MITF Downregulation Induces Death in Human Mast Cell Leukemia Cells and Impairs IgE-Dependent Degranulation
Abstract
1. Introduction
2. Results
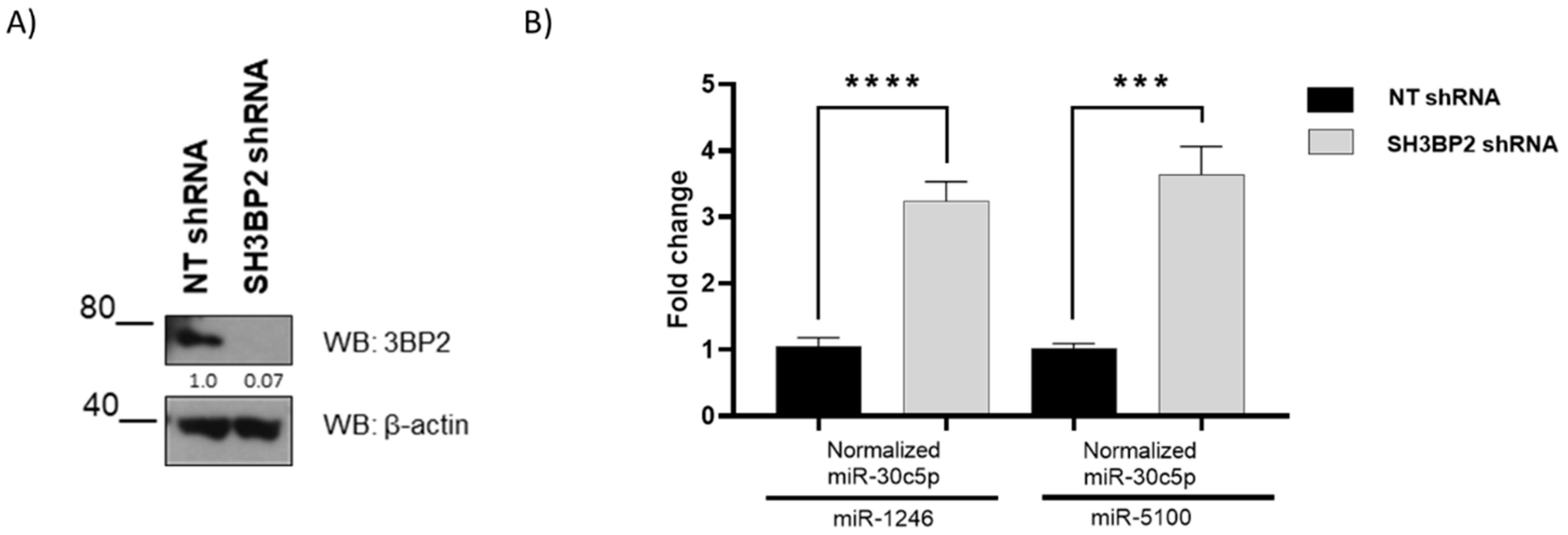
2.1. Analysis of Up-Regulated Mirnas That Target MITF Validation in Hmc-1 Cell Lines
