Telomere Length as a New Risk Marker of Early-Onset Colorectal Cancer
Abstract
:1. Introduction
2. Results
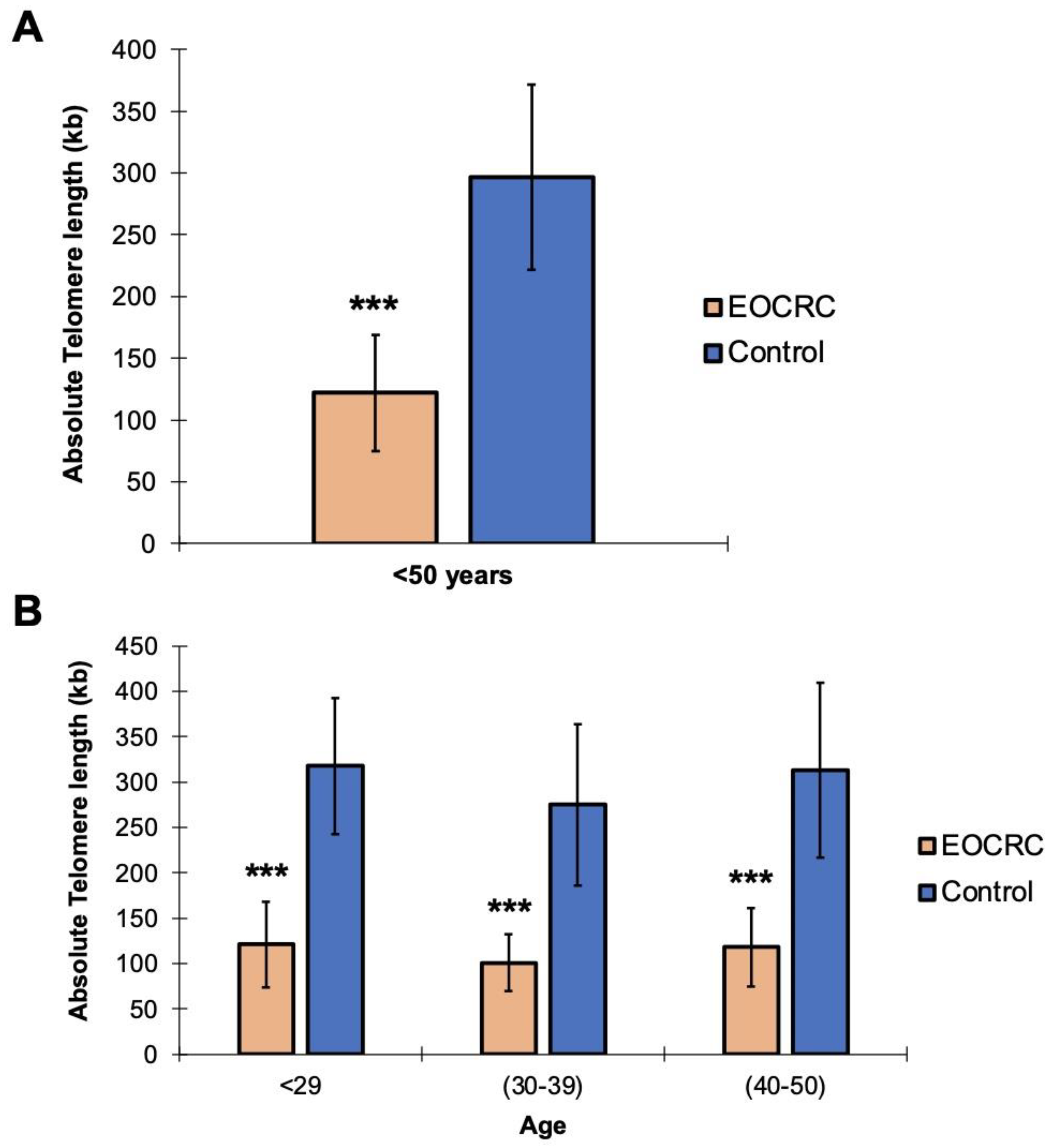
2.1. Telomeric Shortening Causes Predisposition to the Development of EOCRC
2.2. hTERT, POT1, TERF2, and TERF2IP Gene Polymorphisms Are Associated with Telomeric Shortening in EOCRC
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. DNA Isolation and Telomere Length Analysis by Real Time Quantitative PCR (RT-qPCR)
4.3. Library Preparation and Sequencing
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sinicrope, F.A. Increasing Incidence of Early-Onset Colorectal Cancer. N. Engl. J. Med. 2022, 386, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.; Linehan, A.; Krstic, A.; Kolch, W.; Sheahan, K.; Winter, D.C.; Mc Dermott, R. Oncotherapeutic Strategies in Early Onset Colorectal Cancer. Cancers 2023, 15, 552. [Google Scholar] [CrossRef]
- Van Leerdam, M.E.; Roos, V.H.; van Hooft, J.E.; Balaguer, F.; Dekker, E.; Kaminski, M.F.; Latchford, A.; Neumann, H.; Ricciardiello, L.; Rupinska, M.; et al. Endoscopic Management of Lynch Syndrome and of Familial Risk of Colorectal Cancer: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2019, 51, 1082–1093. [Google Scholar] [CrossRef] [PubMed]
- Vertecchi, E.; Rizzo, A.; Salvati, E. Telomere Targeting Approaches in Cancer: Beyond Length Maintenance. Int. J. Mol. Sci. 2022, 23, 3784. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Paolisso, G. Telomerase Activation: A Potential Key Modulator for Human Healthspan and Longevity. Ageing Res. Rev. 2014, 15, 1–5. [Google Scholar] [CrossRef]
- Cui, Y.; Cai, Q.; Qu, S.; Chow, W.H.; Wen, W.; Xiang, Y.B.; Wu, J.; Rothman, N.; Yang, G.; Shu, X.O.; et al. Association of Leukocyte Telomere Length with Colorectal Cancer Risk: Nested Case-Control Findings from the Shanghai Women’s Health Study. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1807–1813. [Google Scholar] [CrossRef] [PubMed]
- Obadina, D.; Haider, H.; Micic, D.; Sakuraba, A. Older Age at First Screening Colonoscopy Is Associated with an Increased Risk of Colorectal Adenomas and Cancer. J. Clin. Gastroenterol. 2022, 10, 1097. [Google Scholar] [CrossRef]
- Joo, J.E.; Clendenning, M.; Wong, E.M.; Rosty, C.; Mahmood, K.; Georgeson, P.; Winship, I.M.; Preston, S.G.; Win, A.K.; Dugué, P.A.; et al. Dna Methylation Signatures and the Contribution of Age-Associated Methylomic Drift to Carcinogenesis in Early-Onset Colorectal Cancer. Cancers 2021, 13, 2589. [Google Scholar] [CrossRef]
- Tomasova, K.; Kroupa, M.; Zinkova, A.; Korabecna, M.; Vymetalkova, V.; Skrobanek, P.; Sojka, L.; Levy, M.; Hemminki, K.; Liska, V.; et al. Monitoring of Telomere Dynamics in Peripheral Blood Leukocytes in Relation to Colorectal Cancer Patients’ Outcomes. Front. Oncol. 2022, 12, 962929. [Google Scholar] [CrossRef]
- García-Martínez, S.; González-Gamo, D.; Tesolato, S.E.; Barabash, A.; de la Serna, S.C.; Domínguez-Serrano, I.; Dziakova, J.; Rivera, D.; Torres, A.J.; Iniesta, P. Telomere Length and Telomerase Activity in Subcutaneous and Visceral Adipose Tissues from Obese and Non-Obese Patients with and without Colorectal Cancer. Cancers 2023, 15, 273. [Google Scholar] [CrossRef]
