TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells
Abstract
1. Introduction
2. Results
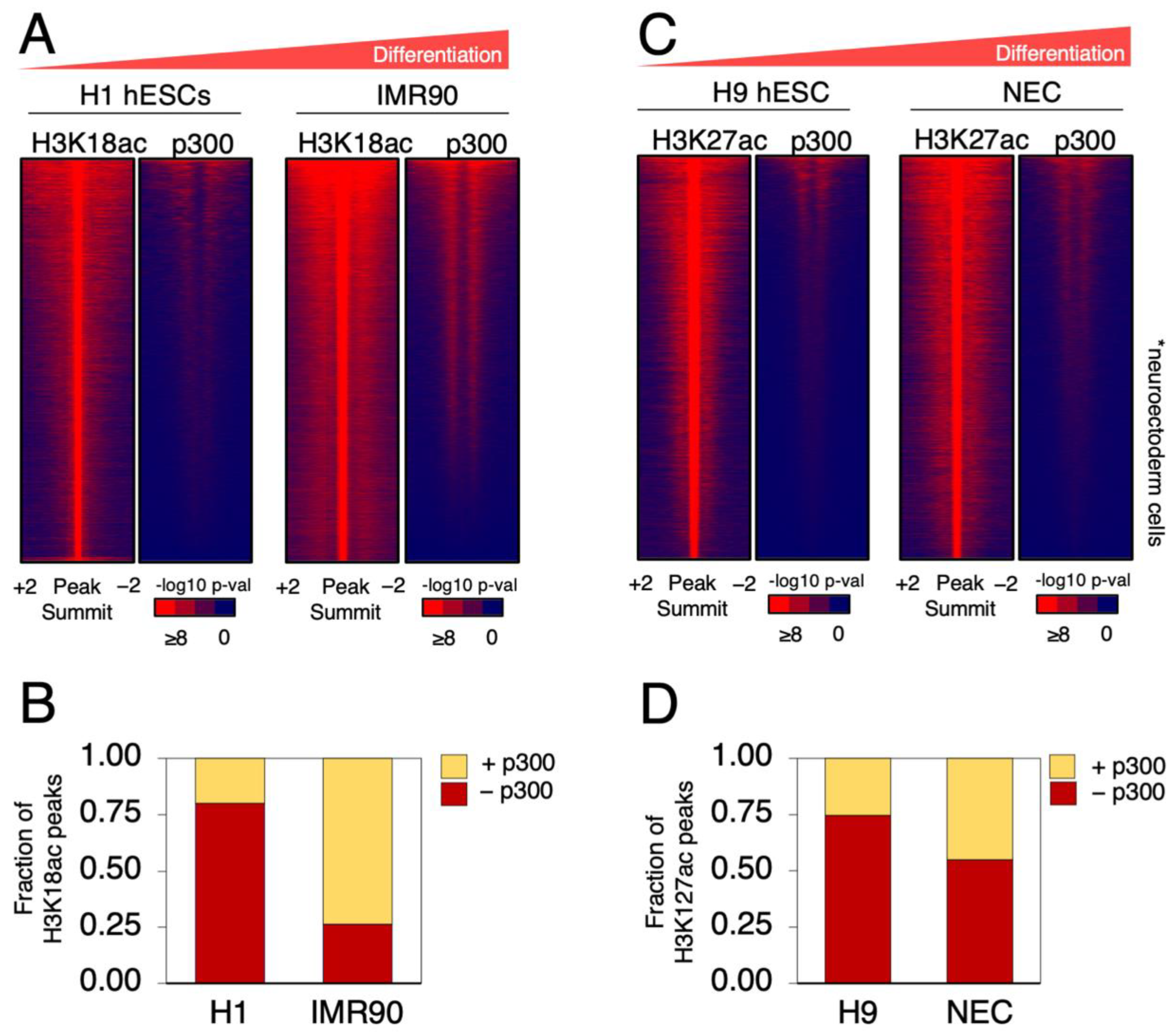
2.1. p300 Occupancy Overlaps with H3K18 and H3K27 Acetylation in Differentiated Cells
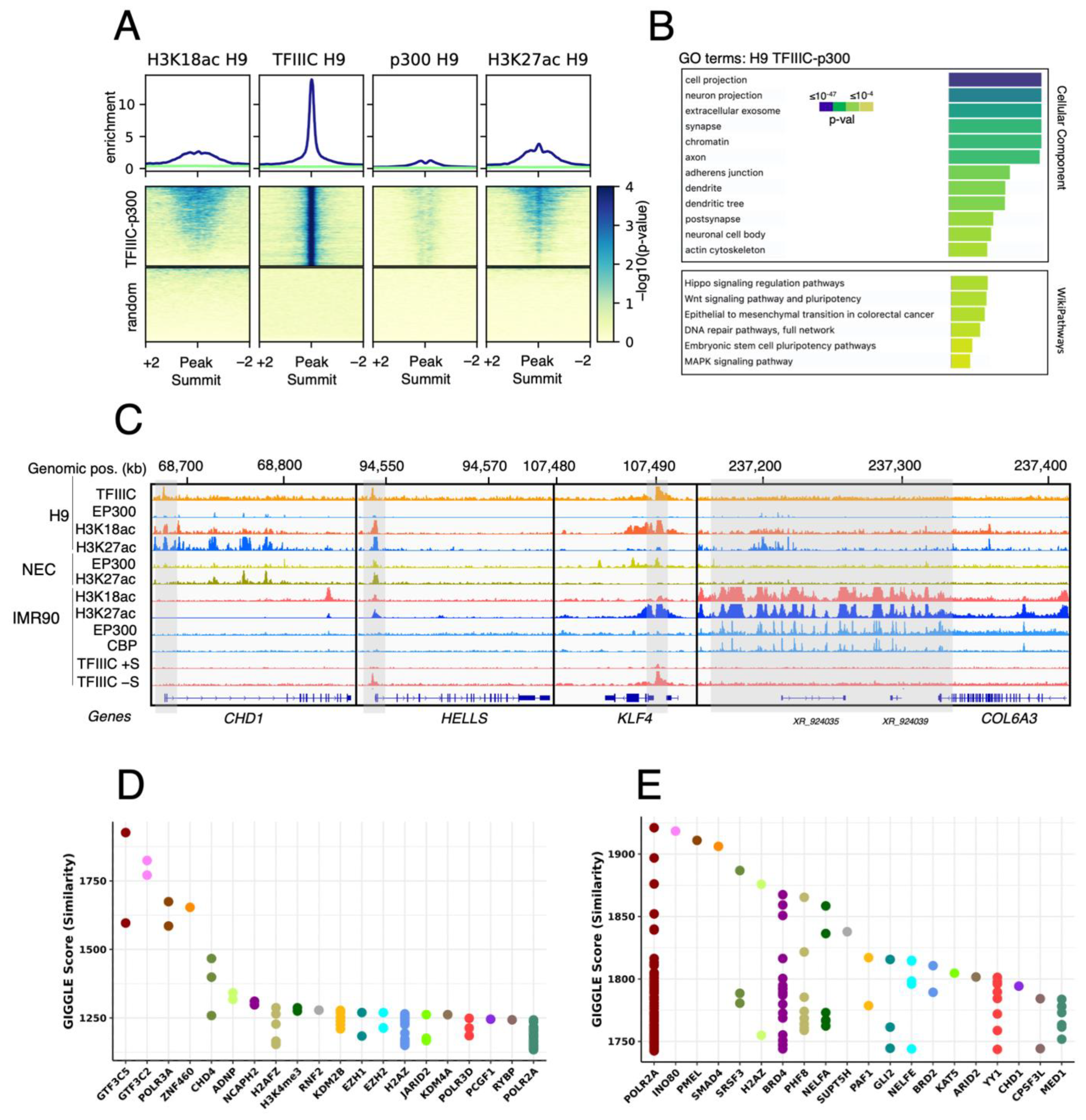
2.2. GTF3C1 as a Putative Novel H3K18ac HAT in hESCs
3. Discussion
4. Materials and Methods
4.1. External Data Sources
4.2. ChIP-Seq Data Analysis
4.3. Bedtools
4.4. GIGGLE Analysis
4.5. Genome Browser Images
4.6. Gene Ontology (GO) Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Puri, M.C.; Nagy, A. Concise review: Embryonic stem cells versus induced pluripotent stem cells: The game is on. Stem. Cells 2012, 30, 10–14. [Google Scholar] [CrossRef]
- Ferrari, R.; Gou, D.; Jawdekar, G.; Johnson, S.A.; Nava, M.; Su, T.; Yousef, A.F.; Zemke, N.R.; Pellegrini, M.; Kurdistani, S.K.; et al. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 2014, 16, 663–676. [Google Scholar] [CrossRef]
- Ferrari, R.; Pellegrini, M.; Horwitz, G.A.; Xie, W.; Berk, A.J.; Kurdistani, S.K. Epigenetic reprogramming by adenovirus e1a. Science 2008, 321, 1086–1088. [Google Scholar] [CrossRef]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef]
- Horwitz, G.A.; Zhang, K.; McBrian, M.A.; Grunstein, M.; Kurdistani, S.K.; Berk, A.J. Adenovirus small e1a alters global patterns of histone modification. Science 2008, 321, 1084–1085. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, A.; Naguibneva, I.; Fritsch, L.; Duquet, A.; Ait-Si-Ali, S.; Robin, P.; Vervisch, A.; Pritchard, L.L.; Cole, P.; Harel-Bellan, A. CBP/p300 and muscle differentiation: No HAT, no muscle. EMBO J. 2001, 20, 6816–6825. [Google Scholar] [CrossRef] [PubMed]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet 2007, 23, 614–622. [Google Scholar] [CrossRef]
