) | |||||||||
| Codominant | TT | 68 (56.7) | 140 (56.7) | 1.00 | 0.998 | 469.9 | 1.00 | 0.928 | 418.5 |
| TC | 44 (36.7) | 91 (36.8) | 1.00 (0.63–1.59) | 1.10 (0.67–1.80) | |||||
| CC | 8 (6.7) | 16 (6.5) | 0.97 (0.40–2.38) | 1.11 (0.43–2.88) | |||||
| Dominant | TT vs. TC-CC | 52 (43.3) | 107 (43.3) | 1.00 (0.64–1.55) | 0.998 | 467.9 | 1.10 (0.68–1.76) | 0.700 | 416.5 |
| Recessive | TT-TC vs. CC | 112 (93.3) | 231 (93.5) | 0.97 (0.40–2.33) | 0.945 | 467.9 | 1.07 (0.42–2.71) | 0.886 | 416.6 |
| Overdominant | TT-CC vs. TC | 76 (63.3) | 156 (63.2) | 1.01 (0.64–1.58) | 0.974 | 467.9 | 1.08 (0.67–1.76) | 0.749 | 416.5 |
| log-Additive | 0,1,2 | 120 (32.7) | 247 (67.3) | 0.99 (0.70–1.42) | 0.977 | 467.9 | 1.07 (0.73–1.57) | 0.714 | 416.5 |
| KDR (rs2071559) | |||||||||
| Codominant | AA | 31 (25.8) | 46 (18.6) | 1.00 | 0.102 | 465.3 | 1.00 | 0.042 | 412.3 |
| AG | 66 (55.0) | 132 (53.4) | 1.35 (0.78–2,32) | 1.41 (0.78–2.55) | |||||
| GG | 23 (19.2) | 69 (27.9) | 2.02 (1.05–3.90) | 2.42 (1.19–4.90) | |||||
| Dominant | AA vs. AG-GG | 89 (74.2) | 201 (81.4) | 1.52 (0.91–2.56) | 0.116 | 465.4 | 1.66 (0.95–2.93) | 0.078 | 413.5 |
| Recessive | AA-AG vs. GG | 97 (80.8) | 178 (72.1) | 1.63 (0.96–2.79) | 0.065 | 464.5 | 1.89 (1.07–3.34) | 0.025 | 411.6 |
| Overdominant | AA-GG vs. AG | 54 (45.0) | 115 (46.6) | 0.94 (0.61–1.46) | 0.779 | 467.8 | 0.89 (0.56–1.42) | 0.625 | 416.4 |
| log-Additive | 0,1,2 | 120 (32.7) | 247 (67.3) | 0.70 (0.51–0.98) | 0.034 | 463.4 | 0.64 (0.45–0.91) | 0.013 | 410.4 |
| KDR (rs1870377) | |||||||||
| Codominant | TT | 62 (51.7) | 123 (49.8) | 1.00 | 0.945 | 469.8 | 1.00 | 0.724 | 418.0 |
| TA | 49 (40.8) | 105 (42.5) | 1.08 (0.68–1.70) | 1.21 (0.74–1.99) | |||||
| AA | 9 (7.5) | 19 (7.7) | 1.06 (0.45–2.49) | 1.24 (0.50–3.06) | |||||
| Dominant | TT vs. TA-AA | 58 (48.3) | 124 (50.2) | 1.08 (0.70–1.67) | 0.737 | 467.8 | 1.21 (0.76–1.95) | 0.422 | 416.0 |
| Recessive | AA-AT vs.TT | 111 (92.5) | 228 (92.3) | 1.03 (0.45–2.35) | 0.948 | 467.9 | 1.13 (0.47–2.71) | 0.784 | 416.5 |
| Overdominant | AA-TT vs. AT | 71 (59.2) | 142 (57.5) | 1.07 (0.69–1.67) | 0.760 | 467.8 | 1.17 (0.73–1.89) | 0.511 | 416.2 |
| log-Additive | 0,1,2 | 120 (32.7) | 247 (67.3) | 1.05 (0.74–1.49) | 0.769 | 467.8 | 1.15 (0.79–1.68) | 0.452 | 416.1 |
| FGF2 (rs308395) | |||||||||
| Codominant | CC | 94 (78.3) | 188 (76.1) | 1.00 | 0.864 | 469.6 | 1.00 | 0.513 | 417.3 |
| CG | 23 (19.2) | 51 (20.6) | 1.11 (0.64–1.92) | 1.36 (0.75–2.46) | |||||
| GG | 3 (2.5) | 8 (3.2) | 1.33 (0.35–5.14) | 1.54 (0.37–6.47) | |||||
| Dominant | CC vs. CG-GG | 26 (21.7) | 59 (23.9) | 1.13 (0.67–1.92) | 0.635 | 467.7 | 1.38 (0.79–2.43) | 0.253 | 415.3 |
