MTHFR c.665C>T and c.1298A>C Polymorphisms in Tailoring Personalized Anti-TNF-α Therapy for Rheumatoid Arthritis
Abstract
1. Introduction
2. Results
2.1. Characteristics of Study Subjects and Response to Therapy
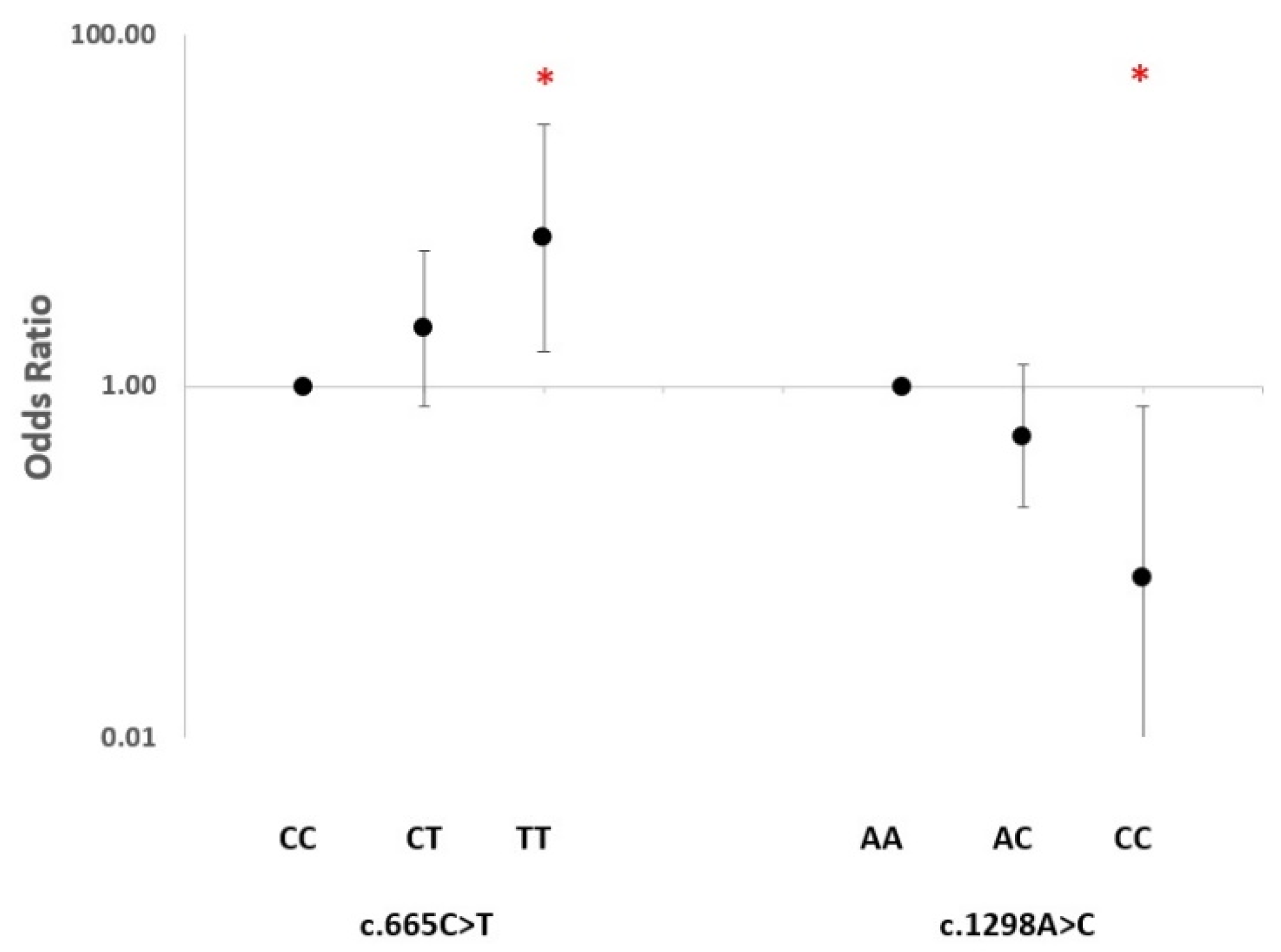
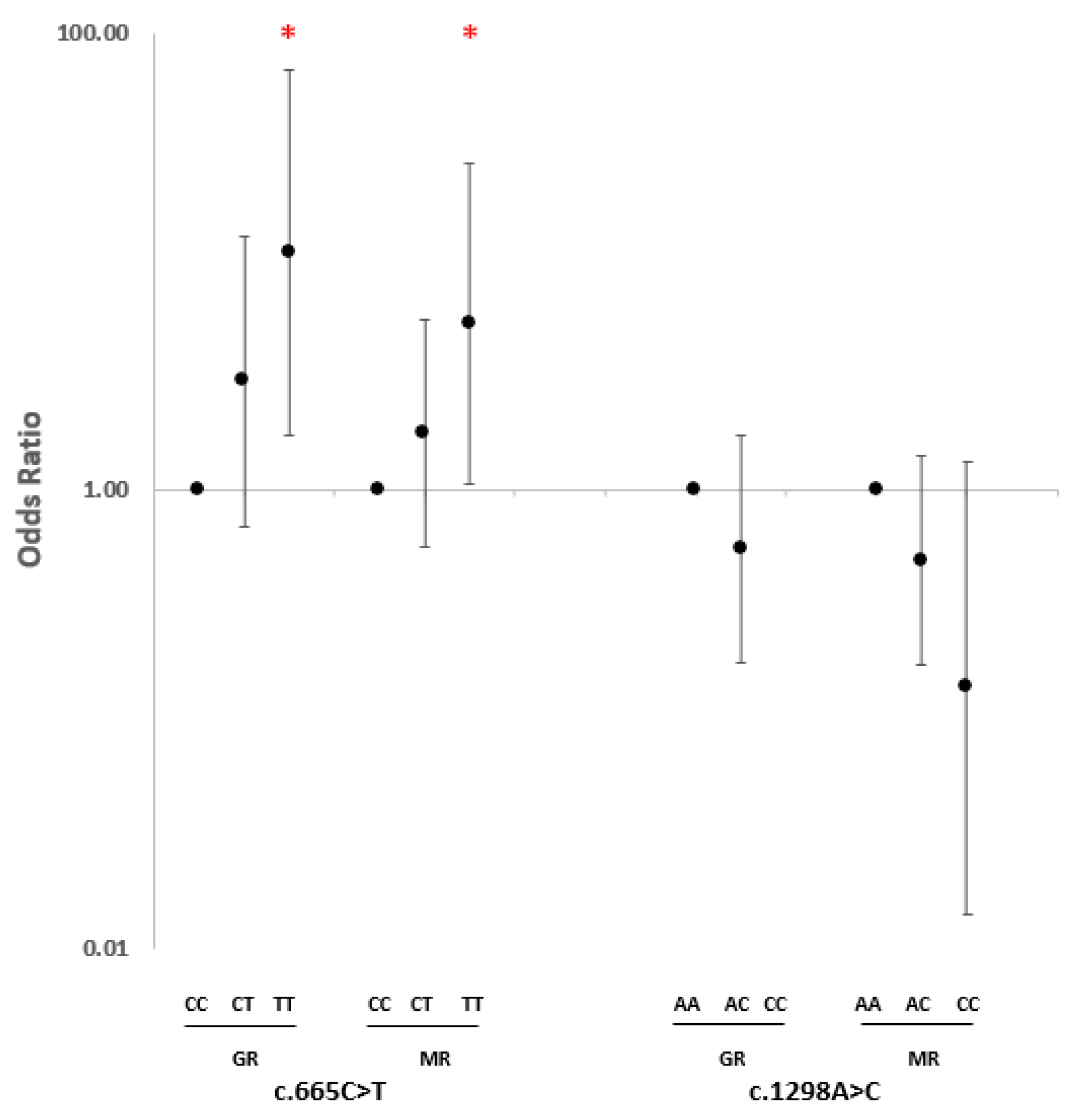
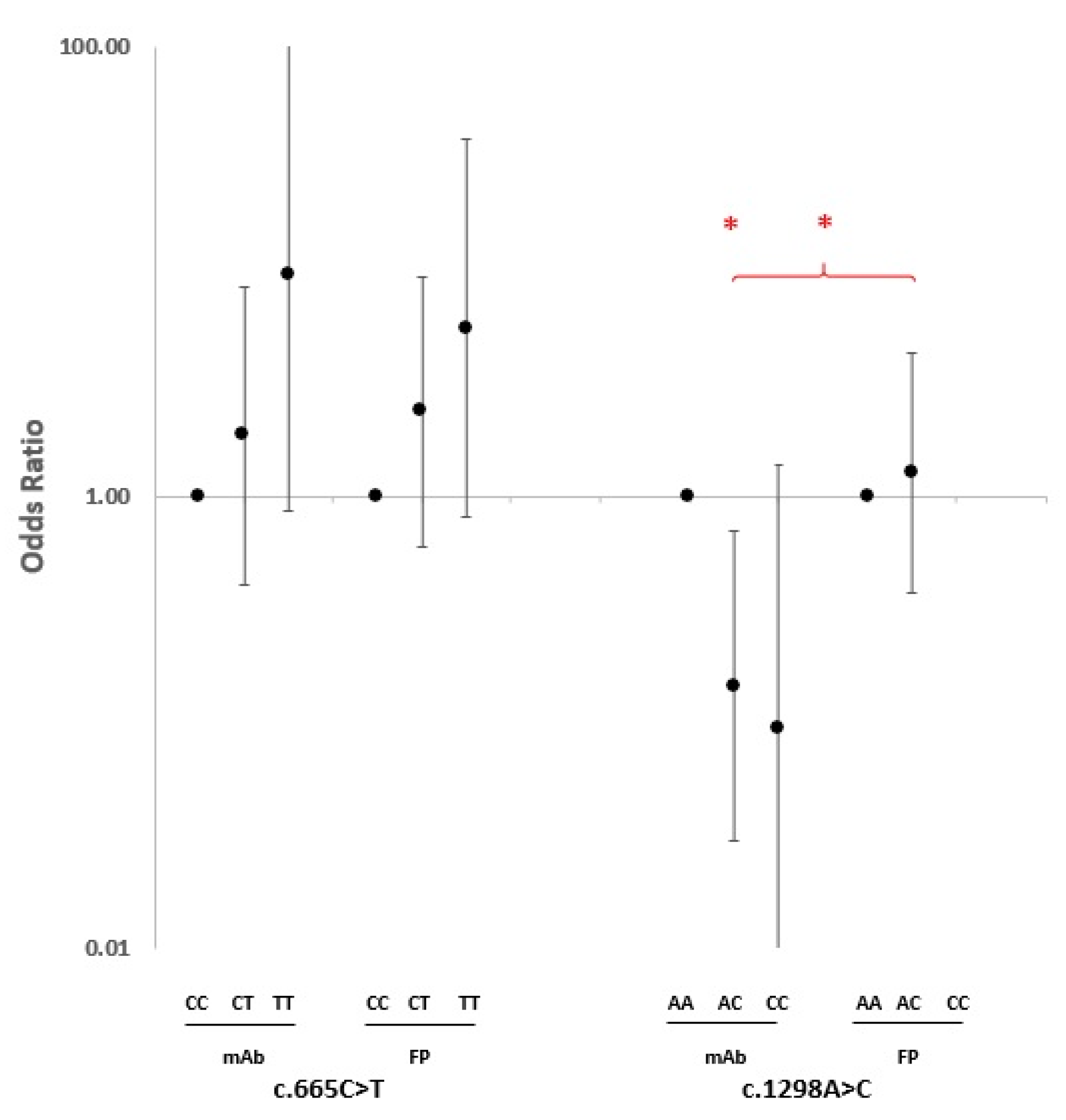
2.2. SNP Association Analysis
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. DNA Extraction and Genotyping
4.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cush, J.J. Rheumatoid Arthritis: Early Diagnosis and Treatment. Med. Clin. N. Am. 2021, 105, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-J.; Anzaghe, M.; Schülke, S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells 2020, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; Barton, A.; Burmester, G.R.; Emery, P.; Firestein, G.S.; Kavanaugh, A.; McInnes, I.B.; Solomon, D.H.; Strand, V.; et al. Rheumatoid Arthritis. Nat. Rev. Dis. Prim. 2018, 4, 1–23. [Google Scholar] [CrossRef]
- Brzustewicz, E.; Henc, I.; Daca, A.; Szarecka, M.; Sochocka-Bykowska, M.; Witkowski, J.; Bryl, E. Autoantibodies, C-Reactive Protein, Erythrocyte Sedimentation Rate and Serum Cytokine Profiling in Monitoring of Early Treatment. Cent. Eur. J. Immunol. 2017, 42, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid Arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- Littlejohn, E.A.; Monrad, S.U. Early Diagnosis and Treatment of Rheumatoid Arthritis. Prim. Care 2018, 45, 237–255. [Google Scholar] [CrossRef]
- Aletaha, D.; Smolen, J.S. Diagnosis and Management of Rheumatoid Arthritis: A Review. JAMA 2018, 320, 1360–1372. [Google Scholar] [CrossRef]
- Smolen, J.S.; Aletaha, D.; Bijlsma, J.W.J.; Breedveld, F.C.; Boumpas, D.; Burmester, G.; Combe, B.; Cutolo, M.; de Wit, M.; Dougados, M.; et al. Treating Rheumatoid Arthritis to Target: Recommendations of an International Task Force. Ann. Rheum. Dis. 2010, 69, 631–637. [Google Scholar] [CrossRef]
- Felson, D.T.; Smolen, J.S.; Wells, G.; Zhang, B.; van Tuyl, L.H.D.; Funovits, J.; Aletaha, D.; Allaart, C.F.; Bathon, J.; Bombardieri, S.; et al. American College of Rheumatology/European League against Rheumatism Provisional Definition of Remission in Rheumatoid Arthritis for Clinical Trials. Ann. Rheum. Dis. 2011, 70, 404–413. [Google Scholar] [CrossRef]