- Basu, M.; Boopathi, R.; Das, S.; Kundu, T.K. The Largest Subunit of Human TFIIIC Complex, TFIIIC220, a Lysine Acetyltransferase Targets Histone H3K18. bioRxiv 2019, 513127. [Google Scholar] [CrossRef]
- Ferrari, R.; de Llobet Cucalon, L.I.; Di Vona, C.; Le Dilly, F.; Vidal, E.; Lioutas, A.; Oliete, J.Q.; Jochem, L.; Cutts, E.; Dieci, G.; et al. TFIIIC Binding to Alu Elements Controls Gene Expression via Chromatin Looping and Histone Acetylation. Mol. Cell 2020, 77, 475–487.e411. [Google Scholar] [CrossRef]
- Ludwig, T.; J, A.T. Defined, feeder-independent medium for human embryonic stem cell culture. Curr. Protoc. Stem. Cell Biol. 2007, 2, Unit 1C 2. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Stamatoyannopoulos, J.A.; Costello, J.F.; Ren, B.; Milosavljevic, A.; Meissner, A.; Kellis, M.; Marra, M.A.; Beaudet, A.L.; Ecker, J.R.; et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010, 28, 1045–1048. [Google Scholar] [CrossRef]
- Hawkins, R.D.; Hon, G.C.; Lee, L.K.; Ngo, Q.; Lister, R.; Pelizzola, M.; Edsall, L.E.; Kuan, S.; Luu, Y.; Klugman, S.; et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem. Cell 2010, 6, 479–491. [Google Scholar] [CrossRef]
- Li, B.; Su, T.; Ferrari, R.; Li, J.Y.; Kurdistani, S.K. A unique epigenetic signature is associated with active DNA replication loci in human embryonic stem cells. Epigenetics 2014, 9, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–283. [Google Scholar] [CrossRef] [PubMed]
- de Llobet Cucalon, L.; Di Vona, C.; Morselli, M.; Vezzoli, M.; Montanini, B.; Teichmann, M.; de la Luna, S.; Ferrari, R. An RNA Polymerase III General Transcription Factor Engages in Cell Type-Specific Chromatin Looping. Int. J. Mol. Sci. 2022, 23, 2260. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Layer, R.M.; Pedersen, B.S.; DiSera, T.; Marth, G.T.; Gertz, J.; Quinlan, A.R. GIGGLE: A search engine for large-scale integrated genome analysis. Nat. Methods 2018, 15, 123–126. [Google Scholar] [CrossRef]
