Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress
Abstract
1. Introduction
2. Results
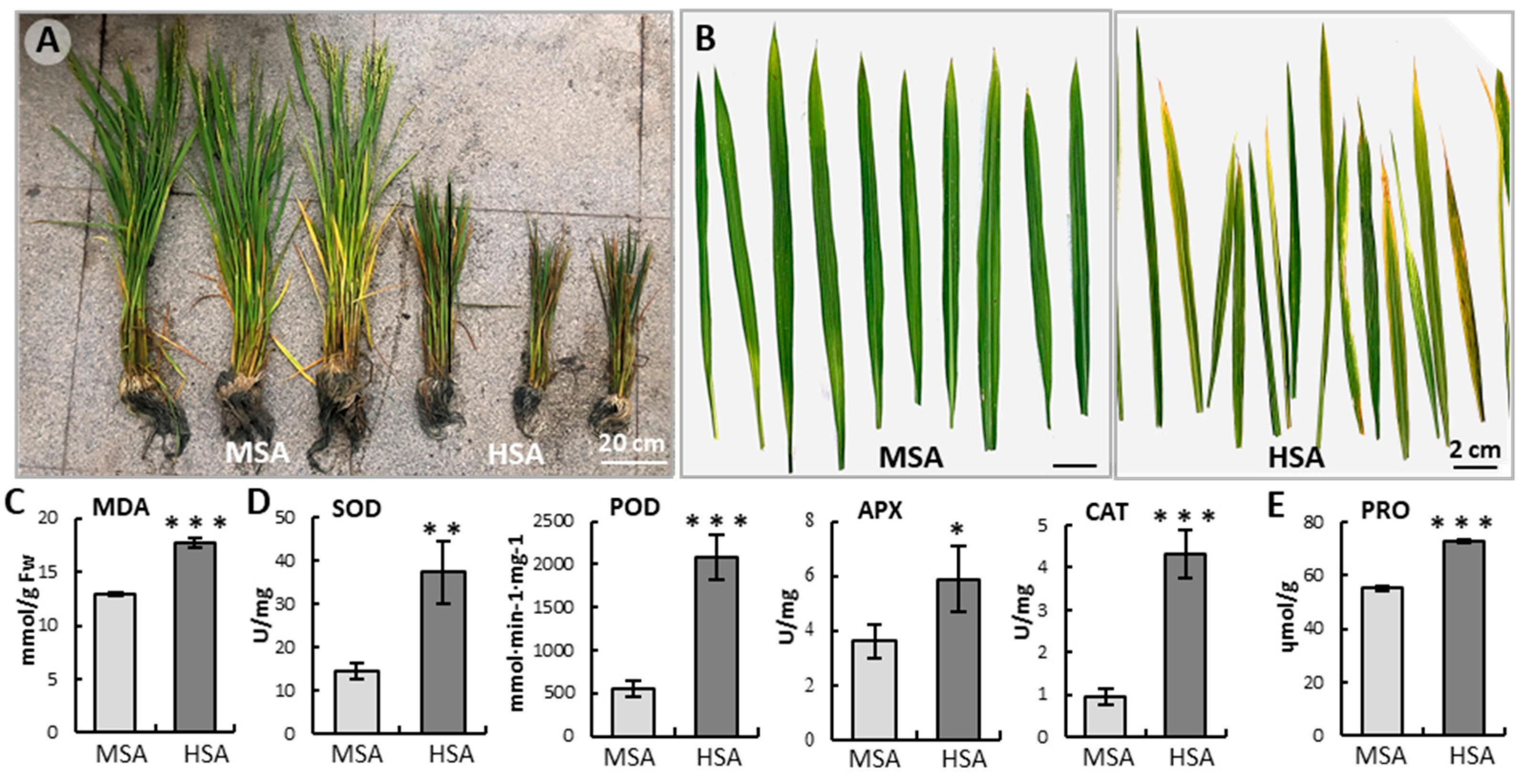
2.1. High Saline–Alkali Stress Inhibited Rice Growth and Development
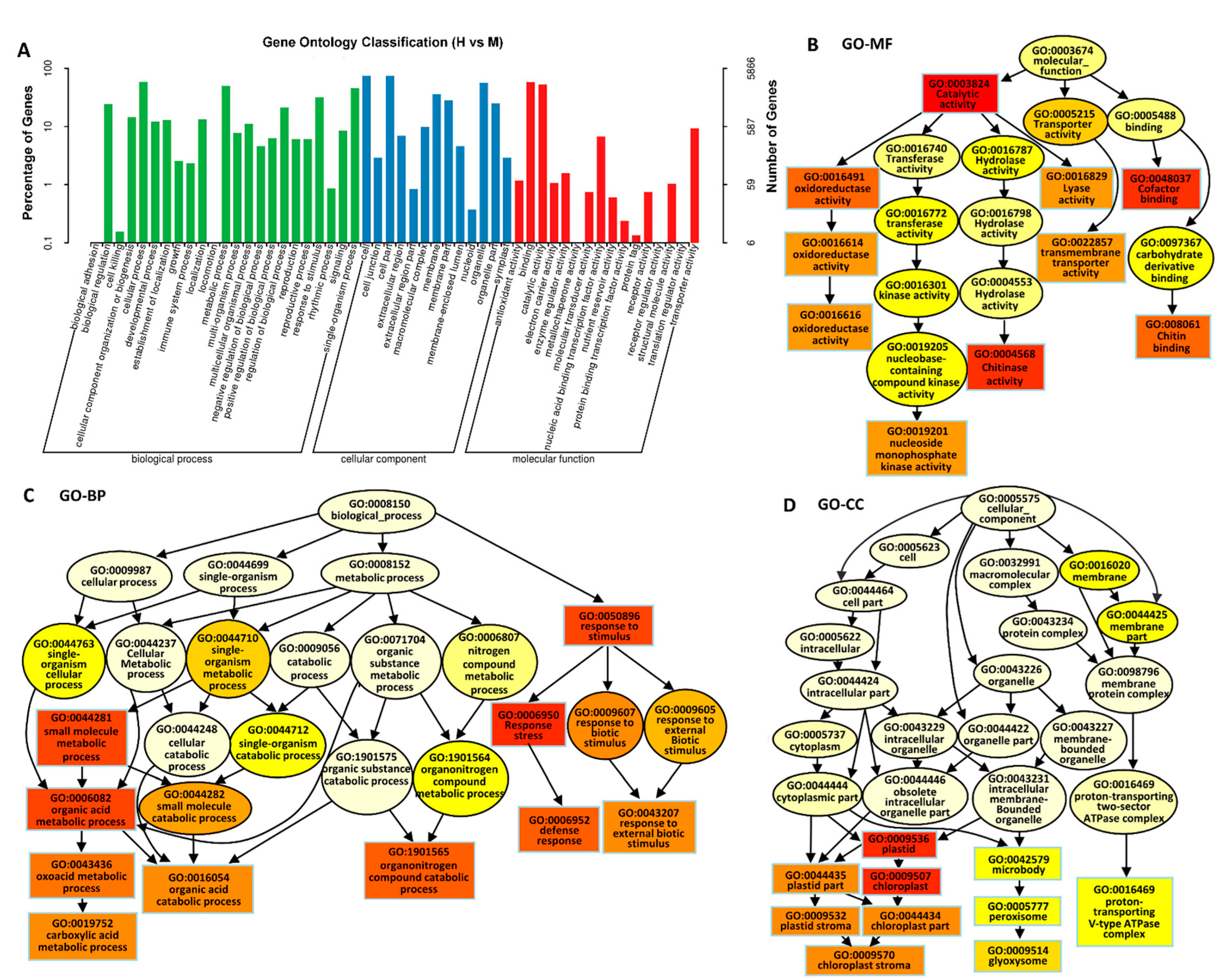
2.2. Transcriptomic Analysis of Rice Leaves
2.3. Identification of DEGs in Response to High Saline–Alkali Stress
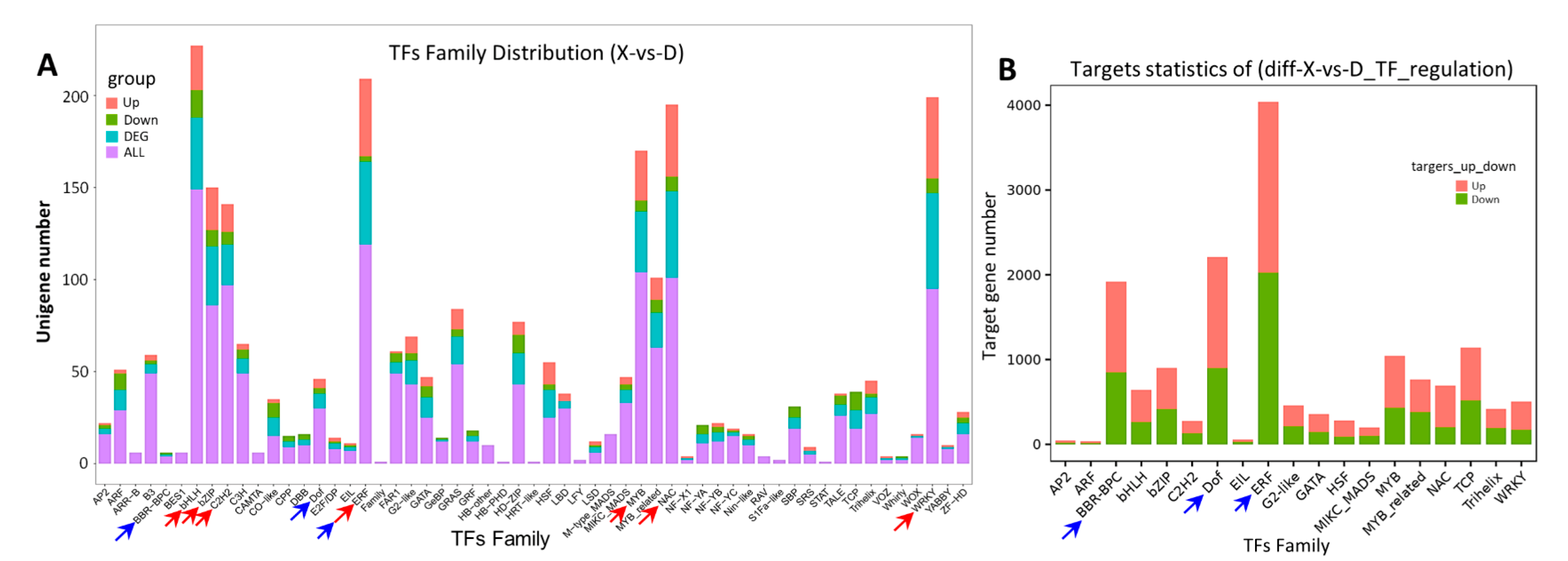
2.4. Transcription Factors Related to High Saline–Alkali Stress
2.5. Non-Targeted Metabolomic Analysis of Differentially Accumulated Metabolites
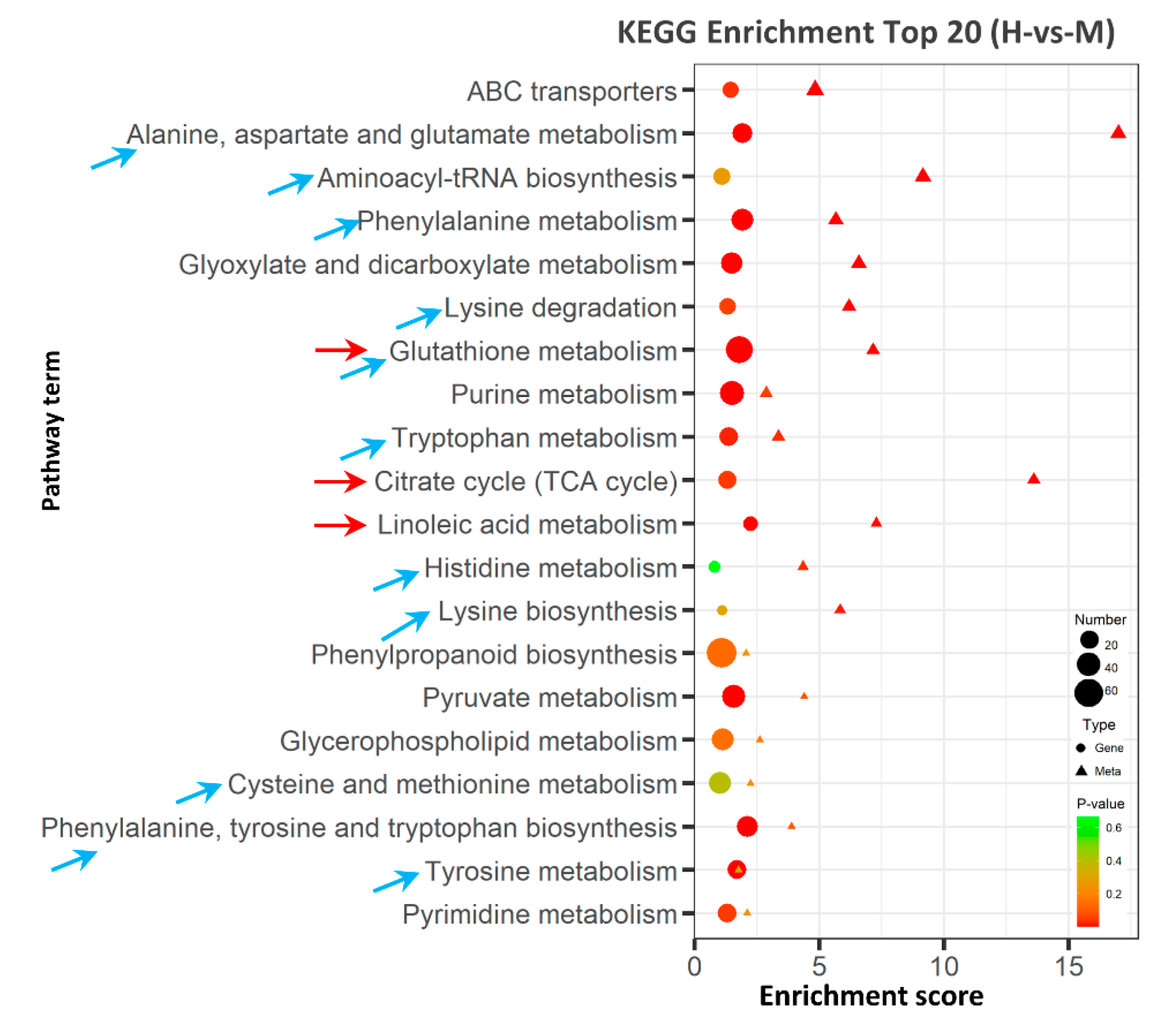
2.6. Integrative Analysis of Transcriptome and Metabolome
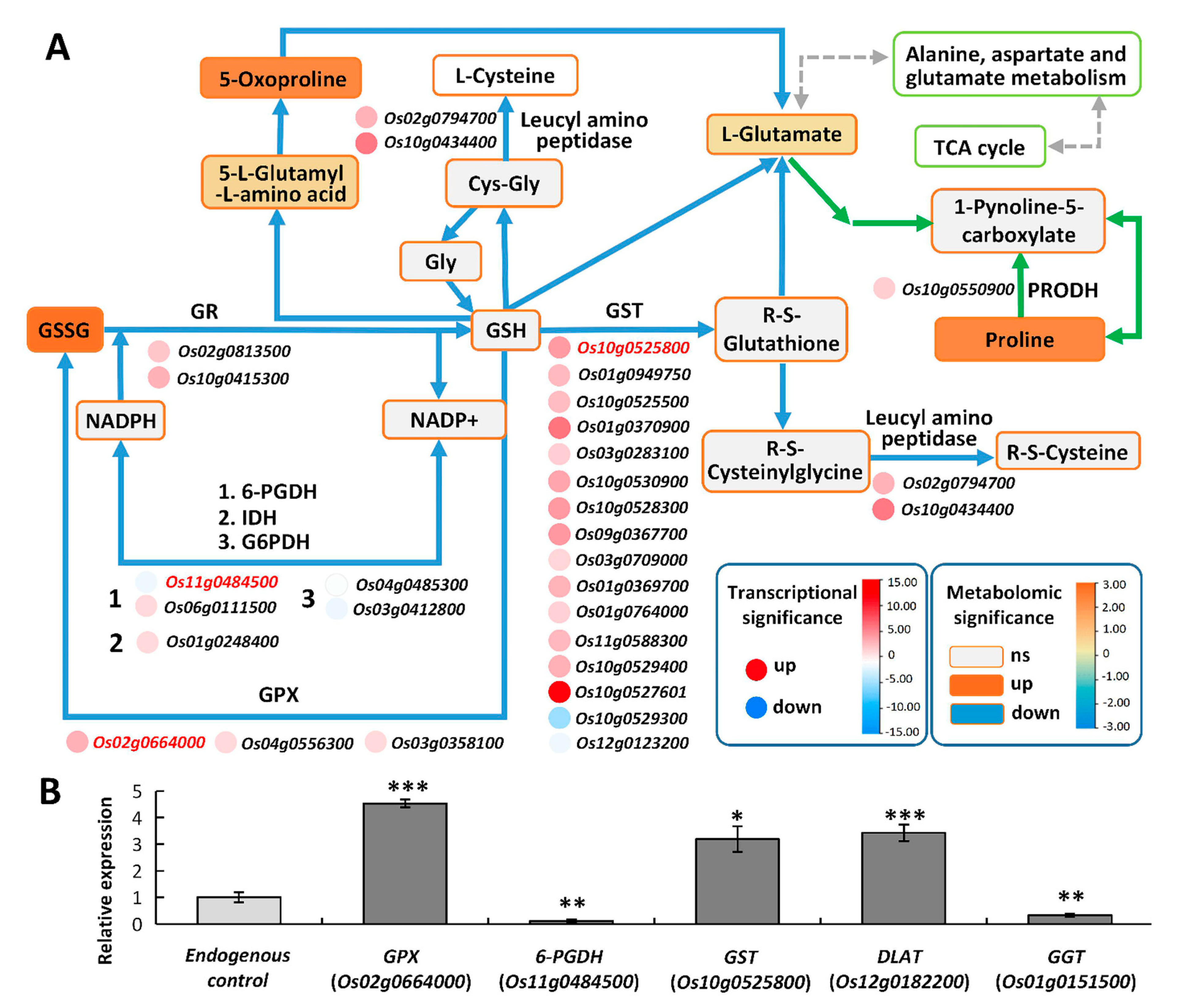
2.6.1. Glutathione Metabolism May Enhance ROS Scavenging under High Saline–Alkali Stress
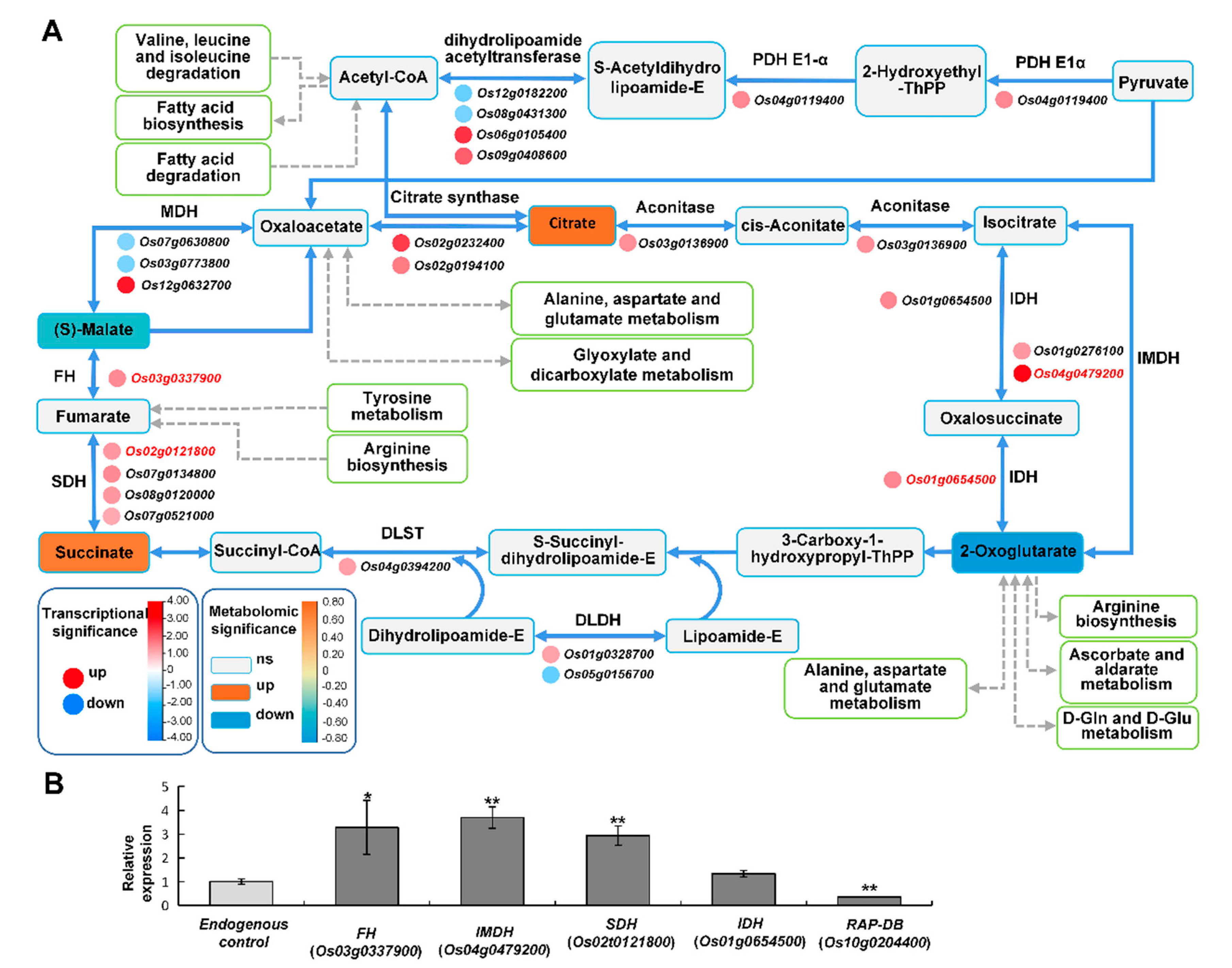
2.6.2. TCA Cycle May Provide Energy to Improve Rice Stress Tolerance
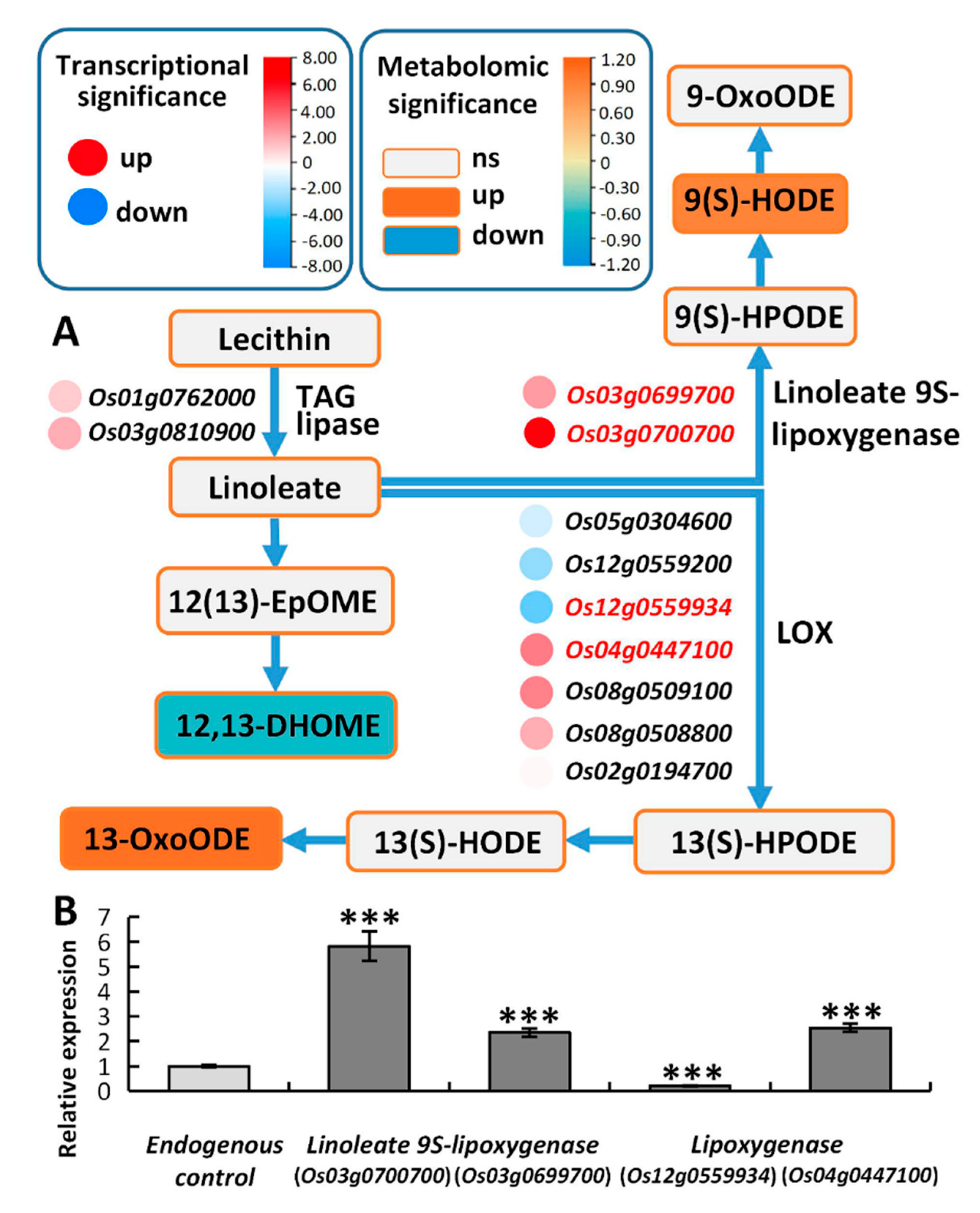
2.6.3. Linoleic Acid Metabolism May Alleviate the Membrane Damage Caused by High Saline–Alkali Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Total RNA Isolation, Library Construction, and Sequencing
4.3. Quantitative Real-time PCR Validation
4.4. Metabolite Extraction
4.5. Metabolite Detection
4.6. Functional Enrichment Analysis of DEGs
4.7. Statistical Analysis
4.8. Physiological Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Seki, K.; Miyazaki, T.; Ishihama, Y. The causes of soil alkalinization in the Songnen Plain of Northeast China. Paddy Water Environ. 2009, 7, 259–270. [Google Scholar] [CrossRef]
- Kaiwen, G.; Zisong, X.; Yuze, H.; Qi, S.; Yue, W.; Yanhui, C.; Jiechen, W.; Wei, L.; Huihui, Z. Effects of salt concentration, pH, and their interaction on plant growth, nutrient uptake, and photochemistry of alfalfa (Medicago sativa) leaves. Plant Signal. Behav. 2020, 15, 1832373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jiang, C.J.; Liang, Z.W. Root Damage under Alkaline Stress Is Associated with Reactive Oxygen Species Accumulation in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef] [PubMed]