- Smolen, J.S.; Landewé, R.; Breedveld, F.C.; Buch, M.; Burmester, G.; Dougados, M.; Emery, P.; Gaujoux-Viala, C.; Gossec, L.; Nam, J.; et al. EULAR Recommendations for the Management of Rheumatoid Arthritis with Synthetic and Biological Disease-Modifying Antirheumatic Drugs: 2013 Update. Ann. Rheum. Dis. 2014, 73, 492–509. [Google Scholar] [CrossRef]
- Inoue, K.; Yuasa, H. Molecular Basis for Pharmacokinetics and Pharmacodynamics of Methotrexate in Rheumatoid Arthritis Therapy. Drug Metab. Pharmacokinet. 2014, 29, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Kästner, P.; Flaxenberg, P.; Währisch, J.; Hanke, P.; Demary, W.; von Hinüber, U.; Rockwitz, K.; Heitz, W.; Pichlmeier, U.; et al. Comparison of the Clinical Efficacy and Safety of Subcutaneous versus Oral Administration of Methotrexate in Patients with Active Rheumatoid Arthritis: Results of a Six-Month, Multicenter, Randomized, Double-Blind, Controlled, Phase IV Trial. Arthritis Rheum. 2008, 58, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Klareskog, L.; van der Heijde, D.; de Jager, J.P.; Gough, A.; Kalden, J.; Malaise, M.; Martín Mola, E.; Pavelka, K.; Sany, J.; Settas, L.; et al. Therapeutic Effect of the Combination of Etanercept and Methotrexate Compared with Each Treatment Alone in Patients with Rheumatoid Arthritis: Double-Blind Randomised Controlled Trial. Lancet 2004, 363, 675–681. [Google Scholar] [CrossRef]
- Lima, A.; Bernardes, M.; Azevedo, R.; Medeiros, R.; Seabra, V. Pharmacogenomics of Methotrexate Membrane Transport Pathway: Can Clinical Response to Methotrexate in Rheumatoid Arthritis Be Predicted? Int. J. Mol. Sci. 2015, 16, 13760–13780. [Google Scholar] [CrossRef]
- Mori, S.; Hirose, J.; Yonemura, K. Contribution of HLA-DRB1*04 Alleles and Anti-Cyclic Citrullinated Antibodies to Development of Resistance to Disease-Modifying Antirheumatic Drugs in Early Rheumatoid Arthritis. Clin. Rheumatol. 2010, 29, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Weinblatt, M.E.; Kaplan, H.; Germain, B.F.; Block, S.; Solomon, S.D.; Merriman, R.C.; Wolfe, F.; Wall, B.; Anderson, L.; Gall, E. Methotrexate in Rheumatoid Arthritis. A Five-Year Prospective Multicenter Study. Arthritis Rheum. 1994, 37, 1492–1498. [Google Scholar] [CrossRef]
- Radner, H.; Aletaha, D. Anti-TNF in Rheumatoid Arthritis: An Overview. Wien. Med. Wochenschr. 2015, 165, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Maini, R.N. Lasker Clinical Medical Research Award. TNF Defined as a Therapeutic Target for Rheumatoid Arthritis and Other Autoimmune Diseases. Nat. Med. 2003, 9, 1245–1250. [Google Scholar] [CrossRef]
