Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female db/db Mice
Abstract
1. Introduction
2. Results
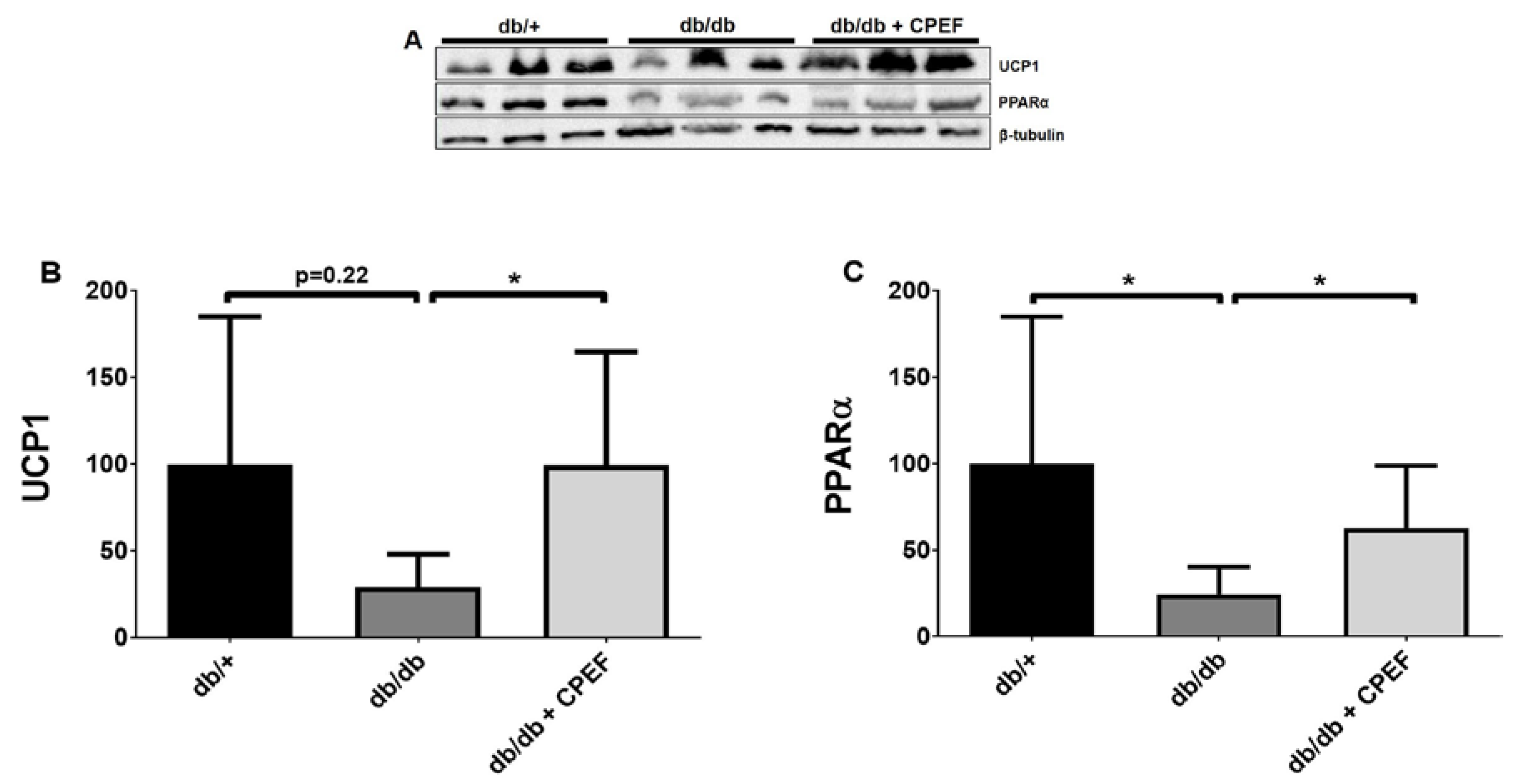
2.1. Treatment with CPEF Increases UCP1 and PPARα Expression in Brown Adipose Tissue
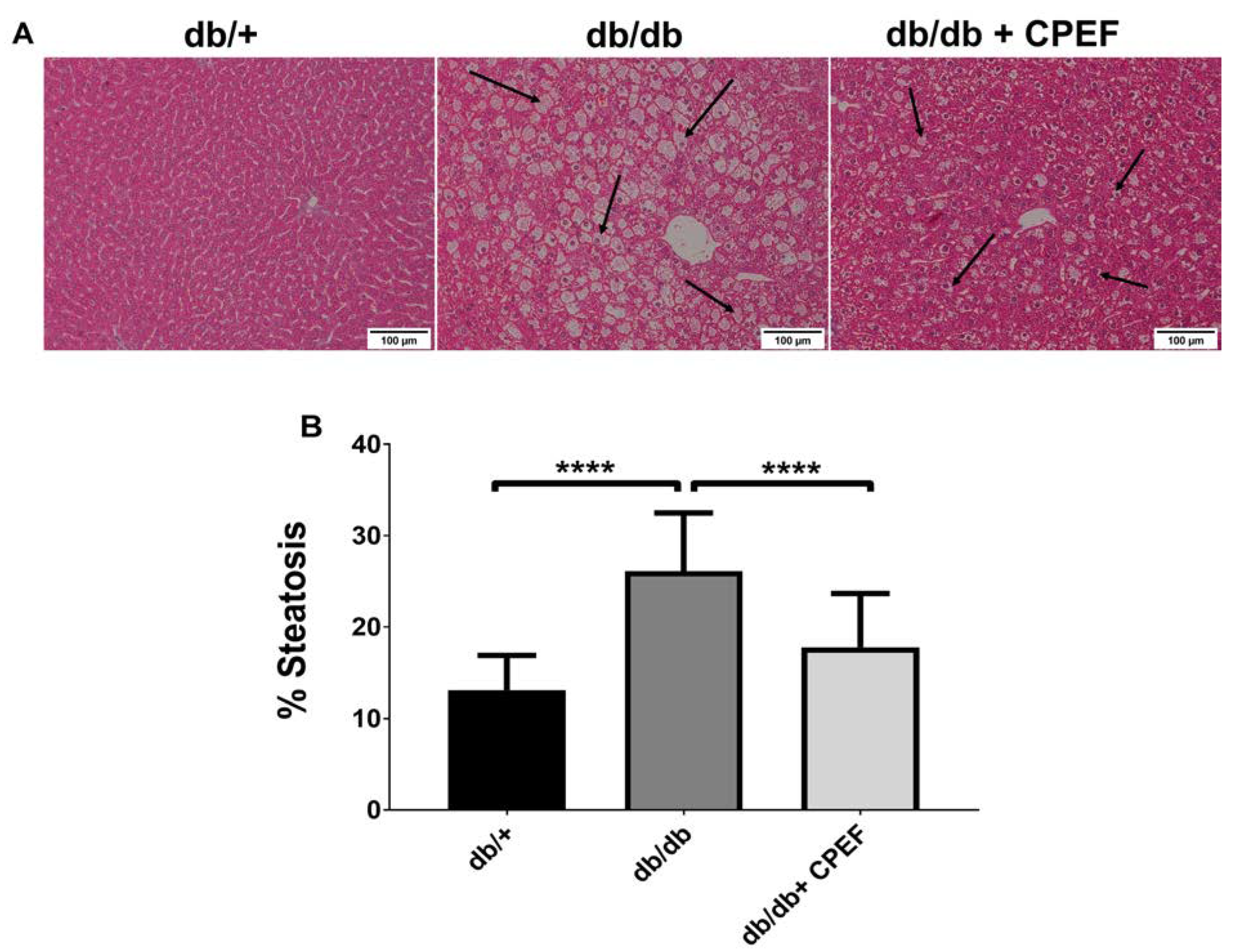
2.2. Treatment with CPEF Decreases Hepatic Lipid Accumulation
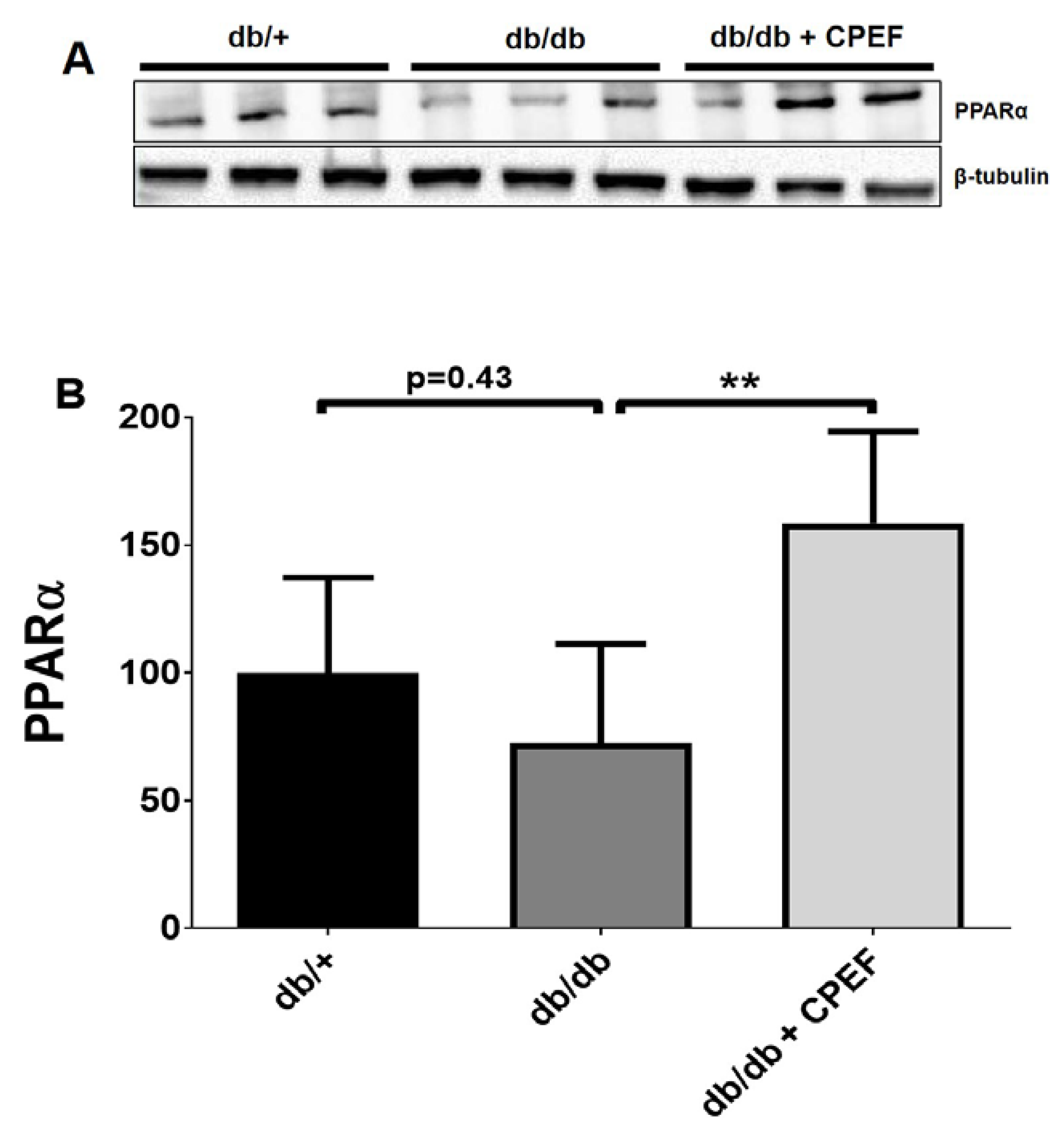
2.3. Treatment with CPEF Increases Hepatic PPARα Expression
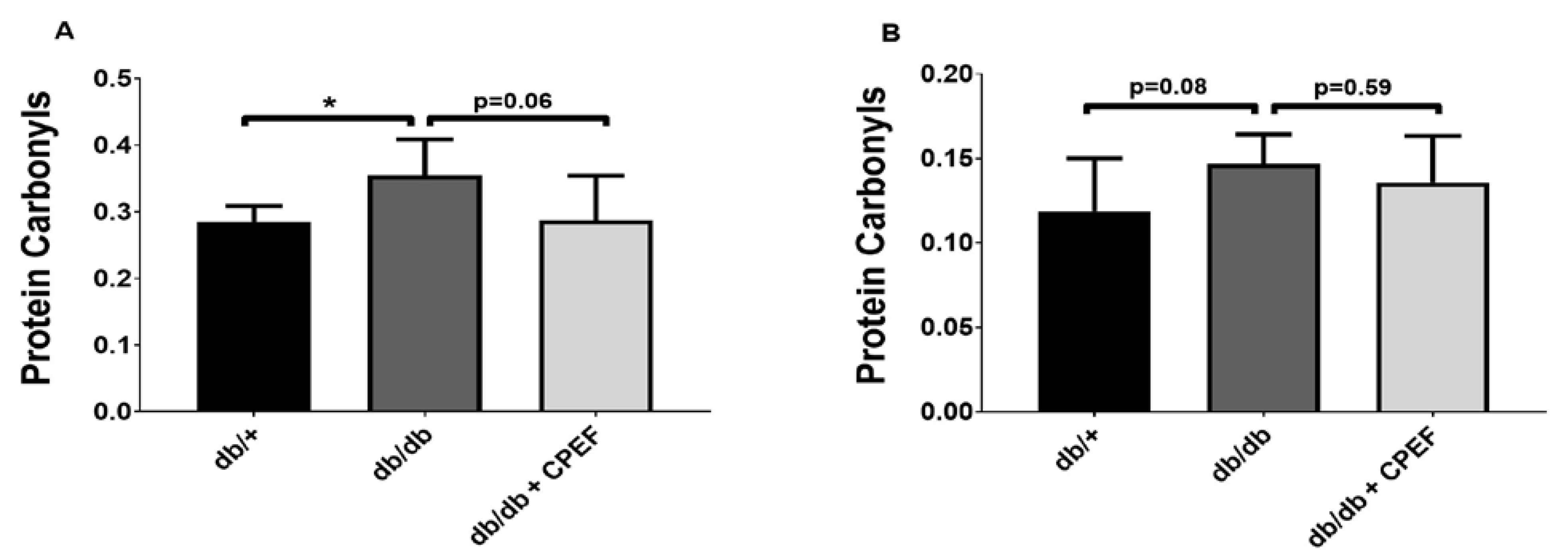