- Fan, C. Genetic mechanisms of salt stress responses in halophytes. Plant Signal Behav. 2020, 15, 1704528. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.H.; Huh, S.M.; Kim, K.M.; Park, W.J.; Seo, J.B.; Cho, K.; Kim, D.Y.; Kim, B.G.; Yoon, I.S. Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci. 2012, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 Interaction Controls Seedling Growth under Salt Stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; Li, F.; Yan, C.; Zhong, X.; Liu, Q.; Xia, X.; Li, H.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Wang, X.; Ren, H.; Wei, Z.; Wang, Y.; Ren, W. Effects of neutral salt and alkali on ion distributions in the roots, shoots, and leaves of two alfalfa cultivars with differing degrees of salt tolerance. J. Integr. Agr. 2017, 16, 1800–1807. [Google Scholar] [CrossRef]
- Xiang, G.; Ma, W.; Gao, S.; Jin, Z.; Yue, Q.; Yao, Y. Transcriptomic and phosphoproteomic profiling and metabolite analyses reveal the mechanism of NaHCO3− induced organic acid secretion in grapevine roots. BMC Plant Biol. 2019, 19, 383. [Google Scholar] [CrossRef]
- Wang, X.; Geng, S.; Ri, Y.; Cao, D.; Liu, J.; Shi, D.; Yang, C. Physiological responses and adaptive strategies of tomato plants to salt and alkali stresses. Sci. Hortic. 2011, 130, 248–255. [Google Scholar] [CrossRef]
- Guo, S.; Niu, Y.; Zhai, H.; Han, N.; Du, Y. Effects of alkaline stress on organic acid metabolism in roots of grape hybrid rootstocks. Sci. Hortic. 2018, 227, 255–260. [Google Scholar] [CrossRef]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Yan, X.; Guo, J. Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance. PLoS ONE 2020, 15, e244365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Tang, J.; Wang, Y.; Kang, H.; Zeng, J. The tolerance to saline-alkaline stress was dependent on the roots in wheat. Physiol. Mol. Biol. Plants 2020, 26, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Shabala, L. Ion transport and osmotic adjustment in plants and bacteria. Biomol. Concepts 2011, 2, 407–419. [Google Scholar] [CrossRef]