- Gerriets, V.; Goyal, A.; Khaddour, K. Tumor Necrosis Factor Inhibitors; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- van Schouwenburg, P.A.; Rispens, T.; Wolbink, G.J. Immunogenicity of Anti-TNF Biologic Therapies for Rheumatoid Arthritis. Nat. Rev. Rheumatol. 2013, 9, 164–172. [Google Scholar] [CrossRef]
- Umiċeviċ Mirkov, M.; Cui, J.; Vermeulen, S.H.; Stahl, E.A.; Toonen, E.J.M.; Makkinje, R.R.; Lee, A.T.; Huizinga, T.W.J.; Allaart, R.; Barton, A.; et al. Genome-Wide Association Analysis of Anti-TNF Drug Response in Patients with Rheumatoid Arthritis. Ann. Rheum. Dis. 2013, 72, 1375–1381. [Google Scholar] [CrossRef]
- Sánchez-Maldonado, J.M.; Cáliz, R.; López-Nevot, M.Á.; Cabrera-Serrano, A.J.; Moñiz-Díez, A.; Canhão, H.; Ter Horst, R.; Quartuccio, L.; Sorensen, S.B.; Glintborg, B.; et al. Validation of GWAS-Identified Variants for Anti-TNF Drug Response in Rheumatoid Arthritis: A Meta-Analysis of Two Large Cohorts. Front. Immunol. 2021, 12, 672255. [Google Scholar] [CrossRef]
- Julià, A.; Marsal, S. Pharmacogenomics of Anti-TNF Response in Psoriasis, Where Are We? Pharmacogenomics 2016, 17, 323–326. [Google Scholar] [CrossRef]
- Cui, J.; Stahl, E.A.; Saevarsdottir, S.; Miceli, C.; Diogo, D.; Trynka, G.; Raj, T.; Mirkov, M.U.; Canhao, H.; Ikari, K.; et al. Genome-Wide Association Study and Gene Expression Analysis Identifies CD84 as a Predictor of Response to Etanercept Therapy in Rheumatoid Arthritis. PLoS Genet. 2013, 9, e1003394. [Google Scholar] [CrossRef] [PubMed]
- O’Rielly, D.D.; Roslin, N.M.; Beyene, J.; Pope, A.; Rahman, P. TNF-Alpha-308 G/A Polymorphism and Responsiveness to TNF-Alpha Blockade Therapy in Moderate to Severe Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Pharm. J. 2009, 9, 161–167. [Google Scholar] [CrossRef]
- Plant, D.; Bowes, J.; Potter, C.; Hyrich, K.L.; Morgan, A.W.; Wilson, A.G.; Isaacs, J.D.; Wellcome Trust Case Control Consortium; British Society for Rheumatology Biologics Register; Barton, A. Genome-Wide Association Study of Genetic Predictors of Anti-Tumor Necrosis Factor Treatment Efficacy in Rheumatoid Arthritis Identifies Associations with Polymorphisms at Seven Loci. Arthritis Rheum. 2011, 63, 645–653. [Google Scholar] [CrossRef]
