Bone Differentiation Ability of CD146-Positive Stem Cells from Human Exfoliated Deciduous Teeth
Abstract
1. Introduction
2. Results
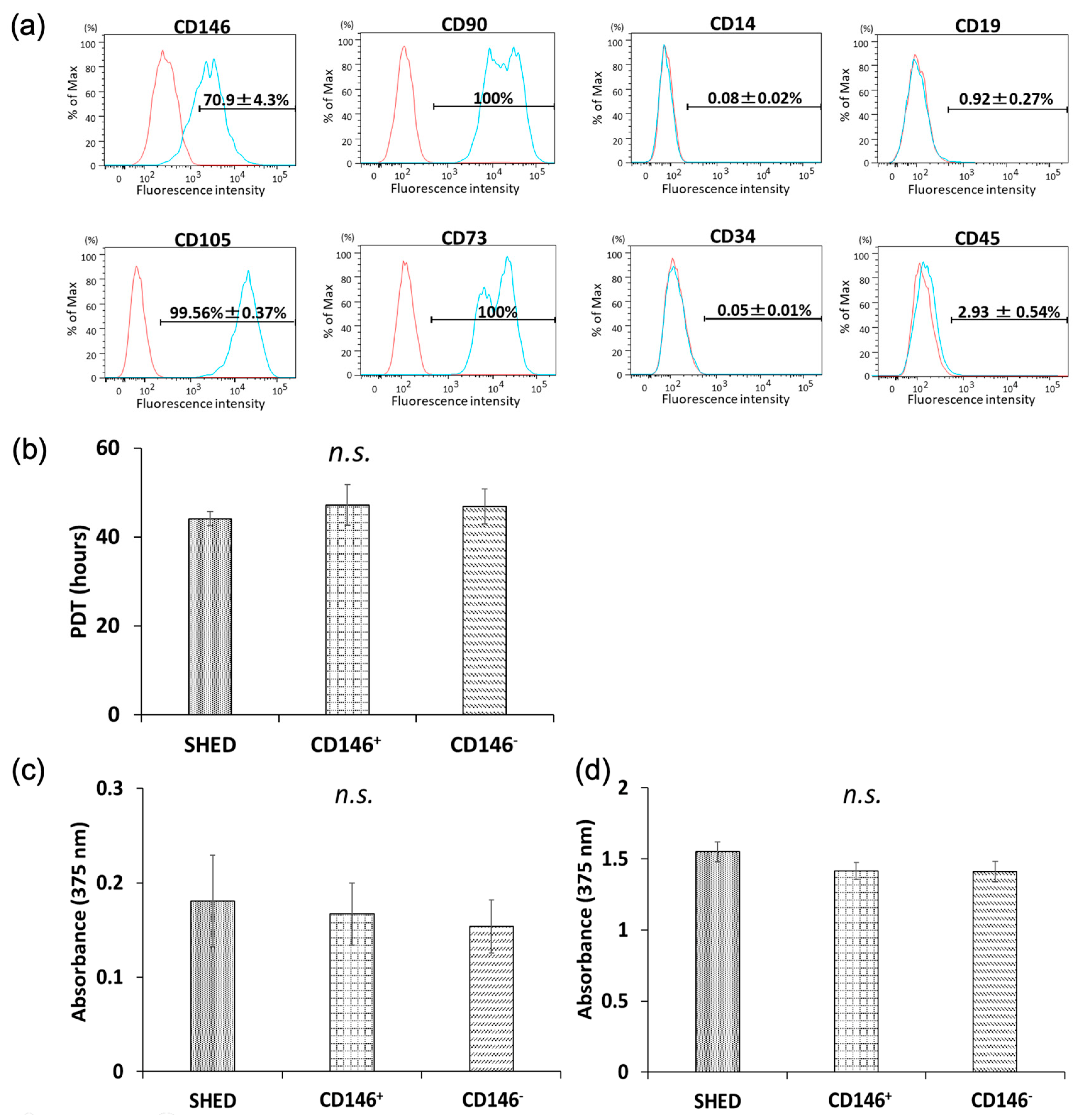
2.1. Surface-Antigen Analyses of Isolated SHED
2.2. Cellular Proliferative Capacity of SHED, CD146 + SHED, and CD146-SHED
2.2.1. PDT
2.2.2. BrdU Proliferation Assay
2.3. Osteogenic Differentiation-Related Gene Expression Analyses in SHED, CD146 + SHED, and CD146-SHED
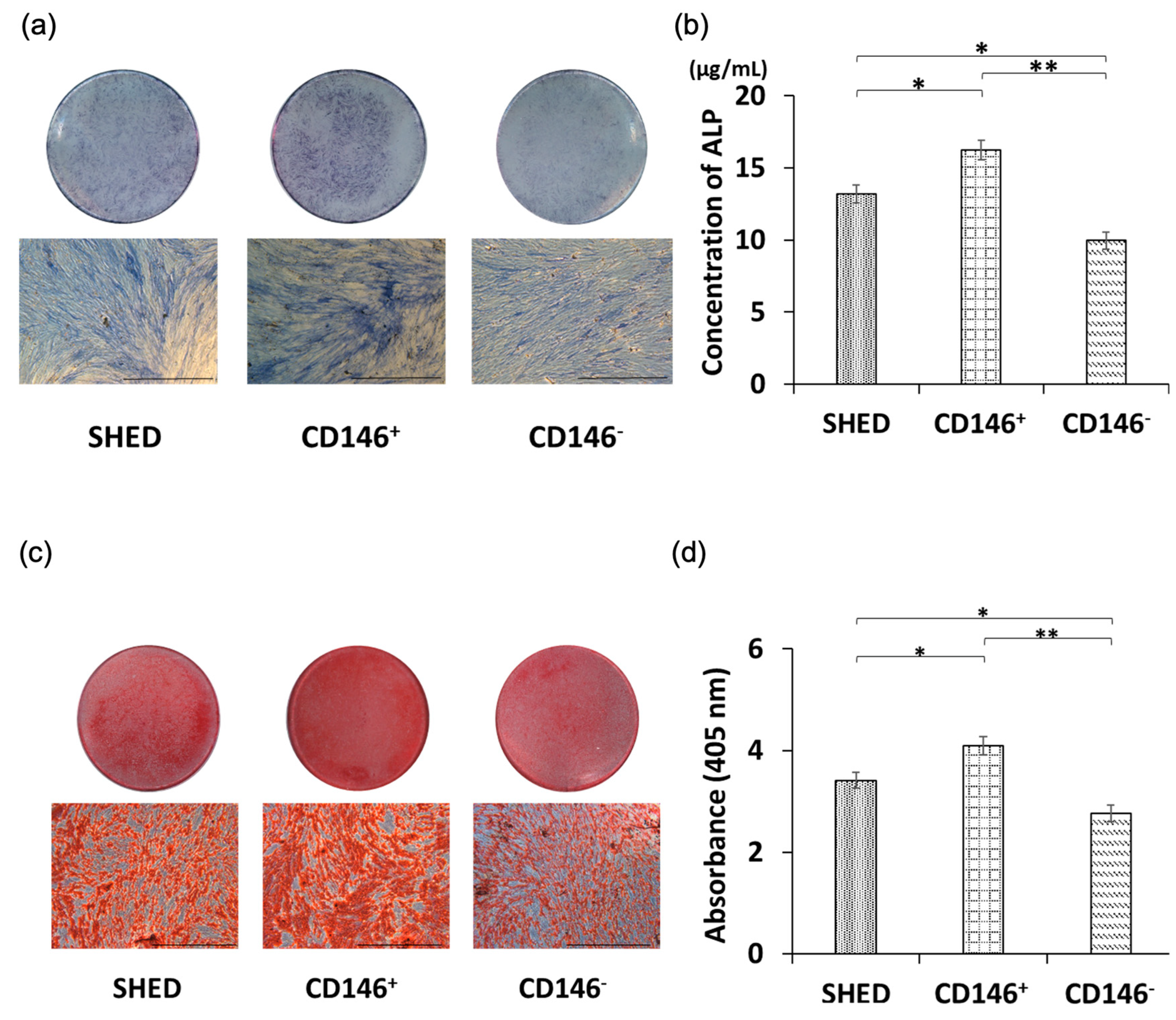
2.4. Osteogenic Differentiation Potential of SHED, CD146 + SHED, and CD146-SHED
2.5. Comparative Analysis of Calcium Deposition among SHED, CD146 + SHED, CD146-SHED
3. Discussion
4. Materials and Methods
4.1. SHED Isolation and Culture
4.2. Fluorescence-Activated Cell Sorting
4.3. Properties of SHED, CD146 + SHED, and CD146-SHED
4.3.1. Cellular Proliferative Capacity of SHED, CD146 + SHED, and CD146-SHED
Population Doubling Time
Bromodeoxyuridine Cell Proliferation Assay
4.3.2. Induction of Osteogenic Differentiation of SHED, CD146 + SHED, and CD146-SHED
4.4. Quantitative Real-Time Polymerase Chain Reaction Analysis
4.5. ALP Staining and Determination of ALP Activity
4.6. Calcium Deposition Analyses (Alizarin Red Staining)
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qi, J.; Yu, T.; Hu, B.; Wu, H.; Ouyang, H. Current Biomaterial-Based Bone Tissue Engineering and Translational Medicine. Int. J. Mol. Sci. 2021, 22, 10233. [Google Scholar] [CrossRef]
- Walmsley, G.G.; Ransom, R.C.; Zielins, E.R.; Leavitt, T.; Flacco, J.S.; Hu, M.S.; Lee, A.S.; Longaker, M.T.; Wan, D.C. Stem Cells in Bone Regeneration. Stem Cell Rev. 2016, 12, 524–529. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Y.; Jiang, Y.; Du, J.; Xu, J.; Liu, Y. Systemic Therapy of MSCs in Bone Regeneration: A Systematic Review and Meta-Analysis. Stem Cell Res. Ther. 2021, 12, 377. [Google Scholar] [CrossRef]
- Edgar, L.; Pu, T.; Porter, B.; Aziz, J.M.; La Pointe, C.; Asthana, A.; Orlando, G. Regenerative Medicine, Organ Bioengineering and Transplantation. Br. J. Surg. 2020, 107, 793–800. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal Stem Cells. J. Orthop. Res. 1991, 9, 641–650. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage Cells from Human Adipose Tissue: Implications for Cell-Based Therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar] [CrossRef]