- Pan, Y.Q.; Guo, H.; Wang, S.M.; Zhao, B.; Zhang, J.L.; Ma, Q.; Yin, H.J.; Bao, A.K. The Photosynthesis, Na(+)/K(+) Homeostasis and Osmotic Adjustment of Atriplex canescens in Response to Salinity. Front. Plant Sci. 2016, 7, 848. [Google Scholar] [CrossRef]
- Shen, Q.; Yu, J.; Fu, L.; Wu, L.; Dai, F.; Jiang, L.; Wu, D.; Zhang, G. Ionomic, metabolomic and proteomic analyses reveal molecular mechanisms of root adaption to salt stress in Tibetan wild barley. Plant Physiol. Biochem. 2018, 123, 319–330. [Google Scholar] [CrossRef]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Park, J.R.; Jang, Y.H.; Kim, E.G.; Kim, K.M. Rice Cultivars Under Salt Stress Show Differential Expression of Genes Related to the Regulation of Na(+)/K(+) Balance. Front. Plant Sci. 2021, 12, 680131. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1; 5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Das Bhowmik, S.; Long, H.; Cheng, Y.; Mundree, S.; Hoang, L.T.M. Rapid Accumulation of Proline Enhances Salinity Tolerance in Australian Wild Rice Oryza australiensis Domin. Plants 2021, 10, 2044. [Google Scholar] [CrossRef]
- Luo, X.; Dai, Y.; Zheng, C.; Yang, Y.; Chen, W.; Wang, Q.; Chandrasekaran, U.; Du, J.; Liu, W.; Shu, K. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021, 229, 950–962. [Google Scholar] [CrossRef]
- Fu, J.; Liu, Z.; Li, Z.; Wang, Y.; Yang, K. Alleviation of the effects of saline-alkaline stress on maize seedlings by regulation of active oxygen metabolism by Trichoderma asperellum. PLoS ONE 2017, 12, e179617. [Google Scholar] [CrossRef]
- Sun, N.; Song, T.; Ma, Z.; Dong, L.; Zhan, L.; Xing, Y.; Liu, J.; Song, J.; Wang, S.; Cai, H. Overexpression of MsSiR enhances alkali tolerance in alfalfa (Medicago sativa L.) by increasing the glutathione content. Plant Physiol Biochem 2020, 154, 538–546. [Google Scholar] [CrossRef]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. TAG. Theoretical and applied genetics. Theor. Angew. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. TAG. Theoretical and applied genetics. Theor. Angew. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, W.; Wang, Y.; Zhou, Y.; Wang, S.; Qi, F.; Wang, N.; Ma, J. Comparative Analysis of Physiological, Hormonal and Transcriptomic Responses Reveal Mechanisms of Saline-Alkali Tolerance in Autotetraploid Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 16146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hu, Y.; Du, A.; Yu, L.; Fu, X.; Wu, C.; Lu, L.; Liu, Y.; Wang, S.; Huang, W.; et al. Cell Wall Matrix Polysaccharides Contribute to Salt-Alkali Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 15019. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Xue, C.Y.; Song, H.D.; Gao, Y.; Feng, L.; Li, Y.; Xuan, Y.H. Tissue-specific activation of DOF11 promotes rice resistance to sheath blight disease and increases grain weight via activation of SWEET14. Plant Biotechnol. J. 2021, 19, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xue, C.Y.; Liu, J.M.; He, Y.; Mei, Q.; Wei, S.; Xuan, Y.H. Sheath blight resistance in rice is negatively regulated by WRKY53 via SWEET2a activation. Biochem. Biophys. Res. Commun. 2021, 585, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Cimini, S.; Locato, V.; Giacinti, V.; Molinari, M.; De Gara, L. A Multifactorial Regulation of Glutathione Metabolism behind Salt Tolerance in Rice. Antioxidants 2022, 11, 1114. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Bedhomme, M.; Lemaire, S.D.; Trost, P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 2012, 185–186, 86–96. [Google Scholar] [CrossRef]
- Kirma, M.; Araujo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef]
- Giaretta, S.; Prasad, D.; Forieri, I.; Vamerali, T.; Trentin, A.R.; Wirtz, M.; Hell, R.; Masi, A. Apoplastic gamma-glutamyl transferase activity encoded by GGT1 and GGT2 is important for vegetative and generative development. Plant Physiol. Biochem. 2017, 115, 44–56. [Google Scholar] [CrossRef]
