Killer Cell Immunoglobulin-like Receptors (KIR) and Human Leucocyte Antigen C (HLA-C) Increase the Risk of Long-Term Chronic Liver Graft Rejection
Abstract
1. Introduction
2. Results
2.1. Transplant Clinical Endpoints
2.2. Impact of KIR Genes and KIR Genotypes in Chronic Rejection
2.3. The iKIR Gene Mismatching Increases the Chronic Rejection Rate
2.4. Impact of HLA-I Allele, HLA-C Allotype, and HLA-C Genotypes on Liver Allograft Chronic Rejection
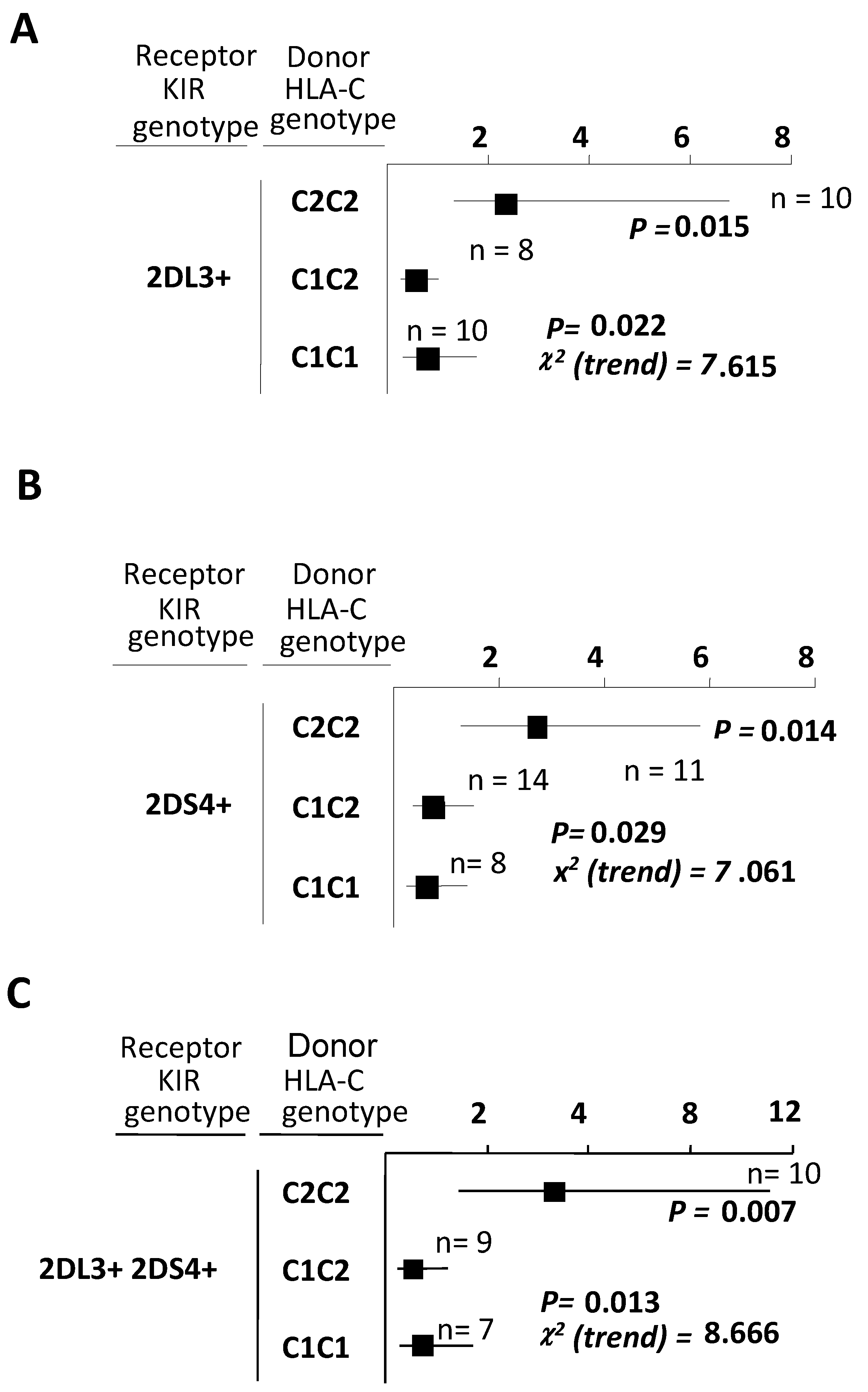
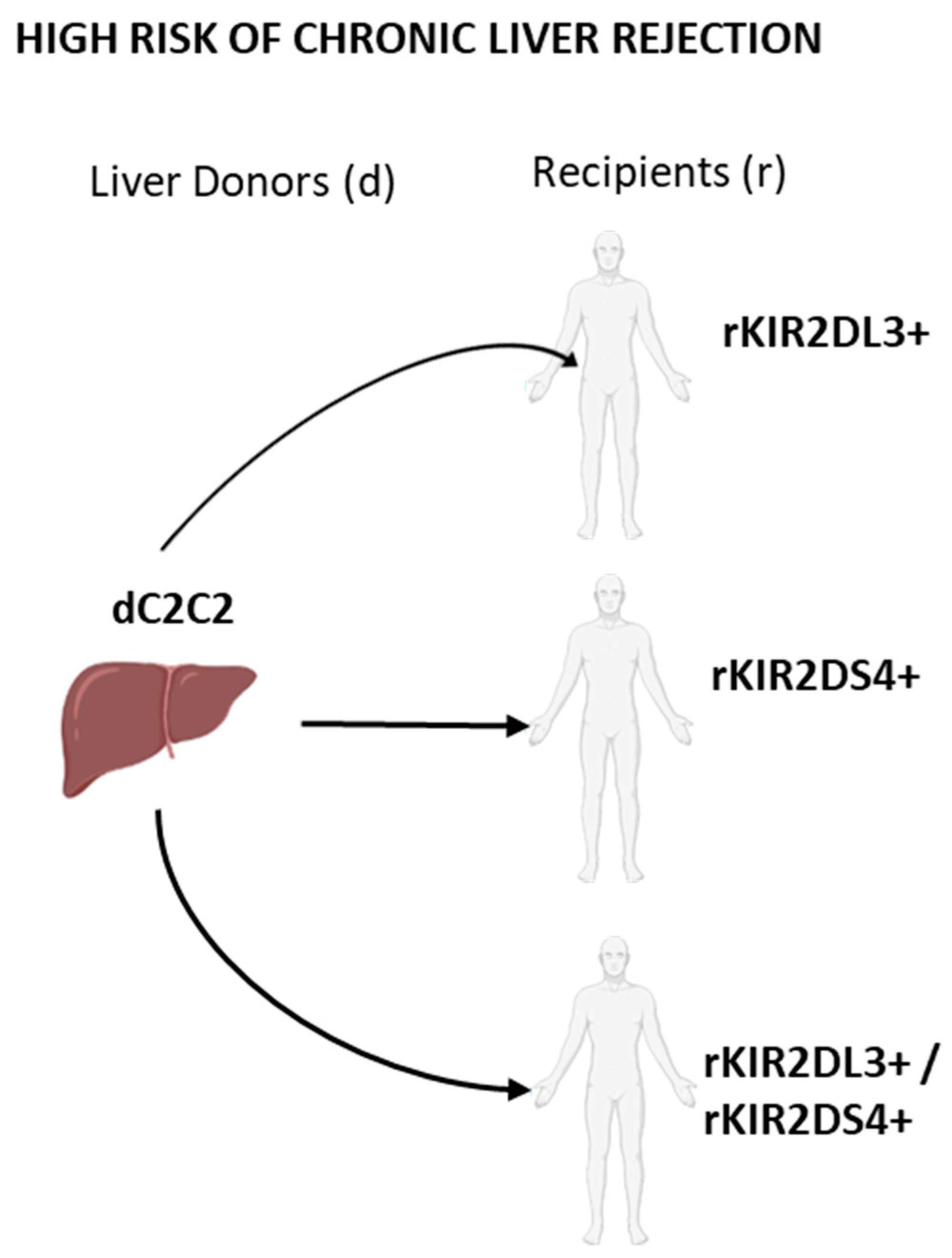
2.5. Combinations between Recipients Bearing Both KIR2DL3 and KIR2DS4 and Liver Donors Having C2-Ligands Predispose to a Higher Chronic Rejection Risk
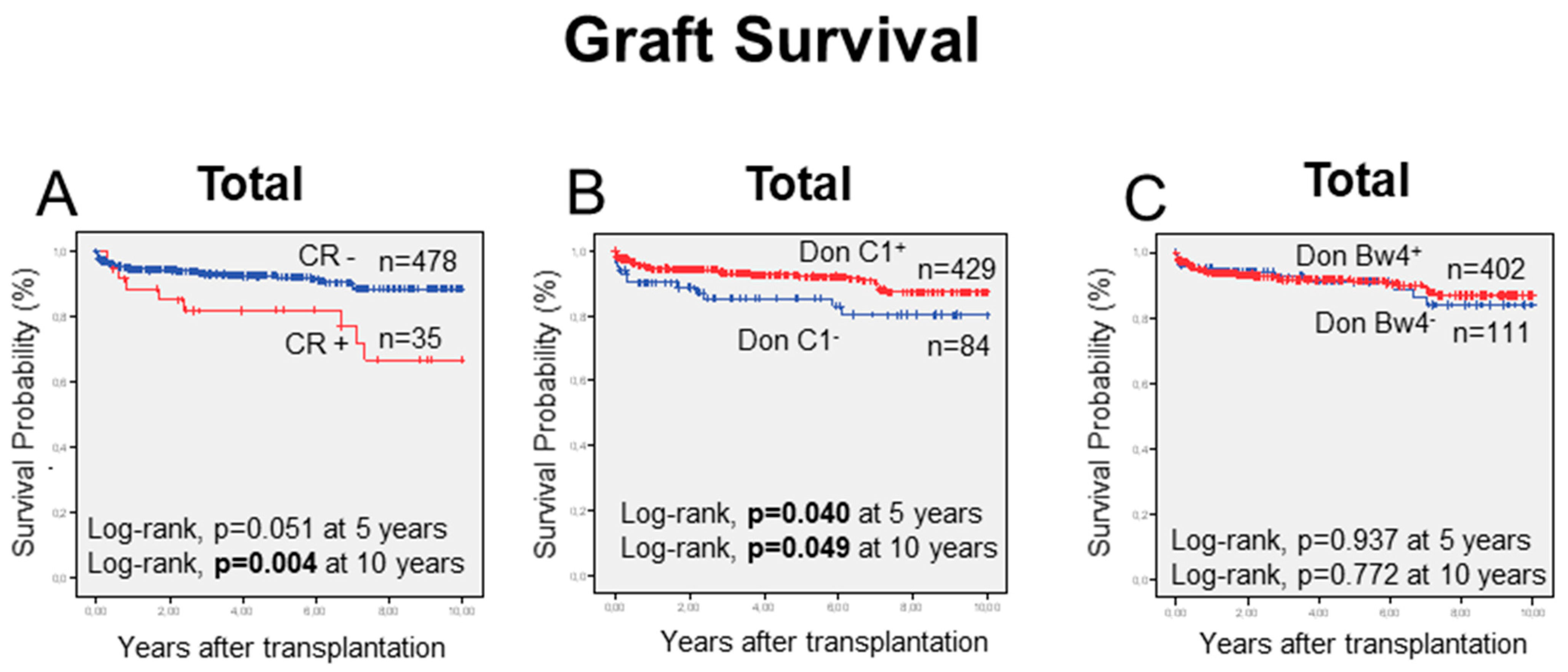
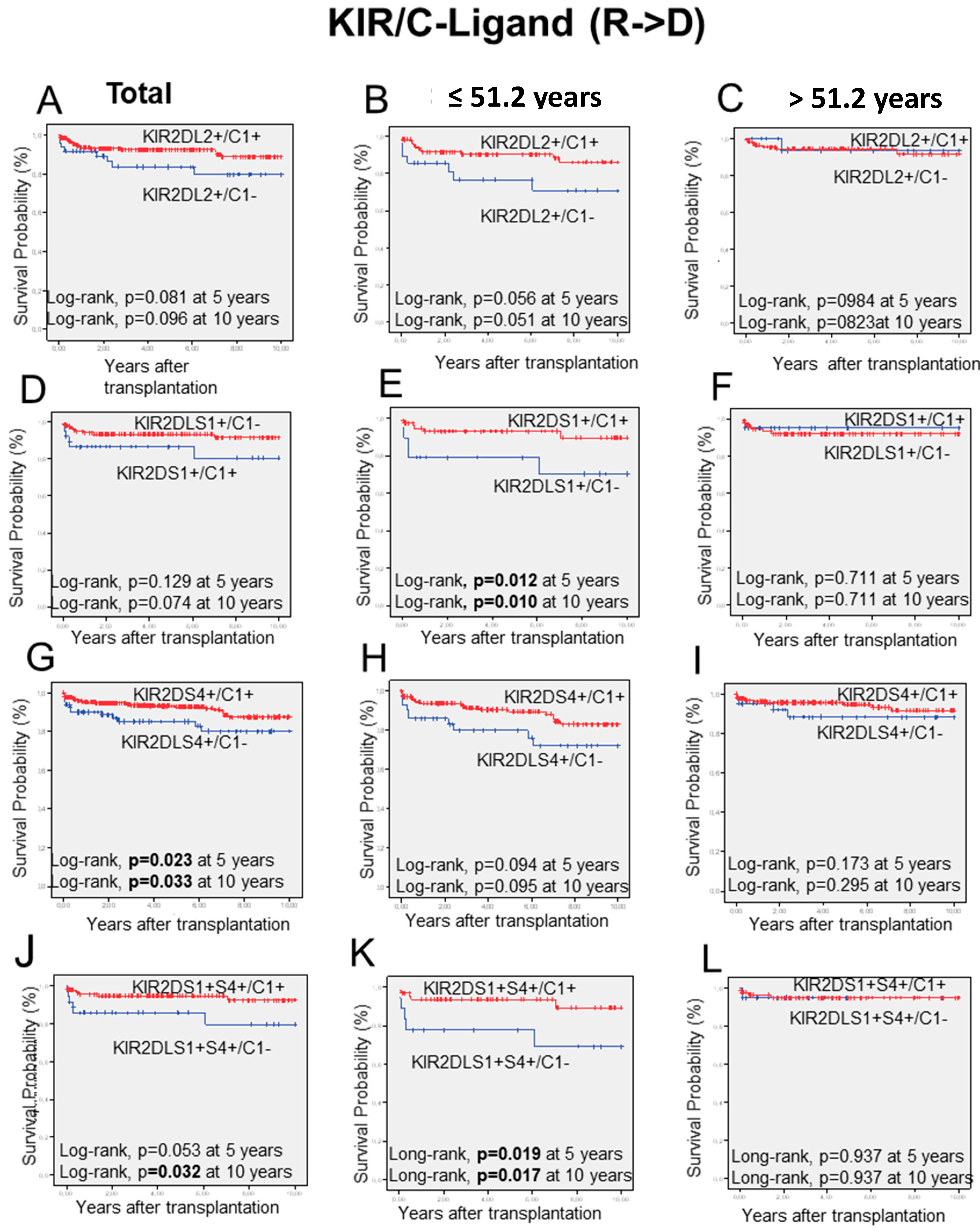
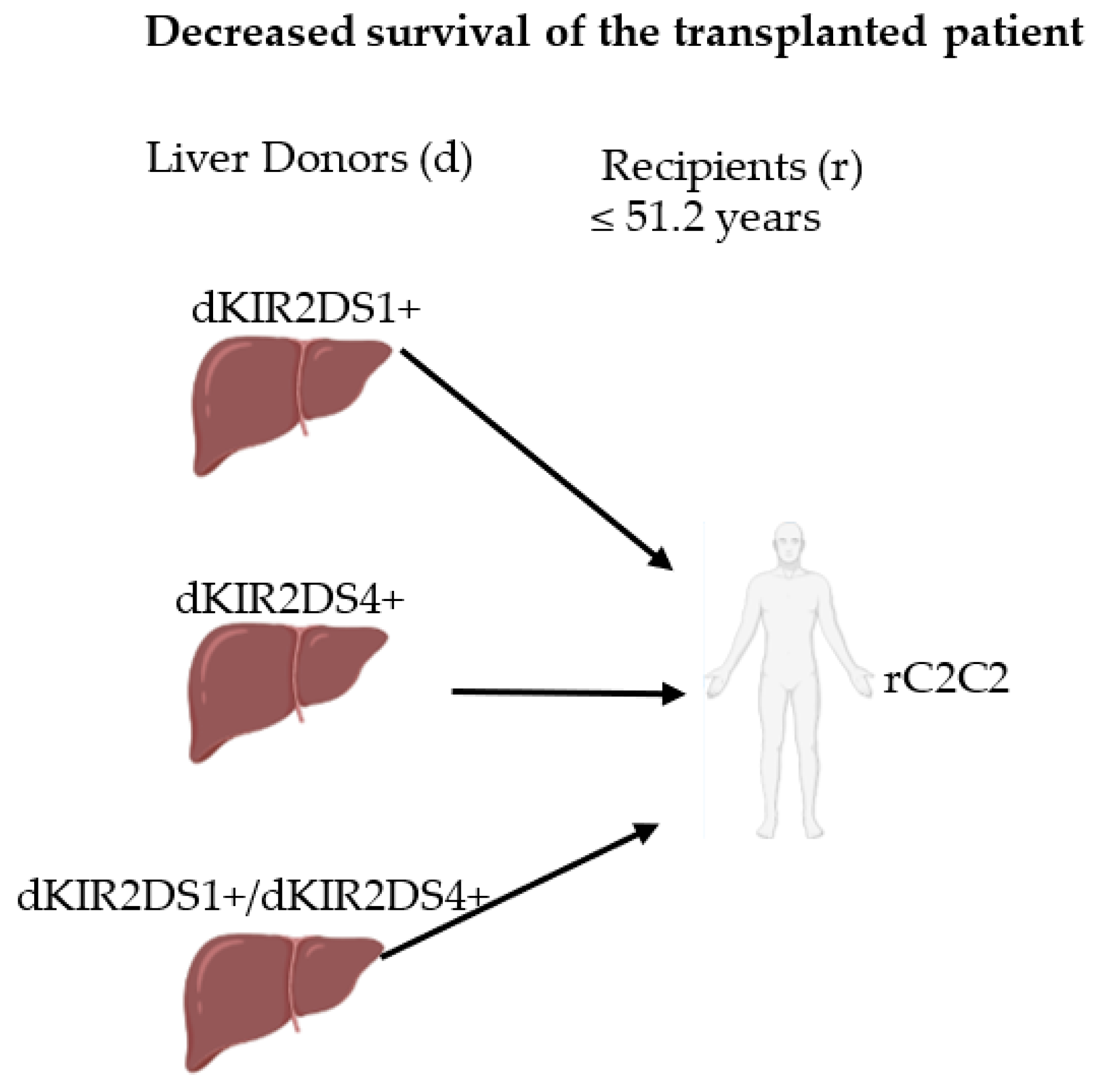
2.6. KIR/HLA-I-Ligand Combinations and Long-Term Graft Outcome
3. Discussion
4. Material and Methods
4.1. Patient Enrollment
4.2. Chronic Liver Rejection Diagnosis
4.3. Viral Pre-Infection Diagnosis
4.4. Immunosuppressive Treatment
4.5. KIR and HLA Typing and Genotype Assignment
4.6. KIR-Ligand Models and NK-Alloreactivity Prediction
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Choudhary, N.S.; Saigal, S.; Bansal, R.K.; Saraf, N.; Gautam, D.; Soin, A.S. Acute and Chronic Rejection After Liver Transplantation: What A Clinician Needs to Know. J. Clin. Exp. Hepatol. 2017, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Angelico, R.; Sensi, B.; Manzia, T.M.; Tisone, G.; Grassi, G.; Signorello, A.; Milana, M.; Lenci, I.; Baiocchi, L. Chronic rejection after liver transplantation: Opening the Pandora’s box. World J. Gastroenterol. 2021, 27, 7771. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Perálvarez, M.; Rico-Juri, J.M.; Tsochatzis, E.; Burra, P.; De la Mata, M.; Lerut, J. Biopsy-proven acute cellular rejection as an efficacy endpoint of randomized trials in liver transplantation: A systematic review and critical appraisal. Transpl. Int. 2016, 29, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Demetris, A.; Adams, D.; Bellamy, C.; Blakolmer, K.; Clouston, A.; Dhillon, A.P.; Fung, J.; Gouw, A.; Gustafsson, B.; Haga, H.; et al. Update of the International Banff Schema for Liver Allograft Rejection: Working recommendations for the histopathologic staging and reporting of chronic rejection. An International Panel. Hepatology 2000, 31, 792–799. [Google Scholar]
- Gruttadauria, S.; Vasta, F.; Mandalà, L.; Cintorino, D.; Piazza, T.; Spada, M.; Verzaro, R.; Gridelli, B. Basiliximab in a triple-drug regimen with tacrolimus and steroids in liver transplantation. Transplant. Proc. 2005, 37, 2611–2613. [Google Scholar] [CrossRef]
- Maluf, D.G.; Stravitz, R.T.; Cotterell, A.H.; Posner, M.P.; Nakatsuka, M.; Sterling, R.K.; Luketic, V.A.; Shiffman, M.L.; Ham, J.M.; Marcos, A.; et al. Adult living donor versus deceased donor liver transplantation: A 6-year single center experience. Am. J. Transplant. 2005, 5, 149–156. [Google Scholar] [CrossRef]
- Wiesner, R.H.; Demetris, A.J.; Belle, S.H.; Seaberg, E.C.; Lake, J.R.; Zetterman, R.K.; Everhart, J.; Detre, K.M. Acute hepatic allograft rejection: Incidence, risk factors, and impact on outcome. Hepatology 1998, 28, 638–645. [Google Scholar] [CrossRef]