- Farragher, T.M.; Plant, D.; Flynn, E.; Eyre, S.; Bunn, D.; Thomson, W.; Symmons, D.; Barton, A. Association of a Rheumatoid Arthritis Susceptibility Variant at the CCL21 Locus with Premature Mortality in Inflammatory Polyarthritis Patients. Arthritis Care Res. 2010, 62, 676–682. [Google Scholar] [CrossRef]
- Shaker, O.G.; Alnoury, A.M.; Hegazy, G.A.; El Haddad, H.E.; Sayed, S.; Hamdy, A. Methylene Tetrahydrofolate Reductase, Transforming Growth Factor-Β1 and Lymphotoxin-α Genes Polymorphisms and Susceptibility to Rheumatoid Arthritis. Rev. Bras. Reum. Engl. Ed. 2016, 56, 414–420. [Google Scholar] [CrossRef]
- Yuan, Y.; Shao, W.; Li, Y. Associations between C677T and A1298C Polymorphisms of MTHFR and Susceptibility to Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Rheumatol. Int. 2017, 37, 557–569. [Google Scholar] [CrossRef]
- Cen, H.; Huang, H.; Zhang, L.-N.; Liu, L.-Y.; Zhou, L.; Xin, X.-F.; Zhuo, R.-J. Associations of Methylenetetrahydrofolate Reductase (MTHFR) C677T and A1298C Polymorphisms with Genetic Susceptibility to Rheumatoid Arthritis: A Meta-Analysis. Clin. Rheumatol. 2017, 36, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, K.; Cheng, G.; Mirabbasi, S.; Khalighi, B.; Wu, Y.; Fan, W. Opposite Impact of Methylene Tetrahydrofolate Reductase C677T and Methylene Tetrahydrofolate Reductase A1298C Gene Polymorphisms on Systemic Inflammation. J. Clin. Lab. Anal. 2018, 32, e22401. [Google Scholar] [CrossRef] [PubMed]
- Vetchinkina, E.A.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Deryagina, T.A.; Alekseeva, E.A.; Bure, I.V.; Zamyatnin, A.A.; Nemtsova, M.V. Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients. J. Pers. Med. 2021, 11, 469. [Google Scholar] [CrossRef]
- Mitoma, H.; Horiuchi, T.; Tsukamoto, H.; Ueda, N. Molecular Mechanisms of Action of Anti-TNF-α Agents—Comparison among Therapeutic TNF-α Antagonists. Cytokine 2018, 101, 56–63. [Google Scholar] [CrossRef]
- Ogino, S.; Wilson, R.B. Genotype and Haplotype Distributions of MTHFR677C>T and 1298A>C Single Nucleotide Polymorphisms: A Meta-Analysis. J. Hum. Genet. 2003, 48, 1–7. [Google Scholar] [CrossRef]
- Steed, M.M.; Tyagi, S.C. Mechanisms of Cardiovascular Remodeling in Hyperhomocysteinemia. Antioxid. Redox Signal. 2011, 15, 1927–1943. [Google Scholar] [CrossRef]
- Fryar-Williams, S. Fundamental Role of Methylenetetrahydrofolate Reductase 677 C → T Genotype and Flavin Compounds in Biochemical Phenotypes for Schizophrenia and Schizoaffective Psychosis. Front. Psychiatry 2016, 7, 172. [Google Scholar] [CrossRef]
