Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome
Abstract
1. Introduction
2. Serological and Urine Biomarkers
2.1. Classical Markers
2.2. New Markers
3. Cellular Biomarkers
3.1. Classical Markers
3.2. New Markers
4. Genomic and Transcriptomic Biomarkers
4.1. Classical Markers
4.2. New Markers
5. Computational Prediction Biomarkers
5.1. Classical Markers
5.2. New Markers
6. Cell-Free DNA Biomarkers
6.1. History
6.2. Is It Useful and Where Is cfDNA Analysis Headed?
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Aldea, P.L.; Rachisan, A.L.; Stanciu, B.I.; Picos, A.; Picos, A.M.; Delean, D.I.; Stroescu, R.; Starcea, M.I.; Borzan, C.M.; Elec, F.I. The Perspectives of Biomarkers in Predicting the Survival of the Renal Graft. Front. Pediatr. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Budde, K.; Osmanodja, B.; Bornemann-Kolatzki, K.; Beck, J.; Schütz, E.; Walson, P.D. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front. Genet. 2022, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mezzolla, V.; Pontrelli, P.; Fiorentino, M.; Stasi, A.; Franzin, R.; Rascio, F.; Grandaliano, G.; Stallone, G.; Infante, B.; Gesualdo, L.; et al. Emerging biomarkers of delayed graft function in kidney transplantation. Transplant. Rev. 2021, 35, 100629. [Google Scholar] [CrossRef]
- DeLuca, D.S.; Blasczyk, R. HistoCheck. Evaluating structural and functional MHC similarities. Methods Mol. Biol. 2007, 409, 395–405. [Google Scholar] [PubMed]
- Duquesnoy, R.J. Reflections on HLA epitope-based matching for transplantation. Front. Immunol. 2016, 7, 469. [Google Scholar] [CrossRef]
- Choudhary, N.S.; Saigal, S.; Bansal, R.K.; Saraf, N.; Gautam, D.; Soin, A.S. Acute and Chronic Rejection After Liver Transplantation: What A Clinician Needs to Know. J. Clin. Exp. Hepatol. 2017, 7, 358–366. [Google Scholar] [CrossRef]
- Pontrelli, P.; Rascio, F.; Castellano, G.; Grandaliano, G.; Gesualdo, L.; Stallone, G. The Role of Natural Killer Cells in the Immune Response in Kidney Transplantation. Front. Immunol. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Lemieux, W.; Fleischer, D.; Yang, A.Y.; Niemann, M.; Oualkacha, K.; Klement, W.; Richard, L.; Polychronakos, C.; Liwski, R.; Claas, F.; et al. Dissecting the impact of molecular T-cell HLA mismatches in kidney transplant failure: A retrospective cohort study. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Boix, F.; Trujillo, C.; Muro, M. Cell-Mediated Immunity (CMI) as the Instrument to Assess the Response Against the Allograft: Present and Future. Curr. Protein Pept. Sci. 2018, 19, 1092–1106. [Google Scholar] [CrossRef]
- Chenouard, A.; Chesneau, M.; Bui Nguyen, L.; Le Bot, S.; Cadoux, M.; Dugast, E.; Paul, C.; Malard-Castagnet, S.; Ville, S.; Guérif, P.; et al. Renal Operational Tolerance Is Associated with a Defect of Blood Tfh Cells That Exhibit Impaired B Cell Help. Am. J. Transplant. 2017, 17, 1490–1501. [Google Scholar] [CrossRef]
- Cano-Romero, F.L.; Laguna Goya, R.; Utrero-Rico, A.; Gómez-Massa, E.; Arroyo-Sánchez, D.; Suárez-Fernández, P.; Lora, D.; Andrés, A.; Castro-Panete, M.J.; Paz-Artal, E. Longitudinal profile of circulating T follicular helper lymphocytes parallels anti-HLA sensitization in renal transplant recipients. Am. J. Transplant. 2019, 19, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Briceño, J. Artificial intelligence and organ transplantation: Challenges and expectations. Curr. Opin. Organ Transplant. 2020, 25, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Neupane, A.S.; Kubes, P. Imaging reveals novel innate immune responses in lung, liver, and beyond. Immunol. Rev. 2022, 306, 244–257. [Google Scholar] [CrossRef]
- Demetris, A.J.; Bellamy, C.; Hübscher, S.G.; O’Leary, J.; Randhawa, P.S.; Feng, S.; Neil, D.; Colvin, R.B.; McCaughan, G.; Fung, J.J.; et al. 2016 comprehensive update of the Banff working group on liver allograft pathology: Introduction of antibody-mediated rejection. Am. J. Transplant. 2016, 16, 2816–2835. [Google Scholar] [CrossRef]