- Ostapcuk, V.; Mohn, F.; Carl, S.H.; Basters, A.; Hess, D.; Iesmantavicius, V.; Lampersberger, L.; Flemr, M.; Pandey, A.; Thoma, N.H.; et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature 2018, 557, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Goodman, J.V.; Yamada, T.; Yang, Y.; Kong, L.; Wu, D.Y.; Zhao, G.; Gabel, H.W.; Bonni, A. The chromatin remodeling enzyme Chd4 regulates genome architecture in the mouse brain. Nat. Commun. 2020, 11, 3419. [Google Scholar] [CrossRef]
- Corley, M.; Kroll, K.L. The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 2015, 359, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, S.; Dai, X.; Ma, J.; Wan, J.; Jiang, H.; Wang, P.; Liu, Z.; Zhang, H. PRC2 regulates RNA polymerase III transcribed non-translated RNA gene transcription through EZH2 and SUZ12 interaction with TFIIIC complex. Nucleic Acids Res. 2015, 43, 6270–6284. [Google Scholar] [CrossRef]
- Shiota, H.; Barral, S.; Buchou, T.; Tan, M.; Coute, Y.; Charbonnier, G.; Reynoird, N.; Boussouar, F.; Gerard, M.; Zhu, M.; et al. Nut Directs p300-Dependent, Genome-Wide H4 Hyperacetylation in Male Germ Cells. Cell Rep. 2018, 24, 3477–3487.e3476. [Google Scholar] [CrossRef]
- Ihry, R.J.; Salick, M.R.; Ho, D.J.; Sondey, M.; Kommineni, S.; Paula, S.; Raymond, J.; Henry, B.; Frias, E.; Wang, Q.; et al. Genome-Scale CRISPR Screens Identify Human Pluripotency-Specific Genes. Cell Rep. 2019, 27, 616–630.e616. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Su, T.; Li, B.; Bonora, G.; Oberai, A.; Chan, Y.; Sasidharan, R.; Berk, A.J.; Pellegrini, M.; Kurdistani, S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012, 22, 1212–1221. [Google Scholar] [CrossRef]