2.4. Effect of CPEF Treatment on Lipid Peroxidation and Protein Oxidation
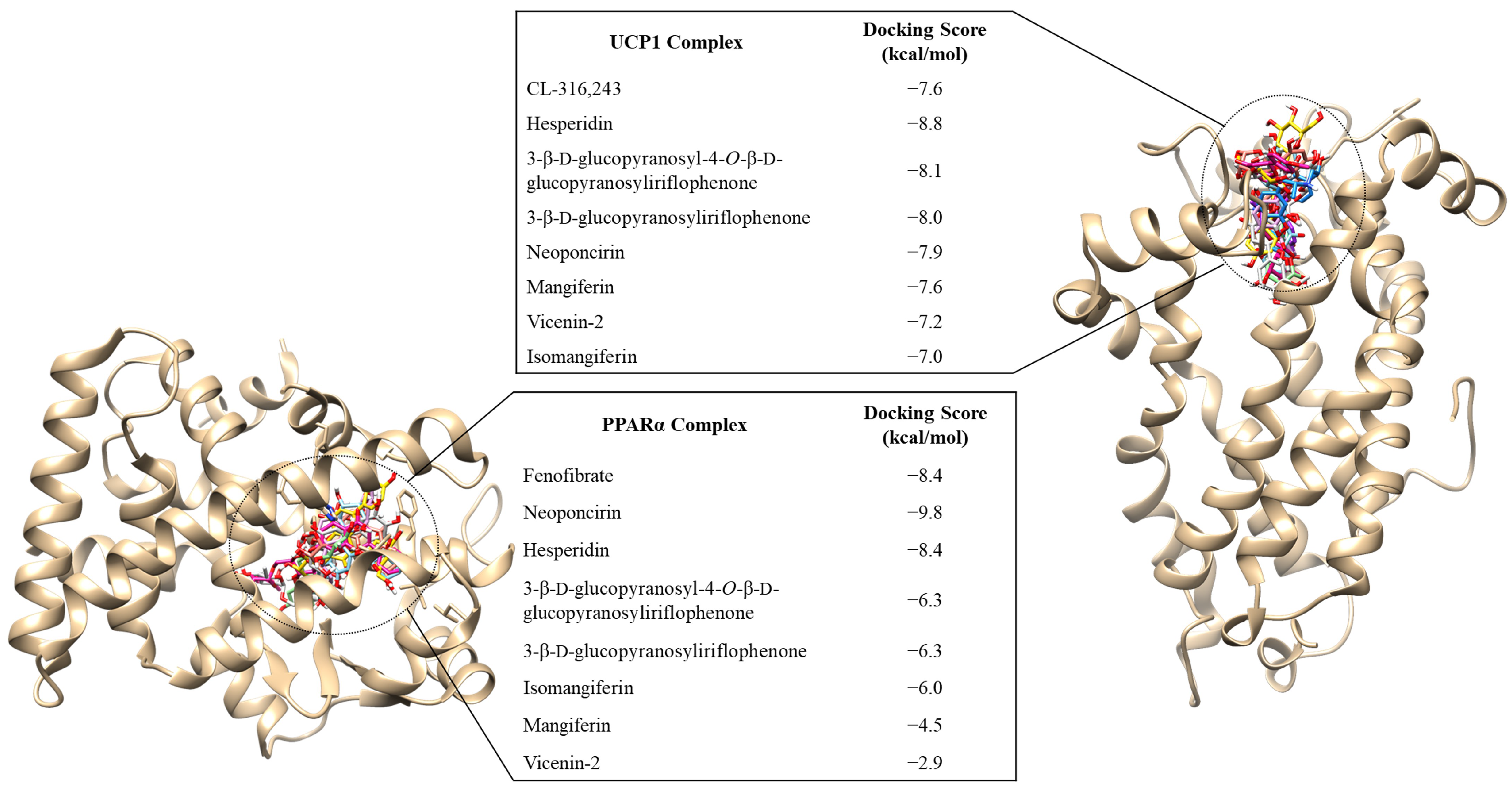
2.5. Prediction of Compounds Responsible for the Increased Expression of UCP1 and PPARα, Elicited through CPEF
3. Discussion
4. Materials and Methods
4.1. Preparation of CPEF
4.2. Animals and Treatment Protocol
4.3. Histology
4.4. Protein Extraction and Western Blot Analysis
4.5. Protein Oxidation
4.6. Lipid Peroxidation
4.7. Molecular Docking
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAT | brown adipose tissue |
| CPEF | crude polyphenol enriched fraction of C. intermedia |
| CVD | cardiovascular disease |
| DMSO | dimethyl sulfoxide |
| H&E | hematoxylin and eosin |
| HSL | hormone-sensitive lipase |
| iBAT | interscapular brown adipose tissue |
| MDA | malondialdehyde |
| PMSF | phenylmethane sulfonyl fluoride |
| PPARα | peroxisome proliferator-activated receptor alpha |
| PPARγ | peroxisome proliferator-activated receptor gamma |
| PVDF | polyvinylidene fluoride |
| SDS | sodium dodecyl sulfate |
| T2D | type 2 diabetes |
| TBARS | thiobarbituric acid reactive substances |
| TBST | tris-buffered saline Tween 20 |
| UCP1 | uncoupling protein 1 |
| UCP3 | uncoupling protein 3 |
| WAT | white adipose tissue |
References
- Hruby, A.; Hu, F.B. The epidemiology of obesity: A big picture. Pharmacoeconomics 2015, 33, 673–689. [Google Scholar] [CrossRef] [PubMed]
- WHO. Obesity and Overweight. Available online: https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 27 November 2019).