- Angelico, R.; Sensi, B.; Manzia, T.M.; Tisone, G.; Grassi, G.; Signorello, A.; Milana, M.; Lenci, I.; Baiocchi, L. Chronic rejection after liver transplantation: Opening the Pandora’s box. World J. Gastroenterol. 2021, 27, 7771. [Google Scholar] [CrossRef] [PubMed]
- Leibler, C.; Matignon, M.; Pilon, C.; Montespan, F.; Bigot, J.; Lang, P.; Carosella, E.D.; Cohen, J.; Rouas-Freiss, N.; Grimbert, P.; et al. Kidney Transplant Recipients Treated with Belatacept Exhibit Increased Naïve and Transitional B Cells. Am. J. Transplant. 2014, 14, 1173–1182. [Google Scholar] [CrossRef]
- Jaikaransingh, V.; Kadambi, P.V. Donor-derived cell-free DNA (ddcf-DNA) and acute antibody-mediated rejection in kidney transplantation. Medicina 2021, 57, 436. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, D.; Jiang, T. The role of donor-derived cell-free DNA in the detection of renal allograft injury. Nephrol. Ther. 2021, 17, 12–17. [Google Scholar] [CrossRef]
- Beck, J.; Oellerich, M.; Schulz, U.; Schauerte, V.; Reinhard, L.; Fuchs, U.; Knabbe, C.; Zittermann, A.; Olbricht, C.; Gummert, J.F.; et al. Donor-Derived Cell-Free DNA Is a Novel Universal Biomarker for Allograft Rejection in Solid Organ Transplantation. Transplant. Proc. 2015, 47, 2400–2403. [Google Scholar] [CrossRef]
- Oellerich, M.; Sherwood, K.; Keown, P.; Schütz, E.; Beck, J.; Stegbauer, J.; Rump, L.C.; Walson, P.D. Liquid biopsies: Donor-derived cell-free DNA for the detection of kidney allograft injury. Nat. Rev. Nephrol. 2021, 17, 591–603. [Google Scholar] [CrossRef]
- Crichton, E.S.; Zeng, S.; la Muraglia, G.M.; Badell, I.R. CXCL13 Is an Indicator of Germinal Center Activity and Alloantibody Formation Following Transplantation. Transplant. Direct 2021, 7, E785. [Google Scholar] [CrossRef] [PubMed]
- Găman, M.A.; Cozma, M.A.; Dobrică, E.C.; Crețoiu, S.M.; Găman, A.M.; Diaconu, C.C. Liquid biopsy and potential liquid biopsy-based biomarkers in philadelphia-negative classical myeloproliferative neoplasms: A systematic review. Life 2021, 11, 677. [Google Scholar] [CrossRef]
- Boix, F.; Millan, O.; San Segundo, D.; Mancebo, E.; Rimola, A.; Fabrega, E.; Fortuna, V.; Mrowiec, A.; Castro-Panete, M.J.; de la Peña, J.; et al. High expression of CD38, CD69, CD95 and CD154 biomarkers in cultured peripheral T lymphocytes correlates with an increased risk of acute rejection in liver allograft recipients. Immunobiology 2016, 221, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Boix, F.; Legaz, I.; Minhas, A.; Alfaro, R.; Jiménez–Coll, V.; Mrowiec, A.; Martínez–Banaclocha, H.; Galián, J.A.; Botella, C.; Moya–Quiles, M.R.; et al. Identification of peripheral CD154+ T cells and HLA-DRB1 as biomarkers of acute cellular rejection in adult liver transplant recipients. Clin. Exp. Immunol. 2021, 203, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Boix, F.; Bolarín, J.M.; Mrowiec, A.; Eguía, J.; Gonzalez-Martinez, G.; de la Peña, J.; Galian, J.A.; Alfaro, R.; Moya-Quiles, M.R.; Legaz, I.; et al. CD28 biomarker quantification and expression level profiles in CD4+ T-lymphocytes in solid organ transplantation. Transpl. Immunol. 2017, 42, 9–17. [Google Scholar] [CrossRef]
- Alfaro, R.; Legaz, I.; González-Martínez, G.; Jimenez-Coll, V.; Martínez-Banaclocha, H.; Galián, J.A.; Botella, C.; de la Peña-Moral, J.; Moya-Quiles, M.R.; Campillo, J.A.; et al. Monitoring of b cell in kidney transplantation: Development of a novel clusters analysis and role of transitional b cells in transplant outcome. Diagnostics 2021, 11, 641. [Google Scholar] [CrossRef] [PubMed]
- Boix-Giner, F.; Millan, O.; San Segundo, D.; Muñoz-Cacho, P.; Mancebo, E.; Llorente, S.; Rafael-Valdivia, L.; Rimola, A.; Fábrega, E.; Mrowiec, A.; et al. High frequency of central memory regulatory T cells allows detection of liver recipients at risk of early acute rejection within the first month after transplantation. Int. Immunol. 2016, 28, 55–64. [Google Scholar] [CrossRef]