- Neil, D.A.H.; Hübscher, S.G. Current views on rejection pathology in liver transplantation. Transpl. Int. 2010, 23, 971–983. [Google Scholar] [CrossRef]
- Demetris, A.J.; Bellamy, C.; Hübscher, S.G.; O’Leary, J.; Randhawa, P.S.; Feng, S.; Neil, D.; Colvin, R.B.; McCaughan, G.; Fung, J.J.; et al. 2016 comprehensive update of the Banff working group on liver allograft pathology: Introduction of antibody-mediated rejection. Am. J. Transplant. 2016, 16, 2816–2835. [Google Scholar]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef]
- Peng, H.; Sun, R. Liver-resident NK cells and their potential functions. Cell. Mol. Immunol. 2017, 14, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Crispe, I.N. The Liver as a Lymphoid Organ. Annu. Rev. Immunol. 2009, 27, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Doherty, D.G.; O’Farrelly, C. Innate and adaptive lymphoid cells in the human liver. Immunol. Rev. 2000, 174, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, Z. Innate lymphocytes: Pathogenesis and therapeutic targets of liver diseases and cancer. Cell. Mol. Immunol. 2021, 18, 57. [Google Scholar] [CrossRef] [PubMed]
- Kansler, E.R.; Li, M.O. Innate lymphocytes—Lineage, localization and timing of differentiation. Cell. Mol. Immunol. 2019, 16, 627. [Google Scholar] [CrossRef]
- De Maria, A.; Moretta, L. Revisited function of human NK cell subsets. Cell Cycle 2011, 10, 1178–1179. [Google Scholar] [CrossRef]
- Legaz, I.; López-Álvarez, M.R.; Campillo, J.A.; Moya-Quiles, M.R.; Bolarín, J.M.; de la Peña, J.; Salgado, G.; Gimeno, L.; García-Alonso, A.M.; Muro, M.; et al. KIR gene mismatching and KIR/C ligands in liver transplantation: Consequences for short-term liver allograft injury. Transplantation 2013, 95, 1037–1044. [Google Scholar] [CrossRef]
- López-Álvarez, M.R.; Campillo, J.A.; Legaz, I.; Blanco-García, R.M.; Salgado-Cecilia, G.; Bolarín, J.M.; Gimeno, L.; Gil, J.; García-Alonso, A.M.; Muro, M.; et al. Divergences in KIR2D+ natural killer and KIR2D+CD8+ T-cell reconstitution following liver transplantation. Hum. Immunol. 2011, 72, 229–237. [Google Scholar] [CrossRef]
- Blanco-García, R.M.; López-Álvarez, M.R.; Garrido, I.P.; Salgado-Cecilia, G.; Campillo, J.A.; Bolarín, J.M.; Legaz, I.; Muro, M.; García-Alonso, A.M.; Martínez-Sánchez, M.V.; et al. CD28 and KIR2D receptors as sensors of the immune status in heart and liver transplantation. Hum. Immunol. 2011, 72, 841–848. [Google Scholar] [CrossRef]
- Hanvesakul, R.; Kubal, C.; Moore, J.; Neil, D.; Cook, M.; Ball, S.; Briggs, D.; Moss, P.; Cockwell, P. Kir and hla-c interactions promote differential dendritic cell maturation and is a major determinant of graft failure following kidney transplantation. PLoS ONE 2011, 6, e23631. [Google Scholar] [CrossRef]
- Jafari, D.; Nafar, M.; Yekaninejad, M.S.; Abdolvahabi, R.; Pezeshki, M.L.; Razaghi, E.; Amirzargar, A.A. Investigation of killer immunoglobulin-like receptor (KIR) and HLA genotypes to predict the occurrence of acute allograft rejection after kidney transplantation. Iran. J. Allergy Asthma Immunol. 2017, 16, 245–255. [Google Scholar] [PubMed]