- Kumari, A.; Das, P.; Parida, A.K.; Agarwal, P.K. Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front. Plant Sci. 2015, 6, 537. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, S.; Min, W.; Ye, J.; Hou, Z. Ionomic and transcriptomic analyses of two cotton cultivars (Gossypium hirsutum L.) provide insights into the ion balance mechanism of cotton under salt stress. PLoS ONE 2019, 14, e226776. [Google Scholar] [CrossRef] [PubMed]
- Borglum, A.D.; Flint, T.; Hansen, L.L.; Kruse, T.A. Refined localization of the pyruvate dehydrogenase E1 alpha gene (PDHA1) by linkage analysis. Hum. Genet. 1997, 99, 80–82. [Google Scholar] [CrossRef]
- Chodok, P.; Eiamsa-ard, P.; Cove, D.J.; Quatrano, R.S.; Kaewsuwan, S. Identification and functional characterization of two Delta12-fatty acid desaturases associated with essential linoleic acid biosynthesis in Physcomitrella patens. J. Ind. Microbiol. Biotechnol. 2013, 40, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Zhang, L.; Hu, X.; Nan, S.; Chen, X.; Fu, H. Cloning and functional analysis of the FAD2 gene family from desert shrub Artemisia sphaerocephala. BMC Plant Biol. 2019, 19, 481. [Google Scholar] [CrossRef]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef]
- Chen, T.; Cohen, D.; Itkin, M.; Malitsky, S.; Fluhr, R. Lipoxygenase functions in 1O2 production during root responses to osmotic stress. Plant Physiol. 2021, 185, 1638–1651. [Google Scholar] [CrossRef]
- Zhang, D.; Tong, J.; He, X.; Xu, Z.; Xu, L.; Wei, P.; Huang, Y.; Brestic, M.; Ma, H.; Shao, H. A Novel Soybean Intrinsic Protein Gene, GmTIP2;3, Involved in Responding to Osmotic Stress. Front. Plant Sci. 2015, 6, 1237. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, X.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Guo, L.; Rui, C.; Zhang, Y.; Cui, R.; et al. Cotton transcriptome analysis reveals novel biological pathways that eliminate reactive oxygen species (ROS) under sodium bicarbonate (NaHCO(3)) alkaline stress. Genomics 2021, 113, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.V.; Davis, D.G. Abiotic stress alters transcript profiles and activity of glutathione S-transferase, glutathione peroxidase, and glutathione reductase in Euphorbia esula. Physiol Plant 2004, 120, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Hao, Y.; Xu, S.; Lyu, Z.; Wang, H.; Kong, L.; Sun, S. Comparative Analysis of the Glutathione S-Transferase Gene Family of Four Triticeae Species and Transcriptome Analysis of GST Genes in Common Wheat Responding to Salt Stress. Int. J. Genom. 