- NCD-RisC. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19.2 million participants. Lancet 2016, 387, 1377–1396. [Google Scholar] [CrossRef] [PubMed]
- Garawi, F.; Devries, K.; Thorogood, N.; Uauy, R. Global differences between women and men in the prevalence of obesity: Is there an association with gender inequality? Eur. J. Clin. Nutr. 2014, 68, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the global burden of disease study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.; Stoenchev, K.; Ashrafian, H.; Teare, J. Current treatments for obesity. Clin. Med. J. R. Coll. Physicians Lond. 2019, 19, 205–212. [Google Scholar] [CrossRef]
- Wadden, T.A.; Tronieri, J.S.; Butryn, M.L. Lifestyle modification approaches for the treatment of obesity in adults. Am. Psychol. 2020, 75, 235–251. [Google Scholar] [CrossRef]
- Joubert, E.; Gelderblom, W.C.A.; Louw, A.; de Beer, D. South African herbal teas: Aspalathus linearis, Cyclopia spp. and Athrixia phylicoides—A review. J. Ethnopharmacol. 2008, 119, 376–412. [Google Scholar] [CrossRef]
- Watt, J.M.; Breyer-Brandwijk, M.G. The Medicinal and Poisonous Plants of Southern Africa, 1st ed.; E & S Livingstone: Edinburgh, UK, 1932; p. 70. [Google Scholar]
- Joubert, E.; de Beer, D.; Malherbe, C.J.; Muller, M.; Louw, A.; Gelderblom, W.C.A. Formal honeybush tea industry reaches 20-year milestone—Progress of product research targeting phenolic composition, quality and bioactivity. S. Afr. J. Bot. 2019, 127, 58–79. [Google Scholar] [CrossRef]
- Dudhia, Z.; Louw, J.; Muller, C.; Joubert, E.; de Beer, D.; Kinnear, C.; Pheiffer, C. Cyclopia maculata and Cyclopia subternata (honeybush tea) inhibits adipogenesis in 3T3-L1 pre-adipocytes. Phytomedicine 2013, 20, 401–408. [Google Scholar] [CrossRef]
- Pheiffer, C.; Dudhia, Z.; Louw, J.; Muller, C.; Joubert, E. Cyclopia maculata (honeybush tea) stimulates lipolysis in 3T3-L1 adipocytes. Phytomedicine 2013, 20, 1168–1171. [Google Scholar] [CrossRef]
- Jack, B.U.; Malherbe, C.J.; Huisamen, B.; Gabuza, K.; Mazibuko-Mbeje, S.; Schulze, A.E.; Joubert, E.; Muller, C.J.F.; Louw, J.; Pheiffer, C. A polyphenol-enriched fraction of Cyclopia intermedia decreases lipid content in 3T3-L1 adipocytes and reduces body weight gain of obese db/db mice. S. Afr. J. Bot. 2017, 110, 216–229. [Google Scholar] [CrossRef]
- Jack, B.U. An Investigation into the Anti-Obesity Properties of Cyclopia. Ph.D. Thesis, University of Stellenbosch, Tygerberg, South Africa, 2016. [Google Scholar]
- Serra, D.; Mera, P.; Malandrino, M.I.; Mir, J.F.; Herrero, L. Mitochondrial fatty acid oxidation in obesity. Antioxid. Redox Signal. 2013, 19, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Yamada, T. UCP1 dependent and independent thermogenesis in brown and beige adipocytes. Front. Endocrinol. 2020, 11, 498. [Google Scholar] [CrossRef]
- Rahman, M.S.; Kim, Y.-S. Mangiferin induces the expression of a thermogenic signature via AMPK signaling during brown-adipocyte differentiation. Food Chem. Toxicol. 2020, 141, 111415. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Kim, Y.-S. PINK1-PRKN mitophagy suppression by mangiferin promotes a brown-fat-phenotype via PKA-P38 MAPK signalling in murine C3H10T1/2 mesenchymal stem cells. Metabolism 2020, 107, 154228. [Google Scholar] [CrossRef]
- Mosqueda-Solís, A.; Sánchez, J.; Portillo, M.P.; Palou, A.; Picó, C. Combination of capsaicin and hesperidin reduces the effectiveness of each compound to decrease the adipocyte size and to induce browning features in adipose tissue of western diet fed rats. J. Agric. Food Chem. 2018, 66, 9679–9689. [Google Scholar] [CrossRef]
- Busiello, R.A.; Savarese, S.; Lombardi, A. Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 2015, 6, 36. [Google Scholar] [CrossRef]
- Pfannenberg, C.; Werner, M.K.; Ripkens, S.; Stef, I.; Deckert, A.; Schmadl, M.; Reimold, M.; Häring, H.-U.; Claussen, C.D.; Stefan, N. Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes 2010, 59, 1789–1793. [Google Scholar] [CrossRef]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Okamatsu-Ogura, Y.; Kameya, T.; Kawai, Y.; Miyagawa, M.; Tsujisaki, M.; Saito, M. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity 2011, 19, 1755–1760. [Google Scholar] [CrossRef]