- Brown, C.; McKee, C.; Bakshi, S.; Walker, K.; Hakman, E.; Halassy, S.; Svinarich, D.; Dodds, R.; Govind, C.K.; Chaudhry, G.R. Mesenchymal Stem Cells: Cell Therapy and Regeneration Potential. J. Tissue Eng. Regen. Med. 2019, 13, 1738–1755. [Google Scholar] [CrossRef]
- Xiang, X.-N.; Zhu, S.-Y.; He, H.-C.; Yu, X.; Xu, Y.; He, C.-Q. Mesenchymal Stromal Cell-Based Therapy for Cartilage Regeneration in Knee Osteoarthritis. Stem. Cell Res. Ther. 2022, 13, 14. [Google Scholar] [CrossRef]
- Shimizu, Y.; Ntege, E.H.; Sunami, H. Current Regenerative Medicine-Based Approaches for Skin Regeneration: A Review of Literature and a Report on Clinical Applications in Japan. Regen. Ther. 2022, 21, 73–80. [Google Scholar] [CrossRef]
- Venkataiah, V.S.; Handa, K.; Njuguna, M.M.; Hasegawa, T.; Maruyama, K.; Nemoto, E.; Yamada, S.; Sugawara, S.; Lu, L.; Takedachi, M.; et al. Periodontal Regeneration by Allogeneic Transplantation of Adipose Tissue Derived Multi-Lineage Progenitor Stem Cells in Vivo. Sci. Rep. 2019, 9, 921. [Google Scholar] [CrossRef]
- Sui, B.; Chen, C.; Kou, X.; Li, B.; Xuan, K.; Shi, S.; Jin, Y. Pulp Stem Cell-Mediated Functional Pulp Regeneration. J. Dent. Res. 2019, 98, 27–35. [Google Scholar] [CrossRef]
- Tassi, S.A.; Sergio, N.Z.; Misawa, M.Y.O.; Villar, C.C. Efficacy of Stem Cells on Periodontal Regeneration: Systematic Review of Pre-Clinical Studies. J. Periodontal Res. 2017, 52, 793–812. [Google Scholar] [CrossRef]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem Cells from Human Exfoliated Deciduous Teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal Human Dental Pulp Stem Cells (DPSCs) in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem Cell Properties of Human Dental Pulp Stem Cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Graziano, A.; d’Aquino, R.; Laino, G.; Papaccio, G. Dental Pulp Stem Cells: A Promising Tool for Bone Regeneration. Stem Cell Rev. 2008, 4, 21–26. [Google Scholar] [CrossRef]
- Király, M.; Porcsalmy, B.; Pataki, A.; Kádár, K.; Jelitai, M.; Molnár, B.; Hermann, P.; Gera, I.; Grimm, W.-D.; Ganss, B.; et al. Simultaneous PKC and CAMP Activation Induces Differentiation of Human Dental Pulp Stem Cells into Functionally Active Neurons. Neurochem. Int. 2009, 55, 323–332. [Google Scholar] [CrossRef]
