Association of FANCM Mutations with Familial and Early-Onset Breast Cancer Risk in a South American Population
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Families
4.2. Controls
4.3. FANCM rs147021911 and rs144567652 Genotyping
4.4. Sanger Sequencing Confirmation
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- McPherson, K.; Steel, C.M.; Dixon, J.M. ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ 2000, 321, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F. Familial risks of breast cancer. Breast Cancer Res. 2002, 4, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Skolnick, M.H.; Cannon-Albright, L.A. Genetic predisposition to breast cancer. Cancer 1992, 70, 1747–1754. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Risch, N.; Thompson, W.D. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am. J. Hum. Genet. 1991, 48, 232–242. [Google Scholar]
- Stratton, M.R.; Rahman, N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008, 40, 17–22. [Google Scholar] [CrossRef]
- Couch, F.J.; Hart, S.N.; Sharma, P.; Toland, A.E.; Wang, X.; Miron, P.; Olson, J.E.; Godwin, A.K.; Pankratz, V.S.; Olswold, C.; et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015, 33, 304–311. [Google Scholar] [CrossRef]
- Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br. J. Cancer 2000, 83, 1301–1308. [Google Scholar] [CrossRef]
- Bogliolo, M.; Bluteau, D.; Lespinasse, J.; Pujol, R.; Vasquez, N.; d’Enghien, C.D.; Stoppa-Lyonnet, D.; Leblanc, T.; Soulier, J.; Surralles, J. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet. Med. 2018, 20, 458–463. [Google Scholar] [CrossRef]
- Nurmi, A.; Muranen, T.A.; Pelttari, L.M.; Kiiski, J.I.; Heikkinen, T.; Lehto, S.; Kallioniemi, A.; Schleutker, J.; Butzow, R.; Blomqvist, C.; et al. Recurrent moderate-risk mutations in Finnish breast and ovarian cancer patients. Int. J. Cancer 2019, 145, 2692–2700. [Google Scholar] [CrossRef]