- Zamir, M.R.; Shahi, A.; Salehi, S.; Amirzargar, A. Natural killer cells and killer cell immunoglobulin-like receptors in solid organ transplantation: Protectors or opponents? Transplant. Rev. 2022, 36, 100723. [Google Scholar] [CrossRef] [PubMed]
- Beilke, J.N.; Gill, R.G. Frontiers in Nephrology: The Varied Faces of Natural Killer Cells in Transplantation—Contributions to Both Allograft Immunity and Tolerance. J. Am. Soc. Nephrol. 2007, 18, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Beilke, J.N.; Kuhl, N.R.; Van Kaer, L.; Gill, R.G. NK cells promote islet allograft tolerance via a perforin-dependent mechanism. Nat. Med. 2005, 11, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, M.A.; Hidalgo, L.G. NK cells in antibody-mediated rejection–Key effector cells in microvascular graft damage. Int. J. Immunogenet. 2021, 48, 110–119. [Google Scholar] [CrossRef]
- Selvakumar, A.; Steffens, U.; Dupont, B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens 1996, 48, 285–294. [Google Scholar] [CrossRef]
- Wende, H.; Colonna, M.; Ziegler, A.; Volz, A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm. Genome 1999, 10, 154–160. [Google Scholar] [CrossRef]
- Vilches, C.; Parham, P. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002, 20, 217–251. [Google Scholar] [CrossRef]
- Moretta, A.; Sivori, S.; Vitale, M.; Pende, D.; Morelli, L.; Augugliaro, R.; Bottino, C.; Moretta, L. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J. Exp. Med. 1995, 182, 875–884. [Google Scholar] [CrossRef]
- Trowsdale, J.; Barten, R.; Haude, A.; Stewart, C.A.; Beck, S.; Wilson, M.J. The genomic context of natural killer receptor extended gene families. Immunol. Rev. 2001, 181, 20–38. [Google Scholar] [CrossRef]
- Faure, M.; Long, E.O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J. Immunol. 2002, 168, 6208–6214. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Long, E.O. KIR2DL4 (CD158d): An activation receptor for HLA-G. Front. Immunol. 2012, 3, 258. [Google Scholar] [CrossRef] [PubMed]
- Parham, P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005, 5, 201–214. [Google Scholar] [CrossRef] [PubMed]
- De Alencar, J.B.; Zacarias, J.M.V.; Moura, B.L. da S.G. de; Braga, M.A.; Visentainer, J.E.L.; Sell, A.M. KIR and HLA ligands demonstrate genetic inheritance diversity in Japanese descendants from Paraná, Brazil. Hum. Immunol. 2018, 79, 191–192. [Google Scholar] [CrossRef]
- Amorim, L.M.; van Tong, H.; Hoan, N.X.; Vargas, L.d.B.; Ribeiro, E.M.d.S.F.; Petzl-Erler, M.L.; Boldt, A.B.W.; Toan, N.L.; Song, L.H.; Velavan, T.P.; et al. KIR-HLA distribution in a Vietnamese population from Hanoi. Hum. Immunol. 2018, 79, 93–100. [Google Scholar] [CrossRef]
- Chewning, J.H.; Gudme, C.N.; Hsu, K.C.; Selvakumar, A.; Dupont, B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J. Immunol. 2007, 179, 854–868. [Google Scholar] [CrossRef]
- Foley, B.A.; De Santis, D.; Van Beelen, E.; Lathbury, L.J.; Christiansen, F.T.; Witt, C.S. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: Implications for patient and donor suitability for haploidentical stem cell transplantations. Blood 2008, 112, 435–443. [Google Scholar] [CrossRef]
- Katz, G.; Markel, G.; Mizrahi, S.; Arnon, T.I.; Mandelboim, O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J. Immunol. 2001, 166, 7260–7267. [Google Scholar] [CrossRef]