- Cortese, C.; Motti, C. MTHFR Gene Polymorphism, Homocysteine and Cardiovascular Disease. Public Health Nutr. 2001, 4, 493–497. [Google Scholar] [CrossRef]
- Friedman, G.; Goldschmidt, N.; Friedlander, Y.; Ben-Yehuda, A.; Selhub, J.; Babaey, S.; Mendel, M.; Kidron, M.; Bar-On, H. A Common Mutation A1298C in Human Methylenetetrahydrofolate Reductase Gene: Association with Plasma Total Homocysteine and Folate Concentrations. J. Nutr. 1999, 129, 1656–1661. [Google Scholar] [CrossRef]
- Weisberg, I.S.; Jacques, P.F.; Selhub, J.; Bostom, A.G.; Chen, Z.; Curtis Ellison, R.; Eckfeldt, J.H.; Rozen, R. The 1298A-->C Polymorphism in Methylenetetrahydrofolate Reductase (MTHFR): In Vitro Expression and Association with Homocysteine. Atherosclerosis 2001, 156, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, Y.; Li, J.; Yang, X.; Zhang, H.; Qin, X.; Hu, Y.; Mo, Z. Serum Homocysteine Concentration Is Significantly Associated with Inflammatory/Immune Factors. PLoS ONE 2015, 10, e0138099. [Google Scholar] [CrossRef] [PubMed]
- Mititelu, R.R.; Pădureanu, R.; Băcănoiu, M.; Pădureanu, V.; Docea, A.O.; Calina, D.; Barbulescu, A.L.; Buga, A.M. Inflammatory and Oxidative Stress Markers-Mirror Tools in Rheumatoid Arthritis. Biomedicines 2020, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elmawla, M.A.; Rizk, S.M.; Youssry, I.; Shaheen, A.A. Impact of Genetic Polymorphism of Methylenetetrahydrofolate Reductase C677T on Development of Hyperhomocysteinemia and Related Oxidative Changes in Egyptian β-Thalassemia Major Patients. PLoS ONE 2016, 11, e0155070. [Google Scholar] [CrossRef]
- Bayarsaihan, D. Epigenetic Mechanisms in Inflammation. J Dent Res 2011, 90, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Linton, M.F.; Yancey, P.G.; Davies, S.S.; Jerome, W.G.; Linton, E.F.; Song, W.L.; Doran, A.C.; Vickers, K.C. The Role of Lipids and Lipoproteins in Atherosclerosis; MDText.com, Inc.: South Dartmouth, MA, USA, 2019. [Google Scholar]
- Popa, C.; Netea, M.G.; van Riel, P.L.C.M.; van der Meer, J.W.M.; Stalenhoef, A.F.H. The Role of TNF-Alpha in Chronic Inflammatory Conditions, Intermediary Metabolism, and Cardiovascular Risk. J. Lipid. Res. 2007, 48, 751–762. [Google Scholar] [CrossRef]
- Fan, S.; Yang, B.; Zhi, X.; Wang, Y.; Zheng, Q.; Sun, G. Combined Genotype and Haplotype Distributions of MTHFR C677T and A1298C Polymorphisms: A Cross-Sectional Descriptive Study of 13,473 Chinese Adult Women. Medicine 2016, 95, e5355. [Google Scholar] [CrossRef]