- Kiiski, J.I.; Tervasmaki, A.; Pelttari, L.M.; Khan, S.; Mantere, T.; Pylkas, K.; Mannermaa, A.; Tengstrom, M.; Kvist, A.; Borg, A.; et al. FANCM mutation c.5791C>T is a risk factor for triple-negative breast cancer in the Finnish population. Breast Cancer Res. Treat 2017, 166, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Figlioli, G.; Kvist, A.; Tham, E.; Soukupova, J.; Kleiblova, P.; Muranen, T.A.; Andrieu, N.; Azzollini, J.; Balmana, J.; Barroso, A.; et al. The Spectrum of FANCM Protein Truncating Variants in European Breast Cancer Cases. Cancer 2020, 12, 292. [Google Scholar] [CrossRef]
- Basbous, J.; Constantinou, A. A tumor suppressive DNA translocase named FANCM. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Peterlongo, P.; Catucci, I.; Colombo, M.; Caleca, L.; Mucaki, E.; Bogliolo, M.; Marin, M.; Damiola, F.; Bernard, L.; Pensotti, V.; et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum. Mol. Genet. 2015, 24, 5345–5355. [Google Scholar] [CrossRef]
- Kiiski, J.I.; Pelttari, L.M.; Khan, S.; Freysteinsdottir, E.S.; Reynisdottir, I.; Hart, S.N.; Shimelis, H.; Vilske, S.; Kallioniemi, A.; Schleutker, J.; et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 15172–15177. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Aznarez, F.J.; Fernandez, V.; Pita, G.; Peterlongo, P.; Dominguez, O.; de la Hoya, M.; Duran, M.; Osorio, A.; Moreno, L.; Gonzalez-Neira, A.; et al. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PLoS ONE 2013, 8, e55681. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; van Luttikhuizen, J.L.; Auber, B.; Schmidt, G.; Hofmann, W.; Penkert, J.; Davenport, C.F.; Hille-Betz, U.; Wendeburg, L.; Bublitz, J.; et al. The identification of pathogenic variants in BRCA1/2 negative, high risk, hereditary breast and/or ovarian cancer patients: High frequency of FANCM pathogenic variants. Int. J. Cancer 2019, 144, 2683–2694. [Google Scholar] [CrossRef]