- Seiler, L.K.; Phung, N.L.; Nikolin, C.; Immenschuh, S.; Erck, C.; Kaufeld, J.; Haller, H.; Falk, C.S.; Jonczyk, R.; Lindner, P.; et al. An Antibody-Aptamer-Hybrid Lateral Flow Assay for Detection of CXCL9 in Antibody-Mediated Rejection after Kidney Transplantation. Diagnostics 2022, 12, 308. [Google Scholar] [CrossRef]
- López-Álvarez, M.R.; Moya-Quiles, M.R.; Minguela, A.; Gil, J.; Miras, M.; Campillo, J.A.; Díaz-Alderete, M.A.; García-Alonso, A.M.; Sánchez-Bueno, F.; Vicario, J.L.; et al. HLA-C matching and liver transplants: Donor-recipient genotypes influence early outcome and CD8+KIR2D+ T-cells recuperation. Transplantation 2009, 88, S54–S61. [Google Scholar] [CrossRef] [PubMed]
- Heyne, N.; Kemmner, S.; Schneider, C.; Nadalin, S.; Königsrainer, A.; Häring, H.U. Urinary neutrophil gelatinase-associated lipocalin accurately detects acute allograft rejection among other causes of acute kidney injury in renal allograft recipients. Transplantation 2012, 93, 1252–1257. [Google Scholar] [CrossRef]
- He, J.; Tsai, L.M.; Leong, Y.A.; Hu, X.; Ma, C.S.; Chevalier, N.; Sun, X.; Vandenberg, K.; Rockman, S.; Ding, Y.; et al. Circulating Precursor CCR7loPD-1hi CXCR5+ CD4+ T Cells Indicate Tfh Cell Activity and Promote Antibody Responses upon Antigen Reexposure. Immunity 2013, 39, 770–781. [Google Scholar] [CrossRef] [PubMed]
- El band, J.E.K.; Llorente, S.; Martinez-Garcia, P.; Alfaro, R.; Jimenez-Coll, V.; Boix, F.; Galián, J.A.; Martinez-Banaclocha, H.; Botella, C.; Moya-Quiles, M.R.; et al. Evaluation of Antibodies Directed Against Two GPCRs, Anti-AT1R and Anti-ETAR, on Kidney Transplant Outcome. Curr. Protein Pept. Sci. 2021, 22, 745–757. [Google Scholar] [CrossRef]
- Alfaro, R.; Jaouad, E.K.E.B.; Llorente, S.; Jimenez-Coll, V.; Martínez-Banaclocha, H.; Galián, J.A.; Botella, C.; Moya-Quiles, M.R.; Peña-Moral, J.D.L.; Minguela, A.; et al. Personalized Medicine for Kidney Transplantation: Association of Graft Survival and Acute Transplant Rejection with Genetic Variation in B Cell Activating Factor System Signaling. OMICS A J. Integr. Biol. 2021, 25, 725–737. [Google Scholar] [CrossRef]
- Alfaro, R.; Legaz, I.; Jimenez-Coll, V.; El Kaaoui El Band, J.; Martínez-Banaclocha, H.; Galián, J.A.; Parrado, A.; Mrowiec, A.; Botella, C.; Moya-Quiles, M.R.; et al. MicroRNA Expression Changes in Kidney Transplant: Diagnostic Efficacy of miR-150-5p as Potential Rejection Biomarker, Pilot Study. J. Clin. Med. 2021, 10, 2748. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef] [PubMed]
- Pattar, S.; Aleinati, M.; Iqbal, F.; Madhu, A.; Blais, S.; Wang, X.; Dallaire, F.; Wang, Y.; Isaac, D.; Fine, N.; et al. Identification of cell-free DNA methylation patterns unique to the human left ventricle as a potential indicator of acute cellular rejection. Clin. Transplant. 2021, 35, e14295. [Google Scholar] [CrossRef]
- Cristoferi, I.; Giacon, T.A.; Boer, K.; van Baardwijk, M.; Neri, F.; Campisi, M.; Kimenai, H.J.A.N.; Clahsen - van Groningen, M.C.; Pavanello, S.; Furian, L.; et al. The applications of DNA methylation as a biomarker in kidney transplantation: A systematic review. Clin. Epigenetics 2022, 14, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Silverman, E.K.; Schmidt, H.H.H.W.; Anastasiadou, E.; Altucci, L.; Angelini, M.; Badimon, L.; Balligand, J.L.; Benincasa, G.; Capasso, G.; Conte, F.; et al. Molecular networks in Network Medicine: Development and applications. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1489. [Google Scholar] [CrossRef] [PubMed]
- Rabant, M.; Amrouche, L.; Lebreton, X.; Aulagnon, F.; Benon, A.; Sauvaget, V.; Bonifay, R.; Morin, L.; Scemla, A.; Delville, M.; et al. Urinary C-X-C Motif Chemokine 10 Independently Improves the Noninvasive Diagnosis of Antibody–Mediated Kidney Allograft Rejection. J. Am. Soc. Nephrol. 2015, 26, 2840–2851. [Google Scholar] [CrossRef] [PubMed]
- Weseslindtner, L.; Hedman, L.; Wang, Y.; Strassl, R.; Helanterä, I.; Aberle, S.W.; Bond, G.; Hedman, K. Longitudinal assessment of the CXCL10 blood and urine concentration in kidney transplant recipients with BK polyomavirus replication—a retrospective study. Transpl. Int. 2020, 33, 555–566. [Google Scholar] [CrossRef]