- González-Galarza, F.F.; Takeshita, L.Y.C.; Santos, E.J.M.; Kempson, F.; Maia, M.H.T.; Da Silva, A.L.S.; Teles E Silva, A.L.; Ghattaoraya, G.S.; Alfirevic, A.; Jones, A.R.; et al. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015, 43, D784–D788. [Google Scholar] [CrossRef]
- Mateo, R.; Cho, Y.; Singh, G.; Stapfer, M.; Donovan, J.; Kahn, J.; Fong, T.-L.; Sher, L.; Jabbour, N.; Aswad, S.; et al. Risk factors for graft survival after liver transplantation from donation after cardiac death donors: An analysis of OPTN/UNOS data. Am. J. Transplant. 2006, 6, 791–796. [Google Scholar] [CrossRef]
- Merion, R.M.; Pelletier, S.J.; Goodrich, N.; Englesbe, M.J.; Delmonico, F.L. Donation after cardiac death as a strategy to increase deceased donor liver availability. Ann. Surg. 2006, 244, 555–562. [Google Scholar] [CrossRef] [PubMed]
- De Vera, M.E.; Lopez-Solis, R.; Dvorchik, I.; Campos, S.; Morris, W.; Demetris, A.J.; Fontes, P.; Marsh, J.W. Liver transplantation using donation after cardiac death donors: Long-term follow-up from a single center. Am. J. Transplant. 2009, 9, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Jay, C.L.; Skaro, A.I.; Ladner, D.P.; Wang, E.; Lyuksemburg, V.; Chang, Y.; Xu, H.; Talakokkla, S.; Parikh, N.; Holl, J.L.; et al. Comparative effectiveness of donation after cardiac death versus donation after brain death liver transplantation: Recognizing who can benefit. Liver Transpl. 2012, 18, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Stewart, Z.A.; Locke, J.E.; Montgomery, R.A.; Singer, A.L.; Cameron, A.M.; Segev, D.L. ABO-incompatible deceased donor liver transplantation in the United States: A national registry analysis. Liver Transpl. 2009, 15, 883–893. [Google Scholar] [CrossRef]
- Adam, R.; Delvart, V.; Karam, V. Reply to Letter Regarding “Compared Efficacy of Preservation Solutions in Liver Transplantation: A Long-Term Graft Outcome Study From the European Liver Transplant Registry”. Am. J. Transplant. 2015, 15, 3274–3275. [Google Scholar] [CrossRef]
- Organización Nacional de Transplantes. Available online: www.ont.es/home/Paginas/default.aspx (accessed on 4 September 2022).
- Yersiz, H.; Shaked, A.; Olthoff, K.; Imagawa, D.; Shackleton, C.; Martin, P.; Busuttil, R.W. Correlation between donor age and the pattern of liver graft recovery after transplantation. Transplantation 1995, 60, 790–794. [Google Scholar] [CrossRef]
- Sahin, U.; Dalva, K.; Gungor, F.; Ustun, C.; Beksac, M. Donor-recipient killer immunoglobulin like receptor (KIR) genotype matching has a protective effect on chronic graft versus host disease and relapse incidence following HLA-identical sibling hematopoietic stem cell transplantation. Ann. Hematol. 2018, 97, 1027–1039. [Google Scholar] [CrossRef]
- Lee, H.; Park, K.H.; Park, H.S.; Ryu, J.H.; Lim, J.; Kim, Y.; Na, G.H.; Kim, D.G.; Oh, E.J. Human Leukocyte Antigen-C Genotype and Killer Immunoglobulin-like Receptor-Ligand Matching in Korean Living Donor Liver Transplantation. Ann. Lab. Med. 2017, 37, 45–52. [Google Scholar] [CrossRef]