2021, 2021, 6289174. [Google Scholar] [CrossRef]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakiere, B.; Vanacker, H.; Miginiac-Maslow, M.; Van Breusegem, F.; Noctor, G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007, 52, 640–657. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Komives, T.; Kiraly, L.; Schroder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Yuan, D.; Chu, C.; Duan, M. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback Regulation of ABA Signaling and Biosynthesis by a bZIP Transcription Factor Targets Drought-Resistance-Related Genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef]
- Yang, C.; Wang, E.; Liu, J. CERK1, more than a co-receptor in plant-microbe interactions. New Phytol. 2022, 234, 1606–1613. [Google Scholar] [CrossRef]
- Espinoza, C.; Liang, Y.; Stacey, G. Chitin receptor CERK 1 links salt stress and chitin−triggered innate immunity in Arabidopsis. Plant J. 2017, 89, 984–995. [Google Scholar] [CrossRef]
- Xu, G.Y.; Rocha, P.S.; Wang, M.L.; Xu, M.L.; Cui, Y.C.; Li, L.Y.; Zhu, Y.X.; Xia, X. A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 2011, 234, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Zou, J.; Kong, L.; Hu, S.; Wang, B.; Yang, J.; Xie, G. OsCCD1, a novel small calcium-binding protein with one EF-hand motif, positively regulates osmotic and salt tolerance in rice. Plant Sci. 2016, 247, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Gu, Z.; Tang, H.; Chen, X.; Liu, F.; Zhang, H. Preliminary Study on Function of Calcineurin B-Like Protein Gene OsCBL8 in Rice. Rice Sci. 2010, 17, 10–18. [Google Scholar] [CrossRef]