- Tatapudi, R.R.; Muthukumar, T.; Dadhania, D.; Ding, R.; Li, B.; Sharma, V.K.; Lozada-Pastorio, E.; Seetharamu, N.; Hartono, C.; Serur, D.; et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004, 65, 2390–2397. [Google Scholar] [CrossRef]
- Lazzeri, E.; Rotondi, M.; Mazzinghi, B.; Lasagni, L.; Buonamano, A.; Rosati, A.; Pradella, F.; Fossombroni, V.; La Villa, G.; Gacci, M.; et al. High CXCL10 expression in rejected kidneys and predictive role of pretransplant serum CXCL10 for acute rejection and chronic allograft nephropathy. Transplantation 2005, 79, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.A.; Kim, E.J.; Begley, B.; Cheeseman, J.; Harden, T.; Perez, S.D.; Thomas, S.; Warshaw, B.; Kirk, A.D. Urinary Chemokines CXCL9 and CXCL10 Are Noninvasive Markers of Renal Allograft Rejection and BK Viral Infection. Am. J. Transplant. 2011, 11, 2228–2234. [Google Scholar] [CrossRef] [PubMed]
- Merhi, B.; Bayliss, G.; Gohh, R.Y. Role for urinary biomarkers in diagnosis of acute rejection in the transplanted kidney. World J. Transplant. 2015, 5, 251. [Google Scholar] [CrossRef] [PubMed]
- Mussap, M.; Plebani, M. Biochemistry and Clinical Role of Human Cystatin C. Crit. Rev. Clin. Lab. Sci. 2008, 41, 467–550. [Google Scholar] [CrossRef]
- Jensen, D.; Kierulf-Lassen, C.; Kristensen, M.L.V.; Nørregaard, R.; Weyer, K.; Nielsen, R.; Christensen, E.I.; Birn, H. Megalin dependent urinary cystatin C excretion in ischemic kidney injury in rats. PLoS ONE 2017, 12, e0178796. [Google Scholar] [CrossRef]
- Szirmay, B.; Kustán, P.; Horváth-Szalai, Z.; Ludány, A.; Lakatos, Á.; Mühl, D.; Wittmann, I.; Miseta, A.; Kovács, G.L.; Koszegi, T. Novel automated immune turbidimetric assay for routine urinary cystatin-C determinations. Bioanalysis 2018, 10, 377–384. [Google Scholar] [CrossRef]
- Spahillari, A.; Parikh, C.R.; Sint, K.; Koyner, J.L.; Patel, U.D.; Edelstein, C.L.; Passik, C.S.; Thiessen-Philbrook, H.; Swaminathan, M.; Shlipak, M.G. Serum Cystatin C– Versus Creatinine-Based Definitions of Acute Kidney Injury Following Cardiac Surgery: A Prospective Cohort Study. Am. J. Kidney Dis. 2012, 60, 922–929. [Google Scholar] [CrossRef]
- Taghizadeh-Afshari, A.; Mohammadi-Fallah, M.; Alizadeh, M.; Abkhiz, S.; Valizadeh, R.; Khadem-Ansari, M.H.; Sayyadi, H.; Kashani, S.A.; Rahimi, M.M. Serum cystatin C versus creatinine in the assessment of allograft function in early periods of kidney transplantation. J. Ren. Inj. Prev. 2017, 7, 11–15. [Google Scholar] [CrossRef]
- Higashi, A.; Dohi, Y.; Uraoka, N.; Sentani, K.; Uga, S.; Kinoshita, H.; Sada, Y.; Kitagawa, T.; Hidaka, T.; Kurisu, S.; et al. The Potential Role of Inflammation Associated with Interaction between Osteopontin and CD44 in a Case of Pulmonary Tumor Thrombotic Microangiopathy Caused by Breast Cancer. Intern. Med. 2015, 54, 2877–2880. [Google Scholar] [CrossRef]
- Millán, O.; Rafael-Valdivia, L.; San Segundo, D.; Boix, F.; Castro-Panete, M.J.; López-Hoyos, M.; Muro, M.; Valero-Hervás, D.; Rimola, A.; Navasa, M.; et al. Should IFN-γ, IL-17 and IL-2 be considered predictive biomarkers of acute rejection in liver and kidney transplant? Results of a multicentric study. Clin. Immunol. 2014, 154, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Askenazi, D.J.; Koralkar, R.; Hundley, H.E.; Montesanti, A.; Parwar, P.; Sonjara, S.; Ambalavanan, N. Urine Biomarkers Predict Acute Kidney Injury in Newborns. J. Pediatr. 2012, 161, 270–275.e1. [Google Scholar] [CrossRef] [PubMed]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Cantaluppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediators Inflamm. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Alchi, B.; Nishi, S.; Kondo, D.; Kaneko, Y.; Matsuki, A.; Imai, N.; Ueno, M.; Iguchi, S.; Sakatsume, M.; Narita, I.; et al. Osteopontin expression in acute renal allograft rejection. Kidney Int. 2005, 67, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, Q.; Qiu, Y.; Xu, M.; Rong, R.; Zhu, T. Osteopontin level correlates with acute cellular renal allograft rejection. J. Surg. Res. 2013, 182, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Guan, Q.; Kwan, C.C.H.; Chen, H.; Gleave, M.E.; Nguan, C.Y.C.; Du, C. Loss of clusterin expression worsens renal ischemia-reperfusion injury. Am. J. Physiol.-Ren. Physiol. 2010, 298, 568–578. [Google Scholar] [CrossRef]