- La Noce, M.; Stellavato, A.; Vassallo, V.; Cammarota, M.; Laino, L.; Desiderio, V.; Del Vecchio, V.; Nicoletti, G.F.; Tirino, V.; Papaccio, G.; et al. Hyaluronan-Based Gel Promotes Human Dental Pulp Stem Cells Bone Differentiation by Activating YAP/TAZ Pathway. Cells. 2021, 26, 2899. [Google Scholar] [CrossRef]
- Seo, B.M.; Sonoyama, W.; Yamaza, T.; Coppe, C.; Kikuiri, T.; Akiyama, K.; Lee, J.S.; Shi, S. SHED Repair Critical-Size Calvarial Defects in Mice. Oral Dis. 2008, 14, 428–434. [Google Scholar] [CrossRef]
- d’Aquino, R.; De Rosa, A.; Lanza, V.; Tirino, V.; Laino, L.; Graziano, A.; Desiderio, V.; Laino, G.; Papaccio, G. Human Mandible Bone Defect Repair by the Grafting of Dental Pulp Stem/Progenitor Cells and Collagen Sponge Biocomplexes. Eur. Cell. Mater. 2009, 18, 75–83. [Google Scholar] [CrossRef]
- Nakajima, K.; Kunimatsu, R.; Ando, K.; Hiraki, T.; Rikitake, K.; Tsuka, Y.; Abe, T.; Tanimoto, K. Success Rates in Isolating Mesenchymal Stem Cells from Permanent and Deciduous Teeth. Sci. Rep. 2019, 9, 16764. [Google Scholar] [CrossRef]
- Nakajima, K.; Kunimatsu, R.; Ando, K.; Ando, T.; Hayashi, Y.; Kihara, T.; Hiraki, T.; Tsuka, Y.; Abe, T.; Kaku, M.; et al. Comparison of the Bone Regeneration Ability between Stem Cells from Human Exfoliated Deciduous Teeth, Human Dental Pulp Stem Cells and Human Bone Marrow Mesenchymal Stem Cells. Biochem. Biophys. Res. Commun. 2018, 497, 876–882. [Google Scholar] [CrossRef]
- Kunimatsu, R.; Nakajima, K.; Awada, T.; Tsuka, Y.; Abe, T.; Ando, K.; Hiraki, T.; Kimura, A.; Tanimoto, K. Comparative Characterization of Stem Cells from Human Exfoliated Deciduous Teeth, Dental Pulp, and Bone Marrow–Derived Mesenchymal Stem Cells. Biochem. Biophys. Res. Commun. 2018, 501, 193–198. [Google Scholar] [CrossRef]
- Wang, X.; Sha, X.J.; Li, G.H.; Yang, F.S.; Ji, K.; Wen, L.Y.; Liu, S.Y.; Chen, L.; Ding, Y.; Xuan, K. Comparative characterization of stem cells from human exfoliated deciduous teeth and dental pulp stem cells. Arch Oral Biol. 2012, 57, 1231–1240. [Google Scholar] [CrossRef]
- Ching, H.S.; Luddin, N.; Rahman, I.A.; Ponnuraj, K.T. Expression of odontogenic and osteogenic markers in DPSCs and SHED: A review. Curr. Stem. Cell Res. Ther. 2017, 12, 71–79. [Google Scholar] [CrossRef]
- Kichenbrand, C.; Velot, E.; Menu, P.; Moby, V. Dental Pulp Stem Cell-Derived Conditioned Medium: An Attractive Alternative for Regenerative Therapy. Tissue Eng. Part B Rev. 2019, 25, 78–88. [Google Scholar] [CrossRef]