- Nguyen-Dumont, T.; Myszka, A.; Karpinski, P.; Sasiadek, M.M.; Akopyan, H.; Hammet, F.; Tsimiklis, H.; Park, D.J.; Pope, B.J.; Slezak, R.; et al. FANCM and RECQL genetic variants and breast cancer susceptibility: Relevance to South Poland and West Ukraine. BMC Med. Genet. 2018, 19, 12. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef]
- Mamrak, N.E.; Shimamura, A.; Howlett, N.G. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017, 31, 93–99. [Google Scholar] [CrossRef]
- Inano, S.; Sato, K.; Katsuki, Y.; Kobayashi, W.; Tanaka, H.; Nakajima, K.; Nakada, S.; Miyoshi, H.; Knies, K.; Takaori-Kondo, A.; et al. RFWD3-Mediated Ubiquitination Promotes Timely Removal of Both RPA and RAD51 from DNA Damage Sites to Facilitate Homologous Recombination. Mol. Cell 2017, 66, 622–634.e8. [Google Scholar] [CrossRef] [PubMed]
- Knies, K.; Inano, S.; Ramirez, M.J.; Ishiai, M.; Surralles, J.; Takata, M.; Schindler, D. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Investig. 2017, 127, 3013–3027. [Google Scholar] [CrossRef] [PubMed]
- Nepal, M.; Che, R.; Ma, C.; Zhang, J.; Fei, P. FANCD2 and DNA Damage. Int. J. Mol. Sci. 2017, 18, 1804. [Google Scholar] [CrossRef] [PubMed]
- Nepal, M.; Che, R.; Zhang, J.; Ma, C.; Fei, P. Fanconi Anemia Signaling and Cancer. Trends Cancer 2017, 3, 840–856. [Google Scholar] [CrossRef]
- Kiiski, J.I.; Fagerholm, R.; Tervasmaki, A.; Pelttari, L.M.; Khan, S.; Jamshidi, M.; Mantere, T.; Pylkas, K.; Bartek, J.; Bartkova, J.; et al. FANCM c.5101C>T mutation associates with breast cancer survival and treatment outcome. Int. J. Cancer 2016, 139, 2760–2770. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Cruz-Coke, R. Ethnic origin and evolution of the Chilean population. Rev. Med. Chil. 1976, 104, 365–368. [Google Scholar]
- Valenzuela, C.Y.; Acuna, M.P.; Harb, Z. Sociogenetic gradient in the Chilean population. Rev. Med. Chil. 1987, 115, 295–299. [Google Scholar]