- Pezeshgi, A.; Azar, S.A.; Ghasemi, H.; Kamali, K.; Esmaeilzadeh, A.; Hajsalimi, B.; Pour-Asghar, S.; Behmanesh, M.R.; Kiafar, M. Role of plasma neutrophil gelatinase-associated lipocalin as an emerging biomarker of acute renal failure following kidney transplantation and its correlation with plasma creatinine. J. Ren. Inj. Prev. 2016, 5, 98. [Google Scholar] [CrossRef]
- Wu, C.Y.; Yang, H.Y.; Chien, H.P.; Tseng, M.H.; Huang, J.L. Urinary clusterin—A novel urinary biomarker associated with pediatric lupus renal histopathologic features and renal survival. Pediatr. Nephrol. 2018, 33, 1189–1198. [Google Scholar] [CrossRef]
- Musiał, K.; Augustynowicz, M.; Miśkiewicz-Migoń, I.; Kałwak, K.; Ussowicz, M.; Zwolińska, D. Clusterin as a New Marker of Kidney Injury in Children Undergoing Allogeneic Hematopoietic Stem Cell Transplantation—A Pilot Study. J. Clin. Med. 2020, 9, 2599. [Google Scholar] [CrossRef] [PubMed]
- Pianta, T.J.; Peake, P.W.; Pickering, J.W.; Kelleher, M.; Buckley, N.A.; Endre, Z.H. Clusterin in kidney transplantation: Novel biomarkers versus serum creatinine for early prediction of delayed graft function. Transplantation 2015, 99, 171–179. [Google Scholar] [CrossRef] [PubMed]
- San Segundo, D.; Millán, O.; Munoz-Cacho, P.; Boix, F.; Paz-Artal, E.; Talayero, P.; Morales, J.M.; Muro, M.; De Cos, M.Á.; Guirado, L.; et al. High proportion of pretransplantation activated regulatory T cells (CD4+ CD25highCD62L+ CD45RO+) predicts acute rejection in kidney transplantation: Results of a multicenter study. Transplantation 2014, 98, 1213–1218. [Google Scholar] [CrossRef]
- Blanco-García, R.M.; López-Álvarez, M.R.; Garrido, I.P.; Salgado-Cecilia, G.; Campillo, J.A.; Bolarín, J.M.; Legaz, I.; Muro, M.; García-Alonso, A.M.; Martínez-Sánchez, M.V.; et al. CD28 and KIR2D receptors as sensors of the immune status in heart and liver transplantation. Hum. Immunol. 2011, 72, 841–848. [Google Scholar] [CrossRef]
- Laguna-Goya, R.; Utrero-Rico, A.; Cano-Romero, F.L.; Gómez-Massa, E.; González, E.; Andrés, A.; Mancebo-Sierra, E.; Paz-Artal, E. Imbalance favoring follicular helper T cells over IL10+ regulatory B cells is detrimental for the kidney allograft. Kidney Int. 2020, 98, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Achour, A.; Simon, Q.; Mohr, A.; Séité, J.F.; Youinou, P.; Bendaoud, B.; Ghedira, I.; Pers, J.O.; Jamin, C. Human regulatory B cells control the TFH cell response. J. Allergy Clin. Immunol. 2017, 140, 215–222. [Google Scholar] [CrossRef]
- O’Halloran, C.; Cullen, K.; Njoroge, J.; Jessop, L.; Smith, J.; Hope, V.; Ncube, F. The extent of and factors associated with self-reported overdose and self-reported receipt of naloxone among people who inject drugs (PWID) in England, Wales and Northern Ireland. Int. Jounal Drug Policy 2017, 46, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, H.; Zhang, D.; Wang, Y.; Wang, Y.; Wang, W.; Hu, X. Identification of a novel peripheral blood signature diagnosing subclinical acute rejection after renal transplantation. Transl. Androl. Urol. 2022, 11, 1399–1409. [Google Scholar] [CrossRef]
- Halloran, P.F.; Reeve, J.; Madill-Thomsen, K.S.; Kaur, N.; Ahmed, E.; Cantos, C.; Al Haj Baddar, N.; Demko, Z.; Liang, N.; Swenerton, R.K.; et al. Combining Donor-derived Cell-free DNA Fraction and Quantity to Detect Kidney Transplant Rejection Using Molecular Diagnoses and Histology as Confirmation. Transplantation 2022, 106. [Google Scholar] [CrossRef]
- Benincasa, G.; Viglietti, M.; Coscioni, E.; Napoli, C. “Transplantomics” for predicting allograft rejection: Real-life applications and new strategies from Network Medicine. Hum. Immunol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Seeto, R.K.; Fleming, J.N.; Dholakia, S.; Dale, B.L. Understanding and using AlloSure donor derived cell-free DNA. Biophys. Rev. 2020, 12, 917–924. [Google Scholar] [CrossRef]