- Zaurin, R.; Ferrari, R.; Nacht, A.S.; Carbonell, J.; Le Dily, F.; Font-Mateu, J.; de Llobet Cucalon, L.I.; Vidal, E.; Lioutas, A.; Beato, M.; et al. A set of accessible enhancers enables the initial response of breast cancer cells to physiological progestin concentrations. Nucleic Acids Res. 2021, 49, 12716–12731. [Google Scholar] [CrossRef]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vezzoli, M.; de Llobet Cucalon, L.I.; Di Vona, C.; Morselli, M.; Montanini, B.; de la Luna, S.; Teichmann, M.; Dieci, G.; Ferrari, R. TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells. Int. J. Mol. Sci. 2023, 24, 3624. https://doi.org/10.3390/ijms24043624
Vezzoli M, de Llobet Cucalon LI, Di Vona C, Morselli M, Montanini B, de la Luna S, Teichmann M, Dieci G, Ferrari R. TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells. International Journal of Molecular Sciences. 2023; 24(4):3624. https://doi.org/10.3390/ijms24043624
Chicago/Turabian StyleVezzoli, Marco, Lara Isabel de Llobet Cucalon, Chiara Di Vona, Marco Morselli, Barbara Montanini, Susana de la Luna, Martin Teichmann, Giorgio Dieci, and Roberto Ferrari. 2023. "TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells" International Journal of Molecular Sciences 24, no. 4: 3624. https://doi.org/10.3390/ijms24043624
APA StyleVezzoli, M., de Llobet Cucalon, L. I., Di Vona, C., Morselli, M., Montanini, B., de la Luna, S., Teichmann, M., Dieci, G., & Ferrari, R. (2023). TFIIIC as a Potential Epigenetic Modulator of Histone Acetylation in Human Stem Cells. International Journal of Molecular Sciences, 24(4), 3624. https://doi.org/10.3390/ijms24043624