- Saeng-ngam, S.C.U.B.; Takpirom, W.C.U.B.; Buaboocha, T.C.U.B.; Chadchawan, S.C.U.B. The role of the OsCam1-1 salt stress sensor in ABA accumulation and salt tolerance in rice. J. Plant Biol. 2012, 55, 198–208. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome Analysis of Salt Stress Responsiveness in the Seedlings of Dongxiang Wild Rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e146242. [Google Scholar] [CrossRef]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef]
- Xiao, J.; Cheng, H.; Li, X.; Xiao, J.; Xu, C.; Wang, S. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013, 163, 1868–1882. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Liang, X.; Wang, Y.; Li, Y.; An, W.; He, X.; Chen, Y.; Shi, Z.; He, J.; Wan, R. Widely-Targeted Metabolic Profiling in Lyciumbarbarum Fruits under Salt-Alkaline Stress Uncovers Mechanism of Salinity Tolerance. Molecules 2022, 27, 1564. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Degenkolbe, T.; Cuadros-Inostroza, A.; Klie, S.; Sulpice, R.; Leisse, A.; Steinhauser, D.; Fernie, A.R.; Willmitzer, L.; Hannah, M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011, 67, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Amir, R.; Galili, G.; Cohen, H. The metabolic roles of free amino acids during seed development. Plant Sci. 2018, 275, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Q.; Zhao, D.S.; Zhang, C.Q.; Wu, H.Y.; Li, Q.F.; Gu, M.H.; Sun, S.S.; Liu, Q.Q. A Connection between Lysine and Serotonin Metabolism in Rice Endosperm. Plant Physiol. 2018, 176, 1965–1980. [Google Scholar] [CrossRef] [PubMed]
- Schenck, C.A.; Maeda, H.A. Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Datta, A.; Bhattacharyya, D.; Singh, S.; Ghosh, A.; Schmidtchen, A.; Malmsten, M.; Bhunia, A. Role of Aromatic Amino Acids in Lipopolysaccharide and Membrane Interactions of Antimicrobial Peptides for Use in Plant Disease Control. J. Biol. Chem. 2016, 291, 13301–13317. [Google Scholar] [CrossRef]