- Han, F.; Sun, Q.; Huang, Z.; Li, H.; Ma, M.; Liao, T.; Luo, Z.; Zheng, L.; Zhang, N.; Chen, N.; et al. Donor plasma mitochondrial DNA is associated with antibody-mediated rejection in renal allograft recipients. Aging (Albany NY) 2021, 13, 8440. [Google Scholar] [CrossRef]
- Loupy, A.; Duong Van Huyen, J.P.; Hidalgo, L.; Reeve, J.; Racapé, M.; Aubert, O.; Venner, J.M.; Falmuski, K.; Cécile Bories, M.; Beuscart, T.; et al. Gene expression profiling for the identification and classification of antibody-mediated heart rejection. Circulation 2017, 135, 917–935. [Google Scholar] [CrossRef] [PubMed]
- Warmuzińska, N.; Łuczykowski, K.; Bojko, B. A Review of Current and Emerging Trends in Donor Graft-Quality Assessment Techniques. J. Clin. Med. 2022, 11, 487. [Google Scholar] [CrossRef]
- Halloran, P.F.; Reeve, J.; Akalin, E.; Aubert, O.; Bohmig, G.A.; Brennan, D.; Bromberg, J.; Einecke, G.; Eskandary, F.; Gosset, C.; et al. Real Time Central Assessment of Kidney Transplant Indication Biopsies by Microarrays: The INTERCOMEX Study. Am. J. Transplant. 2017, 17, 2851–2862. [Google Scholar] [CrossRef] [PubMed]
- Eller, K.; Böhmig, G.A.; Banas, M.C.; Viklicky, O. Editorial: Advances in the diagnosis and treatment in kidney transplantation. Front. Med. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Pereira, A.B.; Chang, J.; Matas, A.; Picton, M.; De Freitas, D.; Bromberg, J.; Serõn, D.; Sellarés, J.; Einecke, G.; et al. Microarray Diagnosis of Antibody-Mediated Rejection in Kidney Transplant Biopsies: An International Prospective Study (INTERCOM). Am. J. Transplant. 2013, 13, 2865–2874. [Google Scholar] [CrossRef]
- Anglicheau, D.; Sharma, V.K.; Ding, R.; Hummel, A.; Snopkowski, C.; Dadhania, D.; Seshan, S.V.; Suthanthiran, M. MicroRNA expression profiles predictive of human renal allograft status. Proc. Natl. Acad. Sci. USA 2009, 106, 5330–5335. [Google Scholar] [CrossRef]
- Ben-Dov, I.Z.; Muthukumar, T.; Morozov, P.; Mueller, F.B.; Tuschl, T.; Suthanthiran, M. MicroRNA Sequence Profiles of Human Kidney Allografts With or Without Tubulointerstitial Fibrosis. Transplantation 2012, 94, 1086–1094. [Google Scholar] [CrossRef]
- Flower, D.R. Immunoinformatics and the in silico prediction of immunogenicity. An introduction. Methods Mol. Biol. 2007, 409, 1–15. [Google Scholar]
- Burlacu, A.; Iftene, A.; Jugrin, D.; Popa, I.V.; Lupu, P.M.; Vlad, C.; Covic, A. Using Artificial Intelligence Resources in Dialysis and Kidney Transplant Patients: A Literature Review. Biomed Res. Int. 2020, 2020. [Google Scholar] [CrossRef]
- Topuz, K.; Zengul, F.D.; Dag, A.; Almehmi, A.; Yildirim, M.B. Predicting graft survival among kidney transplant recipients: A Bayesian decision support model. Decis. Support Syst. 2018, 106, 97–109. [Google Scholar] [CrossRef]
- Shaikhina, T.; Lowe, D.; Daga, S.; Briggs, D.; Higgins, R.; Khovanova, N. Decision tree and random forest models for outcome prediction in antibody incompatible kidney transplantation. Biomed. Signal Process. Control 2019, 52, 456–462. [Google Scholar] [CrossRef]
- Segev, D.L.; Gentry, S.E.; Warren, D.S.; Reeb, B.; Montgomery, R.A. Kidney Paired Donation and Optimizing the Use of Live Donor Organs. JAMA 2005, 293, 1883–1890. [Google Scholar] [CrossRef]
- Patterson, E.A.; Whelan, M.P. A framework to establish credibility of computational models in biology. Prog. Biophys. Mol. Biol. 2017, 129, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Narayan, A.; Liu, Z.; Bergquist, J.A.; Charlebois, C.; Rampersad, S.; Rupp, L.; Brooks, D.; White, D.; Tate, J.; MacLeod, R.S. UncertainSCI: Uncertainty quantification for computational models in biomedicine and bioengineering. Comput. Biol. Med. 2023, 152, 106407. [Google Scholar] [CrossRef]