- Nowak, I.; Magott-Procelewska, M.; Kowal, A.; Miazga, M.; Wagner, M.; Niepiekło-Miniewska, W.; Kamińska, M.; Wiśniewski, A.; Majorczyk, E.; Klinger, M.; et al. Killer Immunoglobulin-like Receptor (KIR) and HLA Genotypes Affect the Outcome of Allogeneic Kidney Transplantation. PLoS ONE 2012, 7, e44718. [Google Scholar] [CrossRef]
- Nowak, I.; Majorczyk, E.; Wiśniewski, A.; Pawlik, A.; Magott-Procelewska, M.; Passowicz-Muszyńska, E.; Malejczyk, J.; Płoski, R.; Giebel, S.; Barcz, E.; et al. Does the KIR2DS5 gene protect from some human diseases? PLoS ONE 2010, 5, e12381. [Google Scholar] [CrossRef]
- Kuśnierczyk, P. Are killer cell immunoglobulin-like receptor genes important for the prediction of kidney graft rejection? Arch. Immunol. Ther. Exp. 2013, 61, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Stringaris, K.; Barrett, A.J. The importance of natural killer cell killer immunoglobulin-like receptor-mismatch in transplant outcomes. Curr. Opin. Hematol. 2017, 24, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Cooley, S.; Weisdorf, D.J.; Guethlein, L.A.; Klein, J.P.; Wang, T.; Le, C.T.; Marsh, S.G.E.; Geraghty, D.; Spellman, S.; Haagenson, M.D.; et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010, 116, 2411–2419. [Google Scholar] [CrossRef] [PubMed]
- López-Alvarez, M.R.; Moya-Quiles, M.R.; Minguela, A.; Gil, J.; Miras, M.; Campillo, J.A.; Díaz-Alderete, M.A.; García-Alonso, A.M.; Sánchez-Bueno, F.; Vicario, J.L.; et al. HLA-C matching and liver transplants: Donor-recipient genotypes influence early outcome and CD8+KIR2D+ T-cells recuperation. Transplantation 2009, 88, S54–S61. [Google Scholar] [CrossRef]
- Parham, P. Immunogenetics of killer cell immunoglobulin-like receptors. Mol. Immunol. 2005, 42, 459–462. [Google Scholar] [CrossRef]
- Schönberg, K.; Fischer, J.C.; Kögler, G.; Uhrberg, M. Neonatal NK-cell repertoires are functionally, but not structurally, biased toward recognition of self HLA class I. Blood 2011, 117, 5152–5156. [Google Scholar] [CrossRef][Green Version]
- Moesta, A.K.; Norman, P.J.; Yawata, M.; Yawata, N.; Gleimer, M.; Parham, P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008, 180, 3969–3979. [Google Scholar] [CrossRef]
- Norman, P.J.; Stephens, H.A.; Verity, D.H.; Chandanayingyong, D.; Vaughan, R.W. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 2001, 52, 195–205. [Google Scholar]
- Husain, Z.; Alper, C.A.; Yunis, E.J.; Dubey, D.P. Complex expression of natural killer receptor genes in single natural killer cells. Immunology 2002, 106, 373–380. [Google Scholar] [CrossRef]
- Graef, T.; Moesta, A.K.; Norman, P.J.; Abi-Rached, L.; Vago, L.; Older Aguilar, A.M.; Gleimer, M.; Hammond, J.A.; Guethlein, L.A.; Bushnell, D.A.; et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A * 11 while diminishing avidity for HLA-C. J. Exp. Med. 2009, 206, 2557–2572. [Google Scholar] [CrossRef]
- Katz, G.; Gazit, R.; Arnon, T.I.; Gonen-Gross, T.; Tarcic, G.; Markel, G.; Gruda, R.; Achdout, H.; Drize, O.; Merims, S.; et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J. Immunol. 2004, 173, 1819–1825. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Piredda, G.; Argiolas, D.; Lai, S.; Congeddu, E.; Ragatzu, P.; Melis, M.; Carta, E.; Michittu, M.B.; Valentini, D.; et al. KIR and their HLA Class I ligands: Two more pieces towards completing the puzzle of chronic rejection and graft loss in kidney transplantation. PLoS ONE 2017, 12, e0180831. [Google Scholar] [CrossRef] [PubMed]