- Mele, L.; Bidault, G.; Mena, P.; Crozier, A.; Brighenti, F.; Vidal-Puig, A.; Rio, D.D. Dietary (poly)phenols, brown adipose tissue activation, and energy expenditure: A narrative review. Adv. Nutr. 2017, 8, 694. [Google Scholar] [CrossRef]
- Azhar, Y.; Parmar, A.; Miller, C.N.; Samuels, J.S.; Rayalam, S. Phytochemicals as novel agents for the induction of browning in white adipose tissue. Nutr. Metab. 2016, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Hondares, E.; Rosell, M.; Díaz-Delfín, J.; Olmos, Y.; Monsalve, M.; Iglesias, R.; Villarroya, F.; Giralt, M. Peroxisome proliferator-activated receptor α (PPARα) induces PPARγ coactivator 1α (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: Involvement of PRDM16. J. Biol. Chem. 2011, 286, 43112–43122. [Google Scholar] [CrossRef]
- Kroon, T.; Harms, M.; Maurer, S.; Bonnet, L.; Alexandersson, I.; Lindblom, A.; Ahnmark, A.; Nilsson, D.; Gennemark, P.; O’Mahony, G.; et al. PPARγ and PPARα synergize to induce robust browning of white fat in vivo. Mol. Metab. 2020, 36, 100964. [Google Scholar] [CrossRef]
- Villarroya, F.; Iglesias, R.; Giralt, M. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2007, 2007, 74364. [Google Scholar] [CrossRef] [PubMed]
- Magliano, D.C.; Bargut, T.C.L.; de Carvalho, S.N.; Aguila, M.B.; Mandarim-de-Lacerda, C.A.; Souza-Mello, V. Peroxisome proliferator-activated receptors-alpha and gamma are targets to treat offspring from maternal diet-induced obesity in mice. PLoS ONE 2013, 8, e64258. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Pettinelli, P.; Del Pozo, T.; Araya, J.; Rodrigo, R.; Araya, A.V.; Smok, G.; Csendes, A.; Gutierrez, L.; Rojas, J.; Korn, O.; et al. Enhancement in liver SREBP-1c/PPAR-alpha ratio and steatosis in obese patients: Correlations with insulin resistance and n-3 long-chain polyunsaturated fatty acid depletion. Biochim. Biophys. Acta 2009, 1792, 1080–1086. [Google Scholar] [CrossRef]
- Videla, L.A.; Pettinelli, P. Misregulation of PPAR functioning and its pathogenic consequences associated with nonalcoholic fatty liver disease in human obesity. PPAR Res. 2012, 2012, 107434. [Google Scholar] [CrossRef]
- Montagner, A.; Polizzi, A.; Fouché, E.; Ducheix, S.; Lippi, Y.; Lasserre, F.; Barquissau, V.; Régnier, M.; Lukowicz, C.; Benhamed, F.; et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 2016, 65, 1202–1214. [Google Scholar] [CrossRef]
- Abd El-Haleim, E.A.; Bahgat, A.K.; Saleh, S. Resveratrol and fenofibrate ameliorate fructose-induced nonalcoholic steatohepatitis by modulation of genes expression. World J. Gastroenterol. 2016, 22, 2931–2948. [Google Scholar] [CrossRef]
- Zhang, N.; Lu, Y.; Shen, X.; Bao, Y.; Cheng, J.; Chen, L.; Li, B.; Zhang, Q. Fenofibrate treatment attenuated chronic endoplasmic reticulum stress in the liver of nonalcoholic fatty liver disease mice. Pharmacology 2015, 95, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Al Zarzour, R.H.; Alshawsh, M.A.; Asif, M.; Al-Mansoub, M.A.; Mohamed, Z.; Ahmad, M.; Majid, A.M.S.A.; Asmawi, M.Z.; Kaur, G.; Al-Dualimi, D.W.; et al. Adipocytokine regulation and antiangiogenic activity underlie the molecular mechanisms of therapeutic effects of Phyllanthus niruri against non-alcoholic fatty liver disease. Nutrients 2018, 10, 1057. [Google Scholar] [CrossRef]
- Cicero, A.F.G.; Colletti, A.; Bellentani, S. Nutraceutical approach to non-alcoholic fatty liver disease (NAFLD): The available clinical evidence. Nutrients 2018, 10, 1153. [Google Scholar] [CrossRef] [PubMed]
- Elvira-Torales, L.I.; Navarro-González, I.; González-Barrio, R.; Martín-Pozuelo, G.; Doménech, G.; Seva, J.; García-Alonso, J.; Periago-Castón, M.J. Tomato juice supplementation influences the gene expression related to steatosis in rats. Nutrients 2018, 10, 1215. [Google Scholar] [CrossRef]
- Guruvaiah, P.; Guo, H.; Li, D.; Xie, Z. Preventive effect of flavonol derivatives abundant sanglan tea on long-term high-fat-diet-induced obesity complications in C57BL/6 mice. Nutrients 2018, 10, 1276. [Google Scholar] [CrossRef]