- Alfaro, R.; Martínez-Banaclocha, H.; Llorente, S.; Jimenez-Coll, V.; Galián, J.A.; Botella, C.; Moya-Quiles, M.R.; Parrado, A.; Muro-Perez, M.; Minguela, A.; et al. Computational Prediction of Biomarkers, Pathways, and New Target Drugs in the Pathogenesis of Immune-Based Diseases Regarding Kidney Transplantation Rejection. Front. Immunol. 2021, 12, 5418. [Google Scholar] [CrossRef]
- Agapito, G.; Cannataro, M. A parallel software pipeline to select relevant genes for pathway enrichment. In Proceedings of the 2022 30th Euromicro International Conference on Parallel, Distributed and Network-based Processing (PDP), Valladolid, Spain, 9–11 March 2022; pp. 219–225. [Google Scholar]
- Maron, B.A.; Wang, R.S.; Shevtsov, S.; Drakos, S.G.; Arons, E.; Wever-Pinzon, O.; Huggins, G.S.; Samokhin, A.O.; Oldham, W.M.; Aguib, Y.; et al. Individualized interactomes for network-based precision medicine in hypertrophic cardiomyopathy with implications for other clinical pathophenotypes. Nat. Commun. 2021, 12, 873. [Google Scholar] [CrossRef] [PubMed]
- Hartzell, S.; Bin, S.; Cantarelli, C.; Haverly, M.; Manrique, J.; Angeletti, A.; Manna, G.L.; Murphy, B.; Zhang, W.; Levitsky, J.; et al. Kidney Failure Associates with T Cell Exhaustion and Imbalanced Follicular Helper T Cells. Front. Immunol. 2020, 11, 2390. [Google Scholar] [CrossRef]
- Wang, L.J.; Ma, X.B.; Xia, H.Y.; Sun, X.; Yu, L.; Yang, Q.; Hu, Z.Q.; Zhao, Y.H.; Hu, W.; Ran, J.H. Identification of Biomarkers for Predicting Allograft Rejection following Kidney Transplantation Based on the Weighted Gene Coexpression Network Analysis. Biomed Res. Int. 2021, 2021. [Google Scholar] [CrossRef]
- Teng, L.; Shen, L.; Zhao, W.; Wang, C.; Feng, S.; Wang, Y.; Bi, Y.; Rong, S.; Shushakova, N.; Haller, H.; et al. SLAMF8 Participates in Acute Renal Transplant Rejection via TLR4 Pathway on Pro-Inflammatory Macrophages. Front. Immunol. 2022, 13, 846695. [Google Scholar] [CrossRef]
- Benincasa, G.; Maron, B.A.; Affinito, O.; D’Alto, M.; Franzese, M.; Argiento, P.; Schiano, C.; Romeo, E.; Bontempo, P.; Golino, P.; et al. Association Between Circulating CD4+ T Cell Methylation Signatures of Network-Oriented SOCS3 Gene and Hemodynamics in Patients Suffering Pulmonary Arterial Hypertension. J. Cardiovasc. Transl. Res. 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Legaz, I.; Bernardo, M.V.; Alfaro, R.; Martínez-Banaclocha, H.; Galián, J.A.; Jimenez-Coll, V.; Boix, F.; Mrowiec, A.; Salmeron, D.; Botella, C.; et al. PCR Array Technology in Biopsy Samples Identifies Up-Regulated mTOR Pathway Genes as Potential Rejection Biomarkers After Kidney Transplantation. Front. Med. 2021, 8, 547849. [Google Scholar] [CrossRef] [PubMed]
- Snyder, T.M.; Khush, K.K.; Valantine, H.A.; Quake, S.R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. USA 2011, 108, 6229–6234. [Google Scholar] [CrossRef] [PubMed]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 31 March 2020).
- Starzl, T.E.; Murase, N.; Ildstad, S.; Ricordi, C.; Demetris, A.J.; Trucco, M. Cell migration, chimerism, and graft acceptance. Lancet 1992, 339, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Tein, M.S.C.; Pang, C.C.P.; Yeung, C.K.; Tong, K.L.; Magnus Hjelm, N. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet 1998, 351, 1329–1330. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, J.; Wang, W.; Zhu, H.; Qian, J.; Wang, S.; Que, S.; Zhang, F.; Yin, S.; Zhou, L.; et al. Integrative Network Analysis Revealed Genetic Impact of Pyruvate Kinase L/R on Hepatocyte Proliferation and Graft Survival after Liver Transplantation. Oxid. Med. Cell. Longev. 2021, 2021. [Google Scholar] [CrossRef]
- Gadi, V.K.; Nelson, J.L.; Boespflug, N.D.; Guthrie, K.A.; Kuhr, C.S. Soluble Donor DNA Concentrations in Recipient Serum Correlate with Pancreas-Kidney Rejection. Clin. Chem. 2006, 52, 379–382. [Google Scholar] [CrossRef]
- Goussous, N.; Xie, W.; Dawany, N.; Scalea, J.R.; Bartosic, A.; Haririan, A.; Kalil, R.; Drachenberg, C.; Costa, N.; Weir, M.R.; et al. Donor-derived Cell-free DNA in Infections in Kidney Transplant Recipients: Case Series. Transplant. Direct 2020, 6, e568. [Google Scholar] [CrossRef]