- Kaymakcalan, Z.; Sakorafas, P.; Bose, S.; Scesney, S.; Xiong, L.; Hanzatian, D.K.; Salfeld, J.; Sasso, E.H. Comparisons of Affinities, Avidities, and Complement Activation of Adalimumab, Infliximab, and Etanercept in Binding to Soluble and Membrane Tumor Necrosis Factor. Clin. Immunol. 2009, 131, 308–316. [Google Scholar] [CrossRef]
- Vos, A.C.W.; Wildenberg, M.E.; Duijvestein, M.; Verhaar, A.P.; van den Brink, G.R.; Hommes, D.W. Anti-Tumor Necrosis Factor-α Antibodies Induce Regulatory Macrophages in an Fc Region-Dependent Manner. Gastroenterology 2011, 140, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Ishii-Watabe, A.; Tada, M.; Kobayashi, T.; Kanayasu-Toyoda, T.; Kawanishi, T.; Yamaguchi, T. Importance of Neonatal FcR in Regulating the Serum Half-Life of Therapeutic Proteins Containing the Fc Domain of Human IgG1: A Comparative Study of the Affinity of Monoclonal Antibodies and Fc-Fusion Proteins to Human Neonatal FcR. J. Immunol. 2010, 184, 1968–1976. [Google Scholar] [CrossRef]
- Biancheri, P.; Brezski, R.J.; Di Sabatino, A.; Greenplate, A.R.; Soring, K.L.; Corazza, G.R.; Kok, K.B.; Rovedatti, L.; Vossenkämper, A.; Ahmad, N.; et al. Proteolytic Cleavage and Loss of Function of Biologic Agents That Neutralize Tumor Necrosis Factor in the Mucosa of Patients with Inflammatory Bowel Disease. Gastroenterology 2015, 149, 1564–1574.e3. [Google Scholar] [CrossRef]
- Palframan, R.; Airey, M.; Moore, A.; Vugler, A.; Nesbitt, A. Use of Biofluorescence Imaging to Compare the Distribution of Certolizumab Pegol, Adalimumab, and Infliximab in the Inflamed Paws of Mice with Collagen-Induced Arthritis. J. Immunol. Methods 2009, 348, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Van den Brande, J.M.H.; Braat, H.; van den Brink, G.R.; Versteeg, H.H.; Bauer, C.A.; Hoedemaeker, I.; van Montfrans, C.; Hommes, D.W.; Peppelenbosch, M.P.; van Deventer, S.J.H. Infliximab but Not Etanercept Induces Apoptosis in Lamina Propria T-Lymphocytes from Patients with Crohn’s Disease. Gastroenterology 2003, 124, 1774–1785. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, A.; Fossati, G.; Bergin, M.; Stephens, P.; Stephens, S.; Foulkes, R.; Brown, D.; Robinson, M.; Bourne, T. Mechanism of Action of Certolizumab Pegol (CDP870): In Vitro Comparison with Other Anti-Tumor Necrosis Factor Alpha Agents. Inflamm. Bowel Dis. 2007, 13, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Scallon, B.; Cai, A.; Solowski, N.; Rosenberg, A.; Song, X.