- Lee, S.H.; Looi, C.Y.; Chong, P.P.; Foo, J.B.; Looi, Q.H.; Ng, C.X.; Ibrahim, Z. Comparison of isolation, expansion and cryopreservation techniques to produce stem cells from human exfoliated deciduous teeth (SHED) with better regenerative potential. Curr. Stem. Cell Res. Ther. 2021, 16, 551–562. [Google Scholar] [CrossRef]
- Uder, C.; Brückner, S.; Winkler, S.; Tautenhahn, H.-M.; Christ, B. Mammalian MSC from Selected Species: Features and Applications: Cross-Species MSC. Cytometry A 2018, 93, 32–49. [Google Scholar] [CrossRef]
- Johnson, K.W.; Dooner, M.; Quesenberry, P.J. Fluorescence Activated Cell Sorting: A Window on the Stem Cell. Curr. Pharm. Biotechnol. 2007, 8, 133–139. [Google Scholar] [CrossRef]
- Harkness, L.; Zaher, W.; Ditzel, N.; Isa, A.; Kassem, M. CD146/MCAM Defines Functionality of Human Bone Marrow Stromal Stem Cell Populations. Stem Cell Res. Ther. 2016, 7, 4. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, X. CD146, a Multi-Functional Molecule beyond Adhesion. Cancer Lett. 2013, 330, 150–162. [Google Scholar] [CrossRef]
- Tsang, W.P.; Shu, Y.; Kwok, P.L.; Zhang, F.; Lee, K.K.H.; Tang, M.K.; Li, G.; Chan, K.M.; Chan, W.-Y.; Wan, C. CD146+ Human Umbilical Cord Perivascular Cells Maintain Stemness under Hypoxia and as a Cell Source for Skeletal Regeneration. PLoS ONE 2013, 8, e76153. [Google Scholar] [CrossRef]
- Ulrich, C.; Abruzzese, T.; Maerz, J.K.; Ruh, M.; Amend, B.; Benz, K.; Rolauffs, B.; Abele, H.; Hart, M.L.; Aicher, W.K. Human Placenta-Derived CD146-Positive Mesenchymal Stromal Cells Display a Distinct Osteogenic Differentiation Potential. Stem. Cells Dev. 2015, 24, 1558–1569. [Google Scholar] [CrossRef]
- Rikitake, K.; Kunimatsu, R.; Yoshimi, Y.; Nakajima, K.; Hiraki, T.; Aisyah Rizky Putranti, N.; Tsuka, Y.; Abe, T.; Ando, K.; Hayashi, Y.; et al. Effect of CD146+ SHED on Bone Regeneration in a Mouse Calvaria Defect Model. Oral Dis. 2021, 29, 725–734. [Google Scholar] [CrossRef]
- Oh, M.; Nör, J.E. The Perivascular Niche and Self-Renewal of Stem Cells. Front. Physiol. 2015, 6, 367. [Google Scholar] [CrossRef]
- Oh, M.; Zhang, Z.; Mantesso, A.; Oklejas, A.E.; Nör, J.E. Endothelial-Initiated Crosstalk Regulates Dental Pulp Stem Cell Self-Renewal. J. Dent. Res. 2020, 99, 1102–1111. [Google Scholar] [CrossRef]