- Widodo; Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Wu, D.; Shen, Q.; Cai, S.; Chen, Z.H.; Dai, F.; Zhang, G. Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 2013, 54, 1976–1988. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, J.; Li, M.; Shi, L. Metabolomics Analysis Reveals the Salt-Tolerant Mechanism in Glycine soja. J. Plant Growth Regul. 2017, 36, 460–471. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, M.; Sang, X.; Li, P.; Ling, Y.; Zhao, F.; Du, D.; Li, Y.; Yang, Z.; He, G. Association between sheath blight resistance and chitinase activity in transgenic rice plants expressing McCHIT1 from bitter melon. Transgenic Res. 2019, 28, 381–390. [Google Scholar] [CrossRef]


| Sample | pH | Organic Matter (g/kg) | alkali-Hydrolyzable Nitrogen (mg/kg) | Available Phosphorus (mg/kg) | Available Potassium (mg/kg) | Available Zinc (mg/kg) | Salt Contents (g/kg) |
|---|---|---|---|---|---|---|---|
| MSA | 8.41 | 5.9 | 34.8 | 7.6 | 99.6 | 1.08 | 0.29 |
| HSA | 9.64 | 12.8 | 46.9 | 10.2 | 185 | 0.42 | 1.24 |
| Sample | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Total Raw Bases (Gb) | Total Clean Bases (Gb) | Valid Bases (%) | Clean Reads Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| MSA | 50.49 | 49.81 | 7.58 | 7.23 | 95.4 | 95.20 | 49.59 |
| HSA | 49.20 | 48.53 | 7.38 | 7.05 | 95.5 | 95.12 | 50.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, G.; Wang, M.; Wang, X.; Liu, K.; Li, Y.; Bu, Y.; Li, L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress. Int. J. Mol. Sci. 2023, 24, 4062. https://doi.org/10.3390/ijms24044062
Qian G, Wang M, Wang X, Liu K, Li Y, Bu Y, Li L. Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress. International Journal of Molecular Sciences. 2023; 24(4):4062. https://doi.org/10.3390/ijms24044062
Chicago/Turabian StyleQian, Guangtao, Mingyu Wang, Xiaoting Wang, Kai Liu, Ying Li, Yuanyuan Bu, and Lixin Li. 2023. "Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress" International Journal of Molecular Sciences 24, no. 4: 4062. https://doi.org/10.3390/ijms24044062
APA StyleQian, G., Wang, M., Wang, X., Liu, K., Li, Y., Bu, Y., & Li, L. (2023). Integrated Transcriptome and Metabolome Analysis of Rice Leaves Response to High Saline–Alkali Stress. International Journal of Molecular Sciences, 24(4), 4062. https://doi.org/10.3390/ijms24044062