-Y.; Shealy, D.; Wagner, C. Binding and Functional Comparisons of Two Types of Tumor Necrosis Factor Antagonists. J. Pharm. Exp. 2002, 301, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Cuppen, B.V.J.; Fu, J.; van Wietmarschen, H.A.; Harms, A.C.; Koval, S.; Marijnissen, A.C.A.; Peeters, J.J.W.; Bijlsma, J.W.J.; Tekstra, J.; van Laar, J.M.; et al. Exploring the Inflammatory Metabolomic Profile to Predict Response to TNF-α Inhibitors in Rheumatoid Arthritis. PLoS ONE 2016, 11, e0163087. [Google Scholar] [CrossRef] [PubMed]
- Min, H.K.; Kim, S.H.; Lee, S.-H.; Kim, H.-R. Baseline Bony Erosions and Time-Averaged DAS28 Predict Discontinuation of TNF Inhibitors in Rheumatoid Arthritis. Sci. Rep. 2022, 12, 19951. [Google Scholar] [CrossRef]
- Krintel, S.B.; Dehlendorff, C.; Hetland, M.L.; Hørslev-Petersen, K.; Andersen, K.K.; Junker, P.; Pødenphant, J.; Ellingsen, T.; Ahlquist, P.; Lindegaard, H.M.; et al. Prediction of Treatment Response to Adalimumab: A Double-Blind Placebo-Controlled Study of Circulating MicroRNA in Patients with Early Rheumatoid Arthritis. Pharm. J. 2016, 16, 141–146. [Google Scholar] [CrossRef]
- Castro-Villegas, C.; Pérez-Sánchez, C.; Escudero, A.; Filipescu, I.; Verdu, M.; Ruiz-Limón, P.; Aguirre, M.A.; Jiménez-Gomez, Y.; Font, P.; Rodriguez-Ariza, A.; et al. Circulating MiRNAs as Potential Biomarkers of Therapy Effectiveness in Rheumatoid Arthritis Patients Treated with Anti-TNFα. Arthritis Res. 2015, 17, 49. [Google Scholar] [CrossRef]
- Millier, M.J.; Fanning, N.C.; Frampton, C.; Stamp, L.K.; Hessian, P.A. Plasma Interleukin-23 and Circulating IL-17A+IFNγ+ Ex-Th17 Cells Predict Opposing Outcomes of Anti-TNF Therapy in Rheumatoid Arthritis. Arthritis Res. 2022, 24, 57. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O.; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann. Rheum. Dis. 2010, 69, 1580–1588. [Google Scholar] [CrossRef]
- van Gestel, A.M.; Prevoo, M.L.; van ’t Hof, M.A.; van Rijswijk, M.H.; van de Putte, L.B.; van Riel, P.L. Development and Validation of the European League Against Rheumatism Response Criteria for Rheumatoid Arthritis. Comparison with the Preliminary American College of Rheumatology and the World Health Organization/International League against Rheumatism Criteria. Arthritis Rheum. 1996, 39, 34–40. [Google Scholar] [CrossRef]