- Kfoury, Y.; Scadden, D.T. Mesenchymal Cell Contributions to the Stem Cell Niche. Cell Stem. Cell 2015, 16, 239–253. [Google Scholar] [CrossRef]
- Leroyer, A.S.; Blin, M.G.; Bachelier, R.; Bardin, N.; Blot-Chabaud, M.; Dignat-George, F. CD146 (Cluster of Differentiation 146): An Adhesion Molecule Involved in Vessel Homeostasis. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1026–1033. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, Q.; Zhang, N.; Du, X.; Xu, G.; Yan, X. CD146, from a Melanoma Cell Adhesion Molecule to a Signaling Receptor. Signal Transduct. Target. Ther. 2020, 5, 148. [Google Scholar] [CrossRef]
- Zeng, Q.; Wu, Z.; Duan, H.; Jiang, X.; Tu, T.; Lu, D.; Luo, Y.; Wang, P.; Song, L.; Feng, J.; et al. Impaired Tumor Angiogenesis and VEGF-Induced Pathway in Endothelial CD146 Knockout Mice. Protein Cell 2014, 5, 445–456. [Google Scholar] [CrossRef]
- Jiang, T.; Zhuang, J.; Duan, H.; Luo, Y.; Zeng, Q.; Fan, K.; Yan, H.; Lu, D.; Ye, Z.; Hao, J.; et al. CD146 Is a Coreceptor for VEGFR-2 in Tumor Angiogenesis. Blood 2012, 120, 2330–2339. [Google Scholar] [CrossRef] [PubMed]
- Espagnolle, N.; Guilloton, F.; Deschaseaux, F.; Gadelorge, M.; Sensébé, L.; Bourin, P. CD146 Expression on Mesenchymal Stem Cells Is Associated with Their Vascular Smooth Muscle Commitment. J. Cell. Mol. Med. 2014, 18, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P. “Mesenchymal” Stem Cells. Annu. Rev. Cell Dev. Biol. 2014, 30, 677–704. [Google Scholar] [CrossRef]
- Crisan, M.; Yap, S.; Casteilla, L.; Chen, C.-W.; Corselli, M.; Park, T.S.; Andriolo, G.; Sun, B.; Zheng, B.; Zhang, L.; et al. A Perivascular Origin for Mesenchymal Stem Cells in Multiple Human Organs. Cell Stem. Cell 2008, 3, 301–313. [Google Scholar] [CrossRef]
- Covas, D.T.; Panepucci, R.A.; Fontes, A.M.; Silva, W.A., Jr.; Orellana, M.D.; Freitas, M.C.C.; Neder, L.; Santos, A.R.D.; Peres, L.C.; Jamur, M.C.; et al. Multipotent Mesenchymal Stromal Cells Obtained from Diverse Human Tissues Share Functional Properties and Gene-Expression Profile with CD146+ Perivascular Cells and Fibroblasts. Exp. Hematol. 2008, 36, 642–654. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human Mesenchymal Stem Cells-Current Trends and Future Prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Nourbakhsh, N.; Soleimani, M.; Taghipour, Z.; Karbalaie, K.; Mousavi, S.-B.; Talebi, A.; Nadali, F.; Tanhaei, S.; Kiyani, G.-A.; Nematollahi, M.; et al. Induced in Vitro Differentiation of Neural-like Cells from Human Exfoliated Deciduous Teeth-Derived Stem Cells. Int. J. Dev. Biol. 2011, 55, 189–195. [Google Scholar] [CrossRef]