- Fuentes, M.; Pulgar, I.; Gallo, C.; Bortolini, M.C.; Canizales-Quinteros, S.; Bedoya, G.; Gonzalez-Jose, R.; Ruiz-Linares, A.; Rothhammer, F. Gene geography of Chile: Regional distribution of American, European and African genetic contributions. Rev. Med. Chil. 2014, 142, 281–289. [Google Scholar] [CrossRef]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–534, 536–537. [Google Scholar]
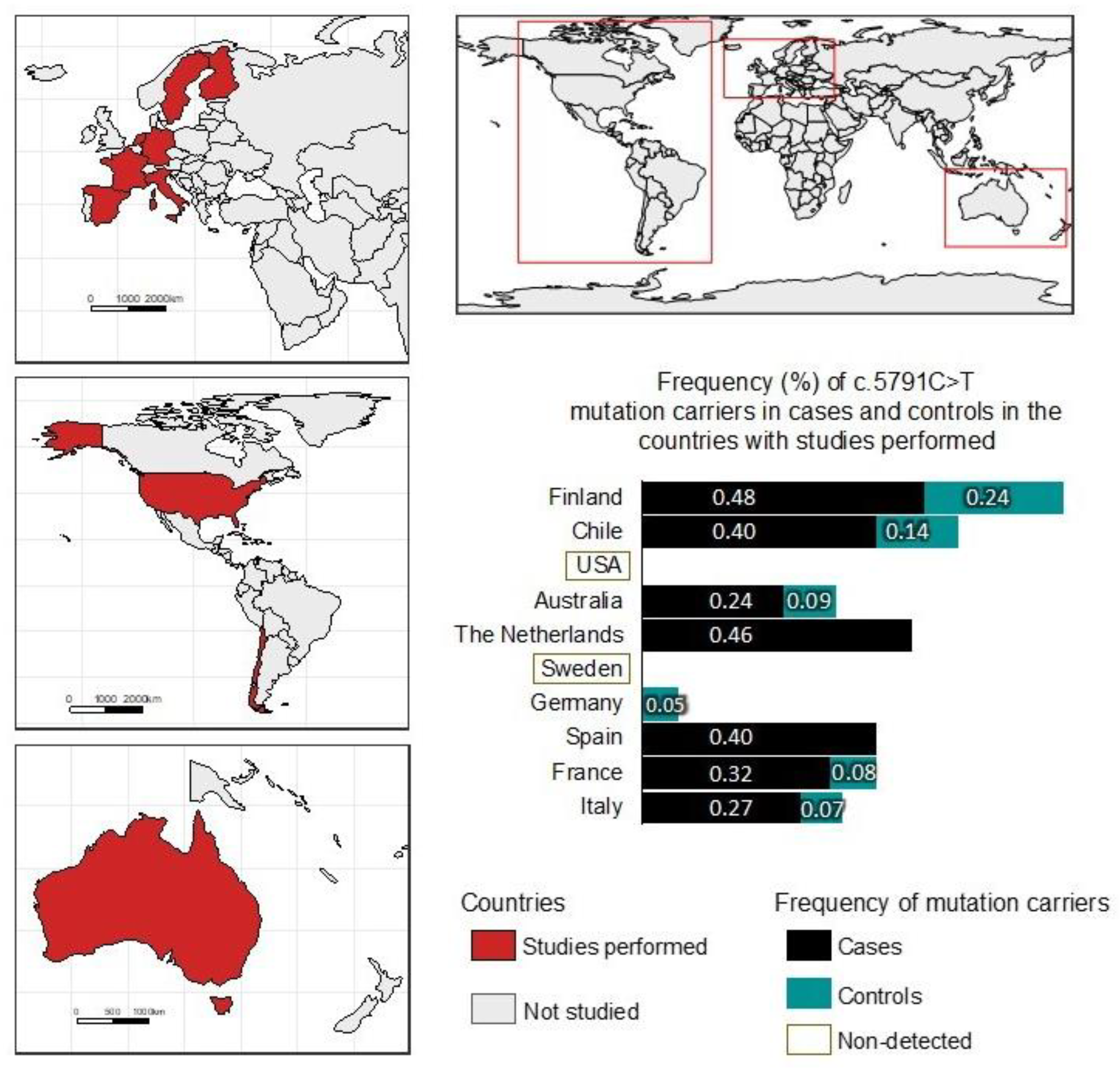
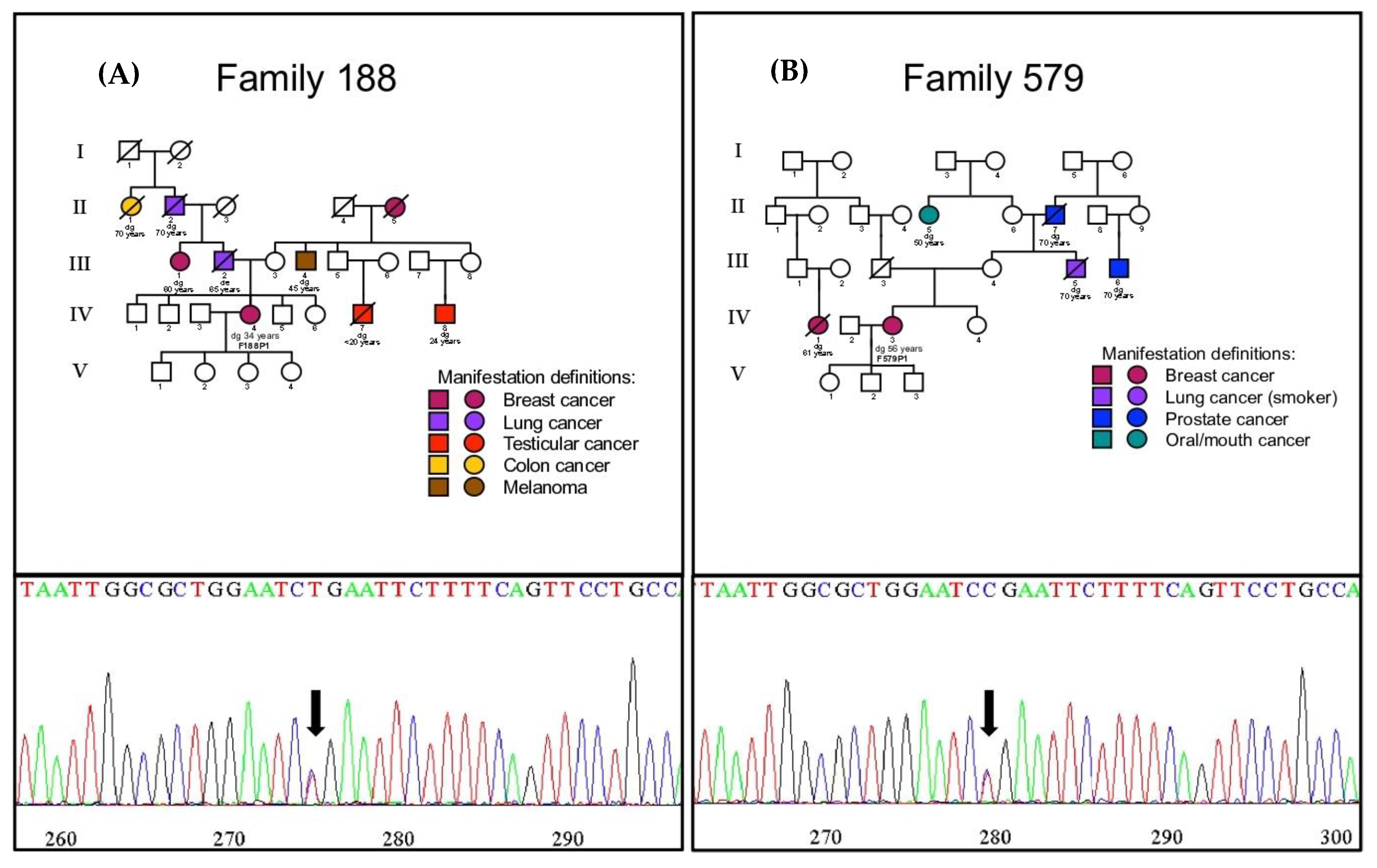
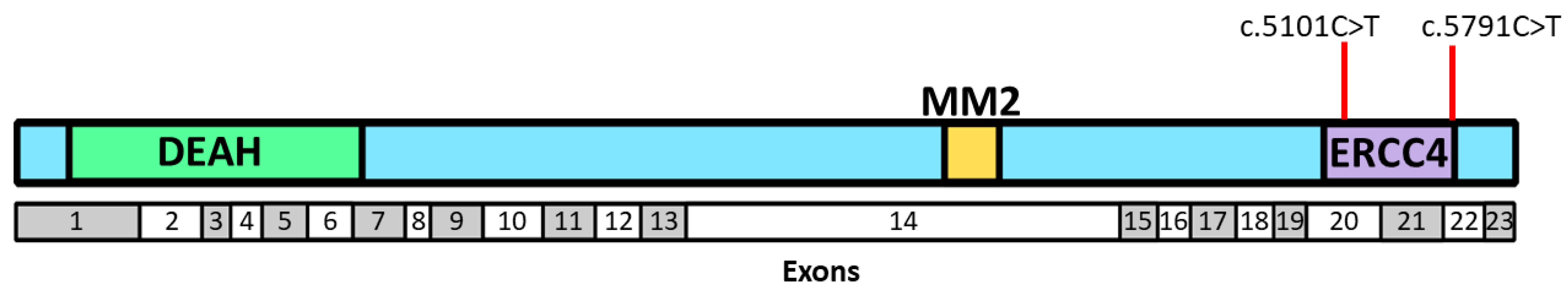
| All BC Cases (n = 492) | Families with ≥2 BC and/or OC Cases (n = 314) | Families with a Single Case, Diagnosis at ≤50 Years of Age (n = 178) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype or Allele | Controls (%) (n = 673) | BC Cases (%) | OR [95% CI] | p-Value a | BC Cases (%) | OR [95% CI] | p-Value a | BC Cases (%) | OR [95% CI] | p-Value a |
| C/C | 672 (99.9) | 490 (99.4) | (ref) | - | 312 (99.4) | (ref) | - | 178 (100) | (ref) | - |
| C/T | 1 (0.1) | 2 (0.4) | 2.7 [0.3–39.8] | 0.5 | 2 (0.6) | 4.3 [0.4–62.5] | 0.2 | 0 (0.0) | - | - |
| T/T | 0 (0.0) | 0 (0.0) | - | - | 0 (0.0) | - | - | 0 (0.0) | - | - |
| C/T + T/T | 1 (0.1) | 2 (0.4) | 2.7 [0.3–38.8] | 0.1 | 2 (0.6) | 4.3 [0.4–62.5] | 0.2 | 0 (0.0) | - | - |
| Allele C | 1345 (99.9) | 982 (99.8) | (ref) | - | 626 (99.7) | (ref) | - | 356 (0.0) | (ref) | - |
| Allele T | 1 (0.1) | 2 (0.2) | 2.7 [0.3–39.7] | 0.5 | 2 (0.3) | 4.2 [0.4–62.3] | 0.2 | 0 (0.0) | - | - |
| Families with 2 BC and/or OC Cases (n = 166) | Families with ≥3 BC and/or OC Cases (n = 148) | ||||||
|---|---|---|---|---|---|---|---|
| Genotype or Allele | Controls (%) (n = 673) | BC Cases (%) | OR [95% CI] | p-Value a | BC Cases (%) | OR [95% CI] | p-Value a |
| C/C | 672 (99.9) | 164 (98.8) | (ref) | - | 148 (100) | (ref) | - |
| C/T | 1 (0.1) | 2 (1.2) | 8.1 [0.9–118.9] | 0.1 | 0 (0.0) | - | - |
| T/T | 0 (0.0) | 0 (0.0) | - | - | 0 (0.0) | - | - |
| C/T + T/T | 1 (0.1) | 2 (1.2) | 8.1 [0.9–118.9] | 0.1 | 0 (0.0) | - | - |