- Lim, J.; Liu, Z.; Apontes, P.; Feng, D.; Pessin, J.E.; Sauve, A.A.; Angeletti, R.H.; Chi, Y. Dual mode action of mangiferin in mouse liver under high fat diet. PLoS ONE 2014, 9, e90137. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Li, D.; Guruvaiah, P.; Xu, N.; Xie, Z. Dietary supplement of large yellow tea ameliorates metabolic syndrome and attenuates hepatic steatosis in db/db Mice. Nutrients 2018, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, Y.Y.; Wang, L.; Teng, T.; Zhou, M.; Wang, S.G.; Tian, Y.Z.; Du, L.; Yin, X.X.; Sun, Y. Mangiferin ameliorates fatty liver via modulation of autophagy and inflammation in high-fat-diet induced mice. Biomed. Pharmacother. 2017, 96, 328–335. [Google Scholar] [CrossRef]
- Wang, X.; Hasegawa, J.; Kitamura, Y.; Wang, Z.; Matsuda, A.; Shinoda, W.; Miura, N.; Kimura, K. Effects of hesperidin on the progression of hypercholesterolemia and fatty liver induced by high-cholesterol diet in rats. J. Pharmacol. Sci. 2011, 117, 129–138. [Google Scholar] [CrossRef]
- Levene, A.P.; Kudo, H.; Armstrong, M.J.; Thursz, M.R.; Gedroyc, W.M.; Anstee, Q.M.; Goldin, R.D. Quantifying hepatic steatosis—More than meets the eye. Histopathology 2012, 60, 971–981. [Google Scholar] [CrossRef]
- Lawal, A.O.; Davids, L.M.; Marnewick, J.L. Rooibos (Aspalathus linearis) and Honeybush (Cyclopia species) modulate the oxidative stress associated injury of diesel exhaust particles in human umbilical vein endothelial cells. Phytomedicine 2019, 59, 152898. [Google Scholar] [CrossRef] [PubMed]
- Marnewick, J.L.; Joubert, E.; Swart, P.; Van Der Westhuizen, F.; Gelderblom, W.C. Modulation of hepatic drug metabolizing enzymes and oxidative status by rooibos (Aspalathus linearis) and honeybush (Cyclopia intermedia), green and black (Camellia sinensis) teas in rats. J. Agric. Food Chem. 2003, 51, 8113–8119. [Google Scholar] [CrossRef]
- Homayouni, F.; Haidari, F.; Hedayati, M.; Zakerkish, M.; Ahmadi, K. Hesperidin supplementation alleviates oxidative DNA damage and lipid peroxidation in type 2 diabetes: A randomized double-blind placebo-controlled clinical trial. Phytother. Res. 2017, 31, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- Pari, L.; Karthikeyan, A.; Karthika, P.; Rathinam, A. Protective effects of hesperidin on oxidative stress, dyslipidaemia and histological changes in iron-induced hepatic and renal toxicity in rats. Toxicol. Rep. 2015, 2, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, E.; Sánchez-Gómez, M.V.; Ruiz, A.; Cavaliere, F.; Ortiz-Sanz, C.; Quintela-López, T.; Capetillo-Zarate, E.; Solé-Domènech, S.; Matute, C. Mangiferin and morin attenuate oxidative stress, mitochondrial dysfunction, and neurocytotoxicity, induced by amyloid beta oligomers. Oxid. Med. Cell. Longev. 2018, 2018, 2856063. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.B.; Sinha, K.; Sil, P.C. Mangiferin, a natural xanthone, protects murine liver in Pb(II) induced hepatic damage and cell death via MAP kinase, NF-ΚB and mitochondria dependent pathways. PLoS ONE 2013, 8, e56894. [Google Scholar] [CrossRef]
- Ferreira, P.S.; Spolidorio, L.C.; Manthey, J.A.; Cesar, T.B. Citrus flavanones prevent systemic inflammation and ameliorate oxidative stress in C57BL/6J mice fed high-fat diet. Food Funct. 2016, 7, 2675–2681. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Ashour, M.B.; Abdel-Moneim, A.; Ahmed, O.M. Hesperidin and naringin attenuate hyperglycemia-mediated oxidative stress and proinflammatory cytokine production in high fat fed/streptozotocin-induced type 2 diabetic rats. J. Diabetes Complicat. 2012, 26, 483–490. [Google Scholar] [CrossRef]
- Jack, B.U.; Malherbe, C.J.; Willenburg, E.L.; de Beer, D.; Huisamen, B.; Joubert, E.; Muller, C.J.F.; Louw, J.; Pheiffer, C. Polyphenol-enriched fractions of Cyclopia intermedia selectively affect lipogenesis and lipolysis in 3T3-L1 adipocytes. Planta Med. 2018, 84, 100–110. [Google Scholar] [CrossRef]
- Cinti, S.; Cancello, R.; Zingaretti, M.C.; Ceresi, E.; De Matteis, R.; Giordano, A.; Himms-Hagen, J.; Ricquier, D. CL316,243 and cold stress induce heterogeneous expression of UCP1 mRNA and protein in rodent brown adipocytes. J. Histochem. Cytochem. 2002, 50, 21–31. [Google Scholar] [CrossRef]