- Kerkis, I.; Caplan, A.I. Stem Cells in Dental Pulp of Deciduous Teeth. Tissue Eng. Part B Rev. 2012, 18, 129–138. [Google Scholar] [CrossRef]
- Fayazi, M.; Salehnia, M.; Ziaei, S. Department of Medical Sciences, Najafabad Branch, Islamic Azad University, Najafabad, Iran; Department of Anatomy, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran; Department of Midwifery, Faculty of Medical Sciences Tarbiat Modares University, Tehran, Iran The Effect of Stem Cell Factor on Proliferation of Human Endometrial CD146+ Cells. Int. J. Reprod. Biomed. (Yazd) 2016, 14, 437–442. [Google Scholar] [CrossRef]
- Zhu, W.; Tan, Y.; Qiu, Q.; Li, X.; Huang, Z.; Fu, Y.; Liang, M. Comparison of the Properties of Human CD146+ and CD146− Periodontal Ligament Cells in Response to Stimulation with Tumour Necrosis Factor α. Arch. Oral Biol. 2013, 58, 1791–1803. [Google Scholar] [CrossRef]
- Paduano, F.; Marrelli, M.; Palmieri, F.; Tatullo, M. CD146 Expression Influences Periapical Cyst Mesenchymal Stem Cell Properties. Stem Cell Rev. 2016, 12, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Endo, I.; Mastumoto, T. Bone and Stem Cells. Regulatory mechanism of mesenchymal stem cell differentiation to osteoblasts. Clin. Calcium 2014, 24, 555–564. [Google Scholar] [PubMed]
- Ohgushi, H.; Tamai, S.; Dohi, Y.; Katuda, T.; Tabata, S.; Suwa, Y. In Vitro Bone Formation by Rat Marrow Cell Culture. J. Biomed. Mater. Res. 1996, 32, 333–340. [Google Scholar] [CrossRef]
- Fakhry, M. Molecular Mechanisms of Mesenchymal Stem Cell Differentiation towards Osteoblasts. World J. Stem Cells 2013, 5, 136. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Olsen, B.R. Osteoblast-Derived VEGF Regulates Osteoblast Differentiation and Bone Formation during Bone Repair. J. Clin. Invest. 2016, 126, 509–526. [Google Scholar] [CrossRef]
- Hu, K.; Olsen, B.R. The Roles of Vascular Endothelial Growth Factor in Bone Repair and Regeneration. Bone 2016, 91, 30–38. [Google Scholar] [CrossRef]