- Kant, S.; Bromberg, J.; Haas, M.; Brennan, D. Donor-derived Cell-free DNA and the Prediction of BK Virus-associated Nephropathy. Transplant. Direct 2020, 6. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2018, 8, 19. [Google Scholar] [CrossRef]
- Jordan, S.C.; Bunnapradist, S.; Bromberg, J.S.; Langone, A.J.; Hiller, D.; Yee, J.P.; Sninsky, J.J.; Woodward, R.N.; Matas, A.J. Donor-derived Cell-free DNA Identifies Antibody-mediated Rejection in Donor Specific Antibody Positive Kidney Transplant Recipients. Transplant. Direct 2018, 4. [Google Scholar] [CrossRef]
- Huang, E.; Sethi, S.; Peng, A.; Najjar, R.; Mirocha, J.; Haas, M.; Vo, A.; Jordan, S.C. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am. J. Transplant. 2019, 19, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
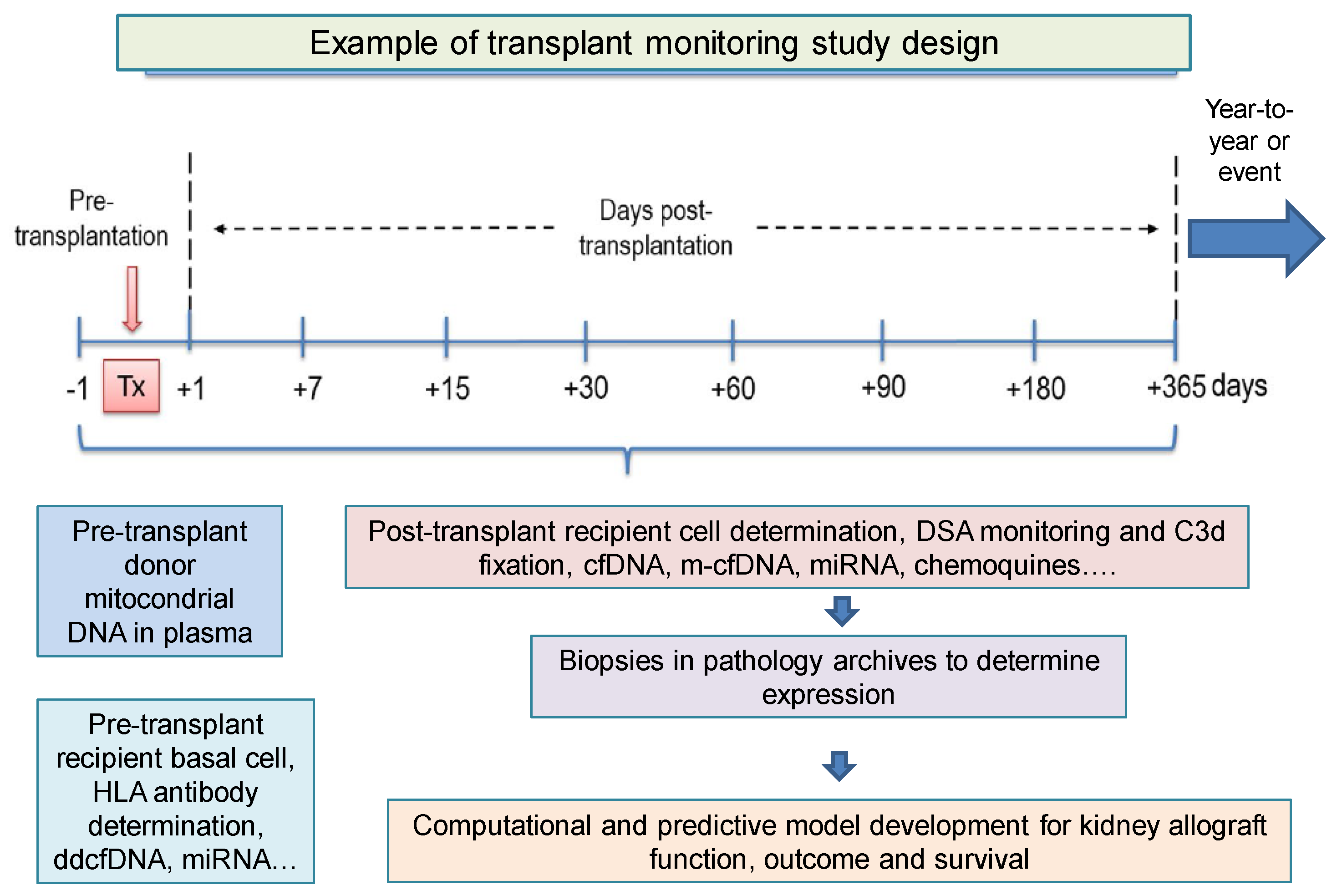
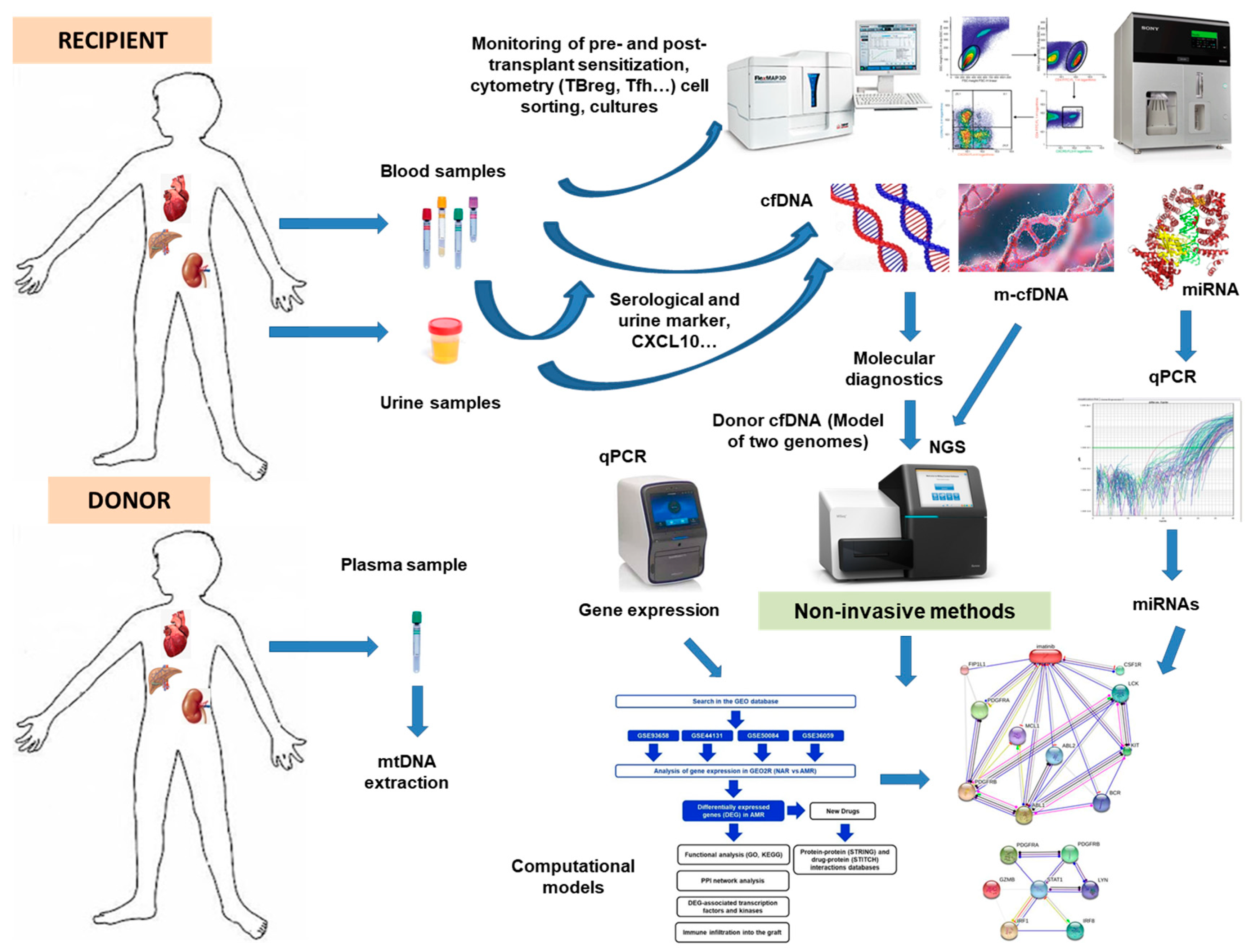
| Serological and Urine Biomarkers | Cellular Biomarkers | Genomic and Transcriptomic Biomarkers | Computational Prediction Biomarkers | Cell-Free DNA Biomarker | ||||
|---|---|---|---|---|---|---|---|---|
| Classical Markers | New Markers | Classical Markers | New Markers | Classical Markers | New Markers | Classical Markers | New Markers | New Markers |
| Physical examination | Neutrophil gelatinase-associated lipocalin in urine (uNGAL) | Expression of costimulatory molecules (CD28, CD69, CD95...) | Monitoring of Tregs and Bregs cells post-transplantation | AlloMap and AlloSure assays | AlloMap gene expression profiling test | Rene Dusquesnoy’s HLAmatchmaker tools | Stepwise logistic regression methods | Donor-specific cell-free DNA (ddcfDNA) |
| Kidney biopsy | Kidney injury molecule-1 (KIM-1) | Production of soluble or intracytoplasmic cytokines | Determination of B cell clusters | SignateraTM and ProsperaTM trials | TruGraf® (Transplant Genomics) | Predicted Indirectly Recognizable HLA Epitopes (PIRCHE-II) algorithm | GEO databases, Weighted Gene Coexpression Network Analysis (WGC-NA) algorithm | |
| Albuminuria | CXCL-10 | ImmuKnow assay (Cylex) to assess changes in ATP production by CD4+ T cells | Circulating B cell follicles (cTfh) | Donor plasma mitochondrial DNA (mtDNA) | Viracor TRAC® (Eurofins Viracor) | STRING database | ||
| Proteinuria | Cystatin C (CysC) | B cell differentiation molecules (BR3/BAFF-R, TACI and APRIL) | OmniGrafTM (Transplant Genomics) | DisGeNET | ||||
| Determination of serum creatinine level | Osteopontin (OPN) | QSant (Nephrosant) assay | Cytoscape software | |||||
| Estimation of glomerular filtration rate (eGFR) | Clusterin (CLU) | miR-142-3p, miR-155, miR-142-5p miR-338-5p miR-150-5p | ||||||
| HLA antibody profiling | CXCL13 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez-Coll, V.; Llorente, S.; Boix, F.; Alfaro, R.; Galián, J.A.; Martinez-Banaclocha, H.; Botella, C.; Moya-Quiles, M.R.; Muro-Pérez, M.; Minguela, A.; et al. Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. Int. J. Mol. Sci. 2023, 24, 3908. https://doi.org/10.3390/ijms24043908
Jimenez-Coll V, Llorente S, Boix F, Alfaro R, Galián JA, Martinez-Banaclocha H, Botella C, Moya-Quiles MR, Muro-Pérez M, Minguela A, et al. Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. International Journal of Molecular Sciences. 2023; 24(4):3908. https://doi.org/10.3390/ijms24043908
Chicago/Turabian StyleJimenez-Coll, Víctor, Santiago Llorente, Francisco Boix, Rafael Alfaro, José Antonio Galián, Helios Martinez-Banaclocha, Carmen Botella, María R. Moya-Quiles, Manuel Muro-Pérez, Alfredo Minguela, and et al. 2023. "Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome" International Journal of Molecular Sciences 24, no. 4: 3908. https://doi.org/10.3390/ijms24043908
APA StyleJimenez-Coll, V., Llorente, S., Boix, F., Alfaro, R., Galián, J. A., Martinez-Banaclocha, H., Botella, C., Moya-Quiles, M. R., Muro-Pérez, M., Minguela, A., Legaz, I., & Muro, M. (2023). Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome. International Journal of Molecular Sciences, 24(4), 3908. https://doi.org/10.3390/ijms24043908