- Hatse, S.; Serena, M.; Vulsteke, C.; Punie, K.; Neven, P.; Smeets, A.; Laenen, A.; Wildiers, H. Impact of Baseline Telomere Length on Survival and Chemotherapy Related Toxicity in Breast Cancer Patients Receiving (Neo)Adjuvant Anthracycline Containing Chemotherapy. Transl. Oncol. 2022, 26, 101551. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Saugar, E.E.; Alamian, A.; Ferreira, T.; Downs, C.A. Changes in Telomere Length and Indicators of Oxidative Stress in Critically Ill Mechanically Ventilated Adults–A Pilot Study. Biol. Res. Nurs. 2022. [Google Scholar] [CrossRef]
- Kimura, M.; Stone, R.C.; Hunt, S.C.; Skurnick, J.; Lu, X.; Cao, X.; Harley, C.B.; Aviv, A. Measurement of Telomere Length by the Southern Blot Analysis of Terminal Restriction Fragment Lengths. Nat. Protoc. 2010, 5, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Razdan, N.; Kaplunov, J.; Mundra, J.J.; Kimura, M.; Aviv, A.; Herbig, U. Stn1 Is Critical for Telomere Maintenance and Long-Term Viability of Somatic Human Cells. Aging Cell 2015, 14, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Boardman, L.A.; Litzelman, K.; Seo, S.; Johnson, R.A.; Vanderboom, R.J.; Kimmel, G.W.; Cunningham, J.M.; Gangnon, R.E.; Engelman, C.D.; Riegert-Johnson, D.L.; et al. The Association of Telomere Length with Colorectal Cancer Differs by the Age of Cancer Onset. Clin. Transl. Gastroenterol. 2014, 5, e52. [Google Scholar] [CrossRef] [PubMed]
- Kroupa, M.; Rachakonda, S.K.; Liska, V.; Srinivas, N.; Urbanova, M.; Jiraskova, K.; Schneiderova, M.; Vycital, O.; Vymetalkova, V.; Vodickova, L.; et al. Relationship of Telomere Length in Colorectal Cancer Patients with Cancer Phenotype and Patient Prognosis. Br. J. Cancer 2019, 121, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Xiu, J.; Lopez, G.Y.; Bentley, R.; Jalali, A.; Heimberger, A.B.; Bainbridge, M.N.; Bondy, M.L.; Walsh, K.M. POT1 Mutation Spectrum in Tumour Types Commonly Diagnosed among POT1-Associated Hereditary Cancer Syndrome Families. J. Med. Genet. 2020, 57, 664–670. [Google Scholar] [CrossRef]
- Shen, J.; Gammon, M.D.; Wu, H.C.; Terry, M.B.; Wang, Q.; Bradshaw, P.T.; Teitelbaum, S.L.; Neugut, A.I.; Santella, R.M. Multiple Genetic Variants in Telomere Pathway Genes and Breast Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2010, 19, 219. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Perea, J.; Marti, M.; Espin, E.; Hernandez-Villafranca, S.; Orihuela, P.; Vidal Tocino, R.; Alcazar, J.A.; Vivas, A.; Narvaez, C.; Prieto, I.; et al. Cohort Profile: The Spanish Early-Onset Colorectal Cancer (SECOC) Cohort: A Multicentre Cohort Study on the Molecular Basis of Colorectal Cancer among Young Individuals in Spain. BMJ Open 2021, 11, e055409. [Google Scholar] [CrossRef]
- Babraham Bioinformatics–FastQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 October 2022).