| Recessive | CC-CG vs.GG | 117 (97.5) | 239 (96.8) | 1.31 (0.34–5.01) | 0.693 | 467.7 | 1.44 (0.35–6.02) | 0.608 | 416.4 |
| Overdominant | CC-GG vs. CG | 97 (80.8) | 196 (79.4) | 1.10 (0.63–1.90) | 0.739 | 467.8 | 1.34 (0.74–2.42) | 0.325 | 415.7 |
| log-Additive | 0,1,2 | 120 (32.7) | 247 (67.3) | 1.13 (0.72–1.75) | 0.595 | 467.6 | 1.31 (0.82–2.12) | 0.256 | 415.3 |
| Characteristic | NO-RIF n = 175 | RIF n = 72 | p |
|---|---|---|---|
| Age (years), mean ± SD | 32.7 ± 3.4 | 34.1 ± 3.7 | 0.005 |
| min–max | 24–39 | 25–39 | |
| BMI (kg/m2), mean ± SD | 23.6 ± 4.3 | 22.7 ± 3.8 | 0.130 |
| Normal or Underweight (BMI ≤ 25), n (%) | 127 (72.6) | 56 (77.8) | 0.491 |
| Overweight or Obese (BMI > 25), n (%) | 48 (27.4) | 16 (22.2) | |
| AMH (pmol/L), median (IQR) | 20.60 (11.4–29.8) | 22.80 (10.7–30.9) | 0.939 |
| Indications for ART, n (%) | |||
| Idiopathic infertility | 84 (48.0) | 35 (48.6%) | 0.763 |
| Male factor | 65 (37.1) | 24 (33.3%) | |
| Female factor (Oviduct + Ovulatory) | 26 (14.9) | 13 (18.1%) | |
| Number of pregnancies, n (%) | 147 (100.0) | 41 (100.0) | |
| 0 | 53 (30.3) | 42 (58.3) | <0.001 |
| 1 | 99 (56.6) | 21 (29.2) | |
| 2 | 21 (12.0) | 7 (9.7) | |
| ≥3 | 2 (1.1) | 2 (2.8) | |
| Embryo transfers, n (%) | 0.699 | ||
| Fresh Embryo Transfer | 44 (29.9) | 11 (26.8) | |
| Frozen Embryo Transfer | 103 (70.1) | 30 (73.2) |
| Characteristic | Fresh Embryo Transfers n = 55 | Frozen Embryo Transfers n = 133 | p |
|---|---|---|---|
| Pregnancy outcome, n (%) | 0.532 | ||
| Miscarriages | 8 (14.5) | 15 (11.3) | |
| Live births | 47 (85.5) | 118 (88.7) | |
| Mode of delivery, n (%) | 0.015 | ||
| Vaginal | 26 (55.3) | 41 (34.7) | |
| C-section | 21 (44.7) | 77 (65.3) | |
| Newborn birthweight (g), mean ± SD | 3315.1 ± 512.6 | 3458.1 ± 412.8 | 0.094 |
| Low (<2500 g), n (%) | 2 (4.3) | 1 (0.8) | 0.435 |
| Normal (2500–4000 g), n (%) | 41 (87.2) | 110 (93.2) | |
| Macrosomic (>4000 g), n (%) | 4 (8.5) | 7 (6.0) | |
| Placenta weight (g), mean ± SD | 605.5 ± 97.5 | 653.5 ± 355.0 | 0.257 |
| Apgar score at 1 min, median (IQR) | 10.0 (10.0–10.0) | 10.0 (10.0–10.0) | 0.269 |
| Apgar score at 5 min, median (IQR) | 10.0 (10.0–10.0) | 10.0 (10.0–10.0) | 0.722 |
| Gestational age (weeks), mean ± SD | 38.2 ± 1.2 | 37.3 ± 5.8 | 0.117 |
| Gene (rs ID) | Genotypes | NO-RIF n = 175 | RIF n = 72 | p Crude | AOR |
|---|---|---|---|---|---|
| VEGFA (rs699947) | CC | 48 (27.4%) | 10 (13.9%) | 0.070 | 0.052 |
| CA | 89 (50.9%) | 42 (58.3%) | |||
| AA | 38 (21.7%) | 20 (27.8%) | |||
| FLT1 (rs722503) | TT | 101 (57.7%) | 39 (54.2%) | 0.751 | 0.722 |
| TC | 62 (35.4%) | 29 (40.3%) | |||
| CC | 12 (6.9%) | 4 (5.6%) | |||
| KDR (rs2071559) | AA | 30 (17.1%) | 16 (22.2%) | 0.431 | 0.609 |
| AG | 98 (56.0%) | 34 (47.2%) | |||