- D’ Alimonte, I.; Nargi, E.; Mastrangelo, F.; Falco, G.; Lanuti, P.; Marchisio, M.; Miscia, S.; Robuffo, I.; Capogreco, M.; Buccella, S.; et al. Vascular Endothelial Growth Factor Enhances in Vitro Proliferation and Osteogenic Differentiation of Human Dental Pulp Stem Cells. J. Biol. Regul. Homeost. Agents 2011, 25, 57–69. [Google Scholar]
- Bai, Y.; Li, P.; Yin, G.; Huang, Z.; Liao, X.; Chen, X.; Yao, Y. BMP-2, VEGF and BFGF Synergistically Promote the Osteogenic Differentiation of Rat Bone Marrow-Derived Mesenchymal Stem Cells. Biotechnol. Lett. 2013, 35, 301–308. [Google Scholar] [CrossRef]
- Muraglia, A.; Cancedda, R.; Quarto, R. Clonal Mesenchymal Progenitors from Human Bone Marrow Differentiate in Vitro According to a Hierarchical Model. J. Cell Sci. 2000, 113, 1161–1166. [Google Scholar] [CrossRef]

| Gene | Sequence (5′→3′) | |
|---|---|---|
| GAPDH | Forward | CCA CTC CTC CAC CTT TGA |
| Reverse | CAC CAC CCT GTT GCT GTA | |
| ALP | Forward | ATG GTG GAC TGC TCA CAA C |
| Reverse | GAC GTA GTT CTG CTC GTG GA | |
| BMP-2 | Forward | AAC ACT GTG CGC AGC TTC C |
| Reverse | CTC CGG GTT GTT TTC CCA C | |
| OCN | Forward | GCA GAG TCC AGG AAA GGG TG |
| Reverse | GTC AGC AAC TCG TCA CAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunimatsu, R.; Rikitake, K.; Yoshimi, Y.; Putranti, N.A.R.; Hayashi, Y.; Tanimoto, K. Bone Differentiation Ability of CD146-Positive Stem Cells from Human Exfoliated Deciduous Teeth. Int. J. Mol. Sci. 2023, 24, 4048. https://doi.org/10.3390/ijms24044048
Kunimatsu R, Rikitake K, Yoshimi Y, Putranti NAR, Hayashi Y, Tanimoto K. Bone Differentiation Ability of CD146-Positive Stem Cells from Human Exfoliated Deciduous Teeth. International Journal of Molecular Sciences. 2023; 24(4):4048. https://doi.org/10.3390/ijms24044048
Chicago/Turabian StyleKunimatsu, Ryo, Kodai Rikitake, Yuki Yoshimi, Nurul Aisyah Rizky Putranti, Yoko Hayashi, and Kotaro Tanimoto. 2023. "Bone Differentiation Ability of CD146-Positive Stem Cells from Human Exfoliated Deciduous Teeth" International Journal of Molecular Sciences 24, no. 4: 4048. https://doi.org/10.3390/ijms24044048
APA StyleKunimatsu, R., Rikitake, K., Yoshimi, Y., Putranti, N. A. R., Hayashi, Y., & Tanimoto, K. (2023). Bone Differentiation Ability of CD146-Positive Stem Cells from Human Exfoliated Deciduous Teeth. International Journal of Molecular Sciences, 24(4), 4048. https://doi.org/10.3390/ijms24044048