| Allele C | 1345 (99.9) | 330 (99.4) | (ref) | - | 296 (100) | (ref) | - |
| Allele T | 1 (0.1) | 2 (0.6) | 8.1 [0.9–118.2] | 0.1 | 0 (0.0) | - | - |
| Inclusion Criteria | Families: n |
|---|---|
| Three or more family members with breast and/or ovarian cancer | 148 (29.8%) |
| Two family members with breast and/or ovarian cancer | 166 (33.6%) |
| Single affected individuals with breast cancer aged ≤35 | 87 (17.9%) |
| Single affected individuals with breast cancer aged 36–50 | 91 (18.7%) |
| TOTAL | 492 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Pison, S.; Morales-González, S.; Fernandez-Ramires, R.; Tapia, J.C.; Maldonado, E.; Calaf, G.M.; Jara, L. Association of FANCM Mutations with Familial and Early-Onset Breast Cancer Risk in a South American Population. Int. J. Mol. Sci. 2023, 24, 4041. https://doi.org/10.3390/ijms24044041
Morales-Pison S, Morales-González S, Fernandez-Ramires R, Tapia JC, Maldonado E, Calaf GM, Jara L. Association of FANCM Mutations with Familial and Early-Onset Breast Cancer Risk in a South American Population. International Journal of Molecular Sciences. 2023; 24(4):4041. https://doi.org/10.3390/ijms24044041
Chicago/Turabian StyleMorales-Pison, Sebastian, Sarai Morales-González, Ricardo Fernandez-Ramires, Julio C. Tapia, Edio Maldonado, Gloria M. Calaf, and Lilian Jara. 2023. "Association of FANCM Mutations with Familial and Early-Onset Breast Cancer Risk in a South American Population" International Journal of Molecular Sciences 24, no. 4: 4041. https://doi.org/10.3390/ijms24044041
APA StyleMorales-Pison, S., Morales-González, S., Fernandez-Ramires, R., Tapia, J. C., Maldonado, E., Calaf, G. M., & Jara, L. (2023). Association of FANCM Mutations with Familial and Early-Onset Breast Cancer Risk in a South American Population. International Journal of Molecular Sciences, 24(4), 4041. https://doi.org/10.3390/ijms24044041