- Demetris, A.J.; Murase, N.; Lee, R.G.; Randhawa, P.; Zeevi, A.; Pham, S.; Duquesnoy, R.; Fung, J.J.; Starzl, T.E. Chronic rejection. A general overview of histopathology and pathophysiology with emphasis on liver, heart and intestinal allografts. Ann. Transplant. 1997, 2, 27–44. [Google Scholar] [PubMed]
- Vilches, C.; Castaño, J.; Gómez-Lozano, N.; Estefanía, E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 2007, 70, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Gourraud, P.-A.; Meenagh, A.; Cambon-Thomsen, A.; Middleton, D. Linkage disequilibrium organization of the human KIR superlocus: Implications for KIR data analyses. Immunogenetics 2010, 62, 729–740. [Google Scholar] [CrossRef]
- Middleton, D.; Christiansen, F.T. 14th International HLA and Immunogenetics Workshop: Report on KIR receptors and their applications. Tissue Antigens 2007, 69 (Suppl. S1), 85–87. [Google Scholar] [CrossRef]
- McQueen, K.L.; Dorighi, K.M.; Guethlein, L.A.; Wong, R.; Sanjanwala, B.; Parham, P. Donor-Recipient Combinations of Group A and B KIR Haplotypes and HLA class I Ligand Affect the Outcome of HLA-Matched, Sibling Donor Hematopoietic Cell Transplantation. Hum. Immunol. 2007, 68, 309–323. [Google Scholar] [CrossRef]
- Pyo, C.W.; Guethlein, L.A.; Vu, Q.; Wang, R.; Abi-Rached, L.; Norman, P.J.; Marsh, S.G.E.; Miller, J.S.; Parham, P.; Geraghty, D.E. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE 2010, 5, e15115. [Google Scholar] [CrossRef]
- Moya-Quiles, M.R.; Torío, A.; Muro, M.; Montes-Ares, O.; Marin, L.; Minguela, A.; Sánchez-Bueno, F.; Garcia-Alonso, A.M.; Parrilla, P.; Alvarez-López, M.R. Impact of HLA-C on acute rejection in liver transplantation. Transplant. Proc. 2003, 35, 1892–1893. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, X.; Liu, K.; Xu, L.; Liu, D. Prognosis after unmanipulated HLA-haploidentical blood and marrow transplantation is correlated to the numbers of KIR ligands in recipients. Eur. J. Haematol. 2007, 78, 338–346. [Google Scholar] [CrossRef]
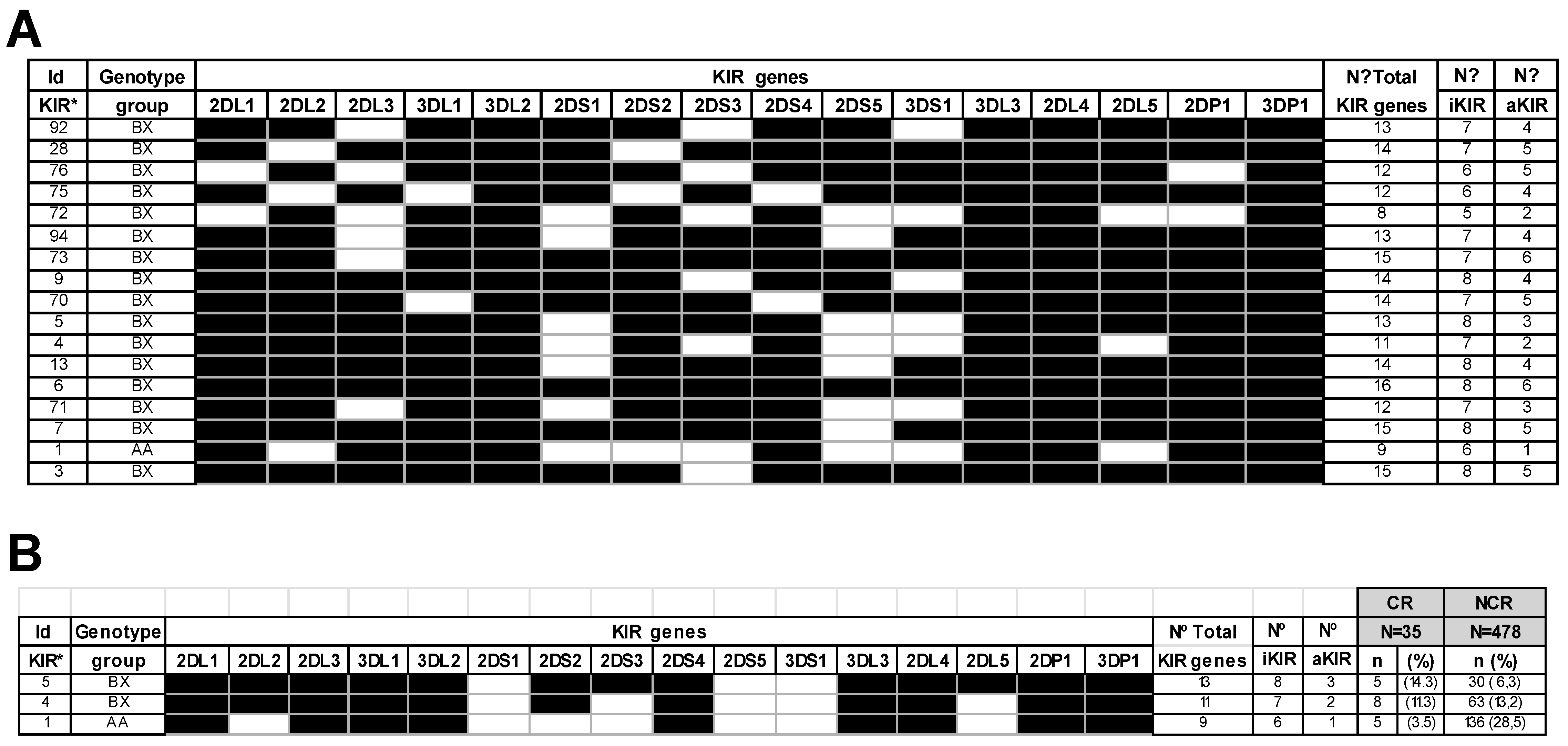
| n (%) | Age (Mean ± SEM) | |
|---|---|---|
| Total of Recipients, n (%) | 513 | 53.0 ± 0.5 |
| CR group | 35 (6.8) | 49.9 ± 2.2 |
| NCR group | 478 (93.2) | 53.3 ± 0.5 |
| Total of donors, n (%) | 513 | 51.2 ± 0.9 |
| CR group | 35 (6.8) | 44.1 ± 3.6 a |
| NCR group | 478 (93.2) | 51.8 ± 0.9 |
| Total of recipients, n (%) | 513 | CR, n (%) |
| Gender | ||
| Male | 382 (74.5) | 26 (6.8) |
| Female | 131 (25.5) | 9 (6.9) |
| Transplant indications, n (%) | ||
| Alcoholic cirrhosis | 169 (32.9) | 17 (10.1) |
| Alcoholic cirrhosis plus HCV and/or HBV | 54 (10.5) | 1 (1.9) |
| Viral chronic hepatitis | 108 (21.1) | 10 (9.3) |
| HCV | 91 (17.7) | 9 (9.9) |
| HBV | 17 (3.3) | 1 (5.9) |
| Hepatocarcinoma | 72 (14.0) | 3 (4.2) |
| Autoimmune diseases | 27 (5.3) | 2 (7.4) |
| Fulminant hepatitis | 16 (3.1) | 0 (0) |
| Others | 67 (13) | 2 (3.5) |
| Virus pre-infected patients *, n (%) | ||
| HCV | 155 (30.2) | 10 (6.5) |
| HBV | 51 (9.9) | 3 (5.9) |
| CMV | 11 (2.1) | 0 (0) |
| Healthy Donors N = 513 | Total Patients N = 513 | Chronic Rejection N = 35 | |||||