| GG | 47 (26.9%) | 22 (30.6%) | |||
| KDR (rs1870377) | TT | 87 (49.7%) | 36 (50.0%) | 0.415 | 0.451 |
| TA | 77 (44.0%) | 28 (38.9%) | |||
| AA | 11 (6.3%) | 8 (11.1%) | |||
| FGF2 (rs308395) | CC | 131 (74.9%) | 57 (79.2%) | 0.770 | 0.790 |
| CG | 38 (21.7%) | 13 (18.1%) | |||
| GG | 6 (3.4%) | 2 (2.8%) |
| Gene | rs No. | Location * | Base Change | MAF ** |
|---|---|---|---|---|
| VEGFA | rs699947 | chr6:43768652 | C > A | A = 0.4950 |
| FLT1 | rs722503 | chr13:28422915 | T > C | C = 0.2435 |
| KDR | rs2071559 | chr4:55126199 | A > G | G = 0.4851 |
| KDR | rs1870377 | chr4:55106807 | T > A | A = 0.2346 |
| FGF2 | rs308395 | chr4:122825787 | C > G | G = 0.1630 |
| Gene | rs No | Primer Sequences | Restriction Enzyme | Fragment Lenght (bp) |
|---|---|---|---|---|
| VEGFA | rs699947 | 5′- GGATGGGGCTGACTAGGTAAGC-3′ 5′- AGCCCCCTTTTCCTCCAAC-3′ | BglII | C 324 A 202, 122 |
| FLT1 | rs722503 | 5′- TCCGCCTGCATTTTGAACAACTAAGTAG-3′ 5′- GGTCTCCTTGGTATTCAAGCACACGTAA-3′ | AvaII | T 368 C 199, 169 |
| KDR | rs2071559 | 5′- CAAACTTTCACTAGGGCTCTTCGT-3′ 5′- AGCCACAAGGGAGAAGCGGATA-3′ | BsmI | A 290 G 174, 116 |
| rs1870377 | 5′- GCCTCACATATTATTGTACCATCC-3′ 5′- CCTCCTGTATCCTGAATGAATCT-3′ | AluI | T 213, 191 A 404 | |
| FGF2 | rs308395 | 5′- TGAGTTATCCGATGTCTGAAATG-3′ 5′- TAACTTGAATTAGACGACGCAGA-3′ | BseNI (BsrI) | C 437 G 369, 69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mrozikiewicz, A.E.; Kurzawińska, G.; Ożarowski, M.; Walczak, M.; Ożegowska, K.; Jędrzejczak, P. Polymorphic Variants of Genes Encoding Angiogenesis-Related Factors in Infertile Women with Recurrent Implantation Failure. Int. J. Mol. Sci. 2023, 24, 4267. https://doi.org/10.3390/ijms24054267
Mrozikiewicz AE, Kurzawińska G, Ożarowski M, Walczak M, Ożegowska K, Jędrzejczak P. Polymorphic Variants of Genes Encoding Angiogenesis-Related Factors in Infertile Women with Recurrent Implantation Failure. International Journal of Molecular Sciences. 2023; 24(5):4267. https://doi.org/10.3390/ijms24054267
Chicago/Turabian StyleMrozikiewicz, Aleksandra E., Grażyna Kurzawińska, Marcin Ożarowski, Michał Walczak, Katarzyna Ożegowska, and Piotr Jędrzejczak. 2023. "Polymorphic Variants of Genes Encoding Angiogenesis-Related Factors in Infertile Women with Recurrent Implantation Failure" International Journal of Molecular Sciences 24, no. 5: 4267. https://doi.org/10.3390/ijms24054267
APA StyleMrozikiewicz, A. E., Kurzawińska, G., Ożarowski, M., Walczak, M., Ożegowska, K., & Jędrzejczak, P. (2023). Polymorphic Variants of Genes Encoding Angiogenesis-Related Factors in Infertile Women with Recurrent Implantation Failure. International Journal of Molecular Sciences, 24(5), 4267. https://doi.org/10.3390/ijms24054267