- Staels, B.; Dallongeville, J.; Auwerx, J.; Schoonjans, K.; Leitersdorf, E.; Fruchart, J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 1998, 98, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Beery, A.K. Inclusion of females does not increase variability in rodent research studies. Curr. Opin. Behav. Sci. 2018, 23, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F.; Arnold, A.P.; Reue, K. A Guide for the design of pre-clinical studies on sex differences in metabolism. Cell Metab. 2017, 25, 1216–1230. [Google Scholar] [CrossRef]
- Burl, R.B.; Ramseyer, V.D.; Rondini, E.A.; Pique-Regi, R.; Lee, Y.H.; Granneman, J.G. Deconstructing adipogenesis induced by Β3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 2018, 28, 300–309. [Google Scholar] [CrossRef]
- Rachid, T.L.; Penna-de-Carvalho, A.; Bringhenti, I.; Aguila, M.B.; Mandarim-de-Lacerda, C.A.; Souza-Mello, V. Fenofibrate (PPARalpha agonist) induces beige cell formation in subcutaneous white adipose tissue from diet-induced male obese mice. Mol. Cell. Endocrinol. 2015, 402, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef]
- Lim, S.; Honek, J.; Xue, Y.; Seki, T.; Cao, Z.; Andersson, P.; Yang, X.; Hosaka, K.; Cao, Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat. Protoc. 2012, 7, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]

| Compound Name | Hydrophobic Interactions | Hydrogen Bond (Length-Å) |
|---|---|---|
| UCP1 | ||
| CL-316,243 | Lys38, Val39, Arg40, Gln44, Gly45, Val139, Arg183, Pro179, Lys237 | Glu46 (2.85), Arg140 (3.07/3.14) |
| Hesperidin | Gly47, Ile241, Arg140, Val39, Gly176, Asn180, Pro179, Phe240 | Arg183 (2.80), Gly45 (2.78), Ala143 (2.81), Gln144 (3.22), Glu168 (2.81), Gln48 (2.88/3.10), Glu46 (2.83/3.01/2.87) |
| 3-β-d-glucopyranosyl-4-O-β-d-glucopyranosyliriflophenone | Ala143, Lys38, Val39, Glu46, Arg40, Gly45, Arg140, Thr172, Glu168, Gly47, Phe240, Ile241 | Thr36 (3.10), Gln48 (3.18), Gln144 (3.13/2.27) |
| 3-β-d-glucopyranosyliriflophenone | Lys38, Val39, Glu46, Gly45, Arg40, Gly45, Ala143, Phe240 | Gln48 (3.12/2.93), Thr36 (2.93), Arg140 (3.02), Asp35 (2.99) |
| Neoponcirin | Lys350, Val139, Pro179, Val39, Phe240, Gly47, Ile241, Leu244, Thr172, Gln247, Lys175, Val39 | Arg183 (3.24), Arg140 (2.89/3.21), Arg40 (2.92), Ala143 (2.96), Glu46 (3.04) |
| PPARα | ||
| Fenofibrate | Thr279, Met320, Leu321, Cys276, Ile354, Phe273, His440, Phe318, Ile317 | Met355 (3.35), Ser280 (3.29), Thr283 (2.79) |
| Neoponcirin | Phe218, Met320, Thr283, Leu321, Ile317, Phe318, Ser280, His440, Tyr314, Ile354, Met355, Leu347, Phe351, Ile272, Phe273, Cyc276, Glu269, Thr279, Met330 | Asn219 (2.80), Glu286 (3.05), Met220 (3.19/3.27) |
| Hesperidin | Phe218, Met320, Ile317, Leu321, Thr283, Ser280, Phe318, Tyr314, Cys276, Phe273, Glu269, Ile354, Leu347, Ile272, Leu344, Phe351, Met355, Met330, Thr279 | Met220 (3.20), Asn219 (2.83), Glu286 (3.05), His440 (3.03) |
| Target Protein Description | Centre (X, Y, Z) | Dimensions of Grid Box (X, Y, Z) |
|---|---|---|
| UCP1 | 27.286, 34.659, 28.111 | 68, 72, 62 |
| PPARα | 12.619, −11.582, −29.582 | 24, 20, 36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jack, B.U.; Ramharack, P.; Malherbe, C.; Gabuza, K.; Joubert, E.; Pheiffer, C. Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female db/db Mice. Int. J. Mol. Sci. 2023, 24, 3868. https://doi.org/10.3390/ijms24043868
Jack BU, Ramharack P, Malherbe C, Gabuza K, Joubert E, Pheiffer C. Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female db/db Mice. International Journal of Molecular Sciences. 2023; 24(4):3868. https://doi.org/10.3390/ijms24043868
Chicago/Turabian StyleJack, Babalwa Unice, Pritika Ramharack, Christiaan Malherbe, Kwazi Gabuza, Elizabeth Joubert, and Carmen Pheiffer. 2023. "Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female db/db Mice" International Journal of Molecular Sciences 24, no. 4: 3868. https://doi.org/10.3390/ijms24043868
APA StyleJack, B. U., Ramharack, P., Malherbe, C., Gabuza, K., Joubert, E., & Pheiffer, C. (2023). Cyclopia intermedia (Honeybush) Induces Uncoupling Protein 1 and Peroxisome Proliferator-Activated Receptor Alpha Expression in Obese Diabetic Female db/db Mice. International Journal of Molecular Sciences, 24(4), 3868. https://doi.org/10.3390/ijms24043868