|---|---|---|---|---|---|---|---|
| KIR Genes | P/A | n (%) | n (%) | P1 | n (%) | P2 | P3 |
| iKIRs | |||||||
| 2DL1+/S1− | + | 311 (60.6) | 306 (59.6) | 0.799 | 22 (7.2) | 0.725 | - |
| − | 202 (39.4) | 207 (40.4) | 13 (6.3) | ||||
| 2DL2 | + | 316 (61.6) | 296 (57.7) | 0.227 | 27 (9.1) | 0.020 b | 0.033 |
| − | 197 (38.4) | 217 (42.3) | 8 (3.7) | ||||
| 2DL3 | + | 449 (87.5) | 454 (88.5) | 0.701 | 28 (6.2) | 0.105 | - |
| − | 64 (12.5) | 59 (11.5) | 7 (11.9) | ||||
| 2DL5 | + | 268 (52.2) | 285 (55.6) | 0.316 | 21 (7.4) | 0.603 | - |
| − | 245 (47.8) | 228 (44.4) | 14 (6.2) | ||||
| 3DL1 | + | 490 (95.5) | 490 (95.5) | 1.000 | 33 (6.7) | 0.665 | - |
| − | 23 (4.5) | 23 (4.5) | 2 (8.7) | ||||
| aKIRs | |||||||
| 2DS1 | + | 192 (37.4) | 204 (39.8) | 0.481 | 12 (5.9) | 0.593 | - |
| − | 321 (62.6) | 309 (60.2) | 23 (7.4) | ||||
| 2DS2 | + | 317 (61.8) | 292 (56.9) | 0.127 | 27 (9.2) | 0.013 c | 0.030 |
| − | 196 (38.2) | 221 (43.1) | 8 (3.6) | ||||
| 2DS3 | + | 169 (32.9) | 164 (32.0) | 0.790 | 17 (10.4) | 0.038 d | 0.037 |
| − | 344 (67.1) | 349 (68.0) | 18 (5.2) | ||||
| 2DS4 | + | 491 (95.7) | 488 (95.1) | 0.766 | 33 (6.8) | 0.685 | - |
| − | 22 (4.3) | 25 (4.9) | 2 (8.0) | ||||
| 2DS5 | + | 143 (27.9) | 174 (33.9) | 0.043 a | 11 (6.3) | 0.854 | - |
| − | 370 (72.1) | 339 (66.1) | 24 (7.1) | ||||
| 3DS1 | + | 143 (27.9) | 215 (41.9) | 0.408 | 13 (6.0) | 0.599 | - |
| − | 370 (72.1) | 298 (58.1) | 22 (7.4) |
| Chronic Rejection | |||||
|---|---|---|---|---|---|
| All KIRs | N | n (%) | OR (95% CI) | P1 | P2 |
| 0 MM | 67 | 3 (4.5) | 1.649 (0.491–5.543) | 0.603 | - |
| ≥1 MM | 446 | 32 (7.2) | |||
| iKIRs a | |||||
| 0 MM | 120 | 3 (2.5) | 3.457 (1.040–11.497) | 0.037 | 0.033 |
| ≥1 MM | 393 | 32 (8.1) | |||
| KIR2DL3 | |||||
| 0 MM | 406 | 21 (5.2) | 2.760 (1.353–5.631) | 0.008 | 0.191 |
| 1 MM | 107 | 14 (13.1) | |||
| aKIRs b | |||||
| 0 MM | 80 | 4 (5.0) | 1.465 (0.503–4.271) | 0.632 | - |
| ≥1 MM | 433 | 31 (7.2) | |||
| Chronic Rejection | |||||
|---|---|---|---|---|---|
| HLA-A Mismatches | N | n | % | OR (95% CI) | P * |
| 2 MM | 328 | 23 | 7 | 1.068 (0.519–2.201) | 1.000 |
| 1 MM | 155 | 10 | 6.5 | 0.910 (0.426–1.944) | 1.000 |
| 0 MM | 27 | 2 | 7.4 | 1.091 (0.248–4.807) | 0.707 |
| HLA-B Mismatches | |||||
| 2 MM | 385 | 25 | 6.5 | 0.895 (0.406–1.972) | 0.837 |
| 1 MM | 111 | 9 | 8.1 | 1.320 (0.597–2.916) | 0.519 |
| 0 MM | 14 | 0 | 0 | 0.931 (0.909–0.954) | 0.614 |
| HLA-C Mismatches | |||||
| 2 MM | 341 | 24 | 7 | 1.108 (0.530–2.319) | 0.855 |
| 1 MM | 141 | 9 | 6.4 | 0.907 (0.414–1.987) | 1.000 |
| 0 MM | 31 | 2 | 6.5 | 0.938 (0.214–4.105) | 1.000 |
| A. Recipient and Donor HLA-C Allotypes and Their Influence in Chronic Rejection Liver | ||||||
| HLA-C Allotypes | Chronic Rejection | |||||
| N = 513 | n | % | P a | |||
| Recipient | C1+ | 409 | 31 | 7.6 | 0.274 | |
| C1− | 104 | 4 | 3.8 | |||
| C2+ | 331 | 22 | 6.6 | 0.856 | ||
| C2− | 182 | 13 | 7.1 | |||
| Donor | C1+ | 429 | 24 | 5.6 | 0.018 b | |
| C1− | 84 | 11 | 13.1 | |||
| C2+ | 340 | 26 | 7.6 | 0.357 | ||
| C2− | 173 | 9 | 5.2 | |||
| B. Recipient and donor HLA-C genotypes and their influence in chronic rejection liver. | ||||||
| Chronic rejection | ||||||
| HLA-C genotypes | N = 513 | n | % | X2 | P a | |