- van Gestel, A.M.; Haagsma, C.J.; van Riel, P.L. Validation of Rheumatoid Arthritis Improvement Criteria That Include Simplified Joint Counts. Arthritis Rheum. 1998, 41, 1845–1850. [Google Scholar] [CrossRef]
| Variable (n) | Stratum | (%) |
|---|---|---|
| Age (81) * | >60 years | 40.74 |
| Sex (81) | Female | 67.90 |
| Tobacco smoking (81) | Current smokers | 24.69 |
| RF (61) | Positive | 78.69 |
| ACPA (59) | Positive | 74.58 |
| Response (85) | ||
| GR (21) | 24.71 | |
| MR (30) | 35.29 | |
| NR (34) | 40 |
| SNP (n) | Genotype | Frequency (%) | Allele | Frequency (%) |
|---|---|---|---|---|
| c.665C>T (81) | CC CT TT | 28 51 21 | C T | 54 46 |
| c.1298A>C (81) | AA AC CC | 46 48 6 | A C | 70 30 |
| Present DAS28 | DAS28 Improvement | ||
|---|---|---|---|
| >1.2 | >0.6 and ≤1.2 | ≤0.6 | |
| ≤3.2 | Good response | Moderate response | No response |
| >3.2 and ≤5.1 | Moderate response | Moderate response | No response |
| >5.1 | Moderate response | No response | No response |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravaei, A.; Pulsatelli, L.; Assirelli, E.; Ciaffi, J.; Meliconi, R.; Salvarani, C.; Govoni, M.; Rubini, M. MTHFR c.665C>T and c.1298A>C Polymorphisms in Tailoring Personalized Anti-TNF-α Therapy for Rheumatoid Arthritis. Int. J. Mol. Sci. 2023, 24, 4110. https://doi.org/10.3390/ijms24044110
Ravaei A, Pulsatelli L, Assirelli E, Ciaffi J, Meliconi R, Salvarani C, Govoni M, Rubini M. MTHFR c.665C>T and c.1298A>C Polymorphisms in Tailoring Personalized Anti-TNF-α Therapy for Rheumatoid Arthritis. International Journal of Molecular Sciences. 2023; 24(4):4110. https://doi.org/10.3390/ijms24044110
Chicago/Turabian StyleRavaei, Amin, Lia Pulsatelli, Elisa Assirelli, Jacopo Ciaffi, Riccardo Meliconi, Carlo Salvarani, Marcello Govoni, and Michele Rubini. 2023. "MTHFR c.665C>T and c.1298A>C Polymorphisms in Tailoring Personalized Anti-TNF-α Therapy for Rheumatoid Arthritis" International Journal of Molecular Sciences 24, no. 4: 4110. https://doi.org/10.3390/ijms24044110
APA StyleRavaei, A., Pulsatelli, L., Assirelli, E., Ciaffi, J., Meliconi, R., Salvarani, C., Govoni, M., & Rubini, M. (2023). MTHFR c.665C>T and c.1298A>C Polymorphisms in Tailoring Personalized Anti-TNF-α Therapy for Rheumatoid Arthritis. International Journal of Molecular Sciences, 24(4), 4110. https://doi.org/10.3390/ijms24044110