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.; O’Connor, B.D. Genomics in the Cloud; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020; 493p. [Google Scholar]
- Geoffroy, V.; Pizot, C.; Redin, C.; Piton, A.; Vasli, N.; Stoetzel, C.; Blavier, A.; Laporte, J.; Muller, J. VaRank: A Simple and Powerful Tool for Ranking Genetic Variants. PeerJ 2015, 3, e796. [Google Scholar] [CrossRef] [PubMed]
- Exome Variant Server. Available online: https://evs.gs.washington.edu/EVS/ (accessed on 23 October 2022).
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
| EOCRC (n = 70) | NHLBI GO-ESP (EA-6500 Samples) | GNOMAD AF_NFE (56885 Samples) | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | REF | ALT | Freq. | Freq. | FDR | Freq. | FDR |
| hTERT | rs79662648 | C | G | 0.086 | 0.036 | 0.0568 | 0.036 | 0.0486 |
| POT1 | rs76436625 | T | C | 0.257 | N/D | NA | 0.108 | 0.0008 |
| POT1 | rs10263573 | A | T | 0.571 | 0.405 | 0.0112 | 0.396 | 0.0050 |
| POT1 | rs3815221 | G | A | 0.571 | 0.405 | 0.0112 | 0.396 | 0.0050 |
| POT1 | rs7794637 | T | C | 0.900 | 0.697 | 0.0006 | 0.690 | 0.0002 |
| POT1 | rs7784168 | T | C | 0.529 | 0.305 | 0.0006 | 0.316 | 0.0007 |
| POT1 | rs4383910 | A | C | 0.829 | N/D | NA | 0.548 | 0.0000 |
| POT1 | rs7782354 | C | T | 0.829 | N/D | NA | 0.573 | 0.0001 |
| TERF2 | rs251796 | A | G | 0.514 | 0.298 | 0.0006 | 0.304 | 0.0007 |
| TERF2 | rs34415214 | G | A | 0.157 | 0.062 | 0.0106 | 0.061 | 0.0050 |
| TERF2IP | rs7205764 | T | C | 0.643 | N/D | NA | 0.506 | 0.0323 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martel-Martel, A.; Corchete, L.A.; Martí, M.; Vidal-Tocino, R.; Hurtado, E.; Álvaro, E.; Jiménez, F.; Jiménez-Toscano, M.; Balaguer, F.; Sanz, G.; et al. Telomere Length as a New Risk Marker of Early-Onset Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 3526. https://doi.org/10.3390/ijms24043526
Martel-Martel A, Corchete LA, Martí M, Vidal-Tocino R, Hurtado E, Álvaro E, Jiménez F, Jiménez-Toscano M, Balaguer F, Sanz G, et al. Telomere Length as a New Risk Marker of Early-Onset Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(4):3526. https://doi.org/10.3390/ijms24043526
Chicago/Turabian StyleMartel-Martel, Abel, Luis A. Corchete, Marc Martí, Rosario Vidal-Tocino, Elena Hurtado, Edurne Álvaro, Fernando Jiménez, Marta Jiménez-Toscano, Francesc Balaguer, Gonzalo Sanz, and et al. 2023. "Telomere Length as a New Risk Marker of Early-Onset Colorectal Cancer" International Journal of Molecular Sciences 24, no. 4: 3526. https://doi.org/10.3390/ijms24043526
APA StyleMartel-Martel, A., Corchete, L. A., Martí, M., Vidal-Tocino, R., Hurtado, E., Álvaro, E., Jiménez, F., Jiménez-Toscano, M., Balaguer, F., Sanz, G., López, I., Hernández-Villafranca, S., Ballestero, A., Vivas, A., Melone, S., Pastor, C., Brandáriz, L., Gómez-Marcos, M. A., Cruz-Hernández, J. J., ... González-Sarmiento, R. (2023). Telomere Length as a New Risk Marker of Early-Onset Colorectal Cancer. International Journal of Molecular Sciences, 24(4), 3526. https://doi.org/10.3390/ijms24043526