| C1C1 | 182 | 13 | 7.1 | 0.856 | ||
| Recipient | C1C2 | 227 | 18 | 7.9 | 0.384 | 0.384 |
| C2C2 | 104 | 4 | 3.8 | 0.274 | ||
| C1C1 | 173 | 9 | 5.2 | 0.357 | ||
| Donor | C1C2 | 256 | 15 | 5.9 | 0.043 | 0.484 |
| C2C2 | 84 | 11 | 13.1 | 0.018 b | ||
| HLA-C Genotypes Combinations | Chronic Rejection | ||||
|---|---|---|---|---|---|
| Donor | Recipient | N = 513 | n | % | P a |
| C1C1 | 65 | 4 | 6.2 | 0.295 | |
| C1C1 | C1C2 | 73 | 5 | 6.8 | |
| C2C2 | 35 | 0 | 0 | ||
| C1C1 | 87 | 4 | 4.6 | 0.806 | |
| C1C2 | C1C2 | 118 | 8 | 6.8 | |
| C2C2 | 51 | 3 | 5.9 | ||
| C1C1 | 30 | 5 | 16.7 | 0.534 | |
| C2C2 | C1C2 | 36 | 5 | 13.9 | |
| C2C2 | 18 | 1 | 5.6 | ||
| Chronic Rejection | |||||||
|---|---|---|---|---|---|---|---|
| Recipient KIRs | Donor HLA-I Ligand | N | n | % | OR (95%CI) | P a | P b |
| iKIRs | |||||||
| KIR2DL1+/S1− | C2+ | 193 | 17 | 8.8 | 2.086 (0.748–5.818) | 0.175 | - |
| C2– | 113 | 5 | 4.4 | ||||
| KIR2DL2+/S2+ | C1+ | 248 | 19 | 7.7 | 0.415 (0.170–1.012) | 0.057 | - |
| C1– | 48 | 8 | 16.7 | ||||
| KIR2DL3+ | C1+ | 374 | 18 | 4.8 | 0.334 (0.148–0.755) | 0.015 | 0.004 |
| C1– | 76 | 10 | 13.2 | ||||
| KIR3DL1+ | Bw4+ | 387 | 27 | 7.0 | 1.213 (0.487–3.020) | 0.826 | - |
| Bw4– | 103 | 6 | 5.8 | ||||
| aKIRs | |||||||
| KIR2DL1+/KIR2DS1+ | C2+ | 145 | 8 | 5.5 | 0.803 (0.232–2.776) | 0.747 | - |
| C2– | 59 | 4 | 6.8 | ||||
| C2+ | 323 | 25 | 7.7 | 1.646 (0.726–3.735) | 0.248 | - | |
| C2– | 165 | 8 | 4.8 | ||||
| KIR2DS4+ | |||||||
| C1+ | 406 | 22 | 5.4 | 0.370 (0.172–0.796) | 0.014 | 0.028 | |
| C1– | 82 | 11 | 13.4 | ||||
| KIR2DL3+/S4+ | C1+ | 353 | 16 | 4.5 | 0.304 (0.132–0.700) | 0.006 | 0.003 |
| C1– | 74 | 10 | 13.5 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legaz, I.; Bolarín, J.M.; Campillo, J.A.; Moya-Quiles, M.R.; Miras, M.; Muro, M.; Minguela, A.; Álvarez-López, M.R. Killer Cell Immunoglobulin-like Receptors (KIR) and Human Leucocyte Antigen C (HLA-C) Increase the Risk of Long-Term Chronic Liver Graft Rejection. Int. J. Mol. Sci. 2022, 23, 12155. https://doi.org/10.3390/ijms232012155
Legaz I, Bolarín JM, Campillo JA, Moya-Quiles MR, Miras M, Muro M, Minguela A, Álvarez-López MR. Killer Cell Immunoglobulin-like Receptors (KIR) and Human Leucocyte Antigen C (HLA-C) Increase the Risk of Long-Term Chronic Liver Graft Rejection. International Journal of Molecular Sciences. 2022; 23(20):12155. https://doi.org/10.3390/ijms232012155
Chicago/Turabian StyleLegaz, Isabel, Jose Miguel Bolarín, Jose Antonio Campillo, María R. Moya-Quiles, Manuel Miras, Manuel Muro, Alfredo Minguela, and María R. Álvarez-López. 2022. "Killer Cell Immunoglobulin-like Receptors (KIR) and Human Leucocyte Antigen C (HLA-C) Increase the Risk of Long-Term Chronic Liver Graft Rejection" International Journal of Molecular Sciences 23, no. 20: 12155. https://doi.org/10.3390/ijms232012155
APA StyleLegaz, I., Bolarín, J. M., Campillo, J. A., Moya-Quiles, M. R., Miras, M., Muro, M., Minguela, A., & Álvarez-López, M. R. (2022). Killer Cell Immunoglobulin-like Receptors (KIR) and Human Leucocyte Antigen C (HLA-C) Increase the Risk of Long-Term Chronic Liver Graft Rejection. International Journal of Molecular Sciences, 23(20), 12155. https://doi.org/10.3390/ijms232012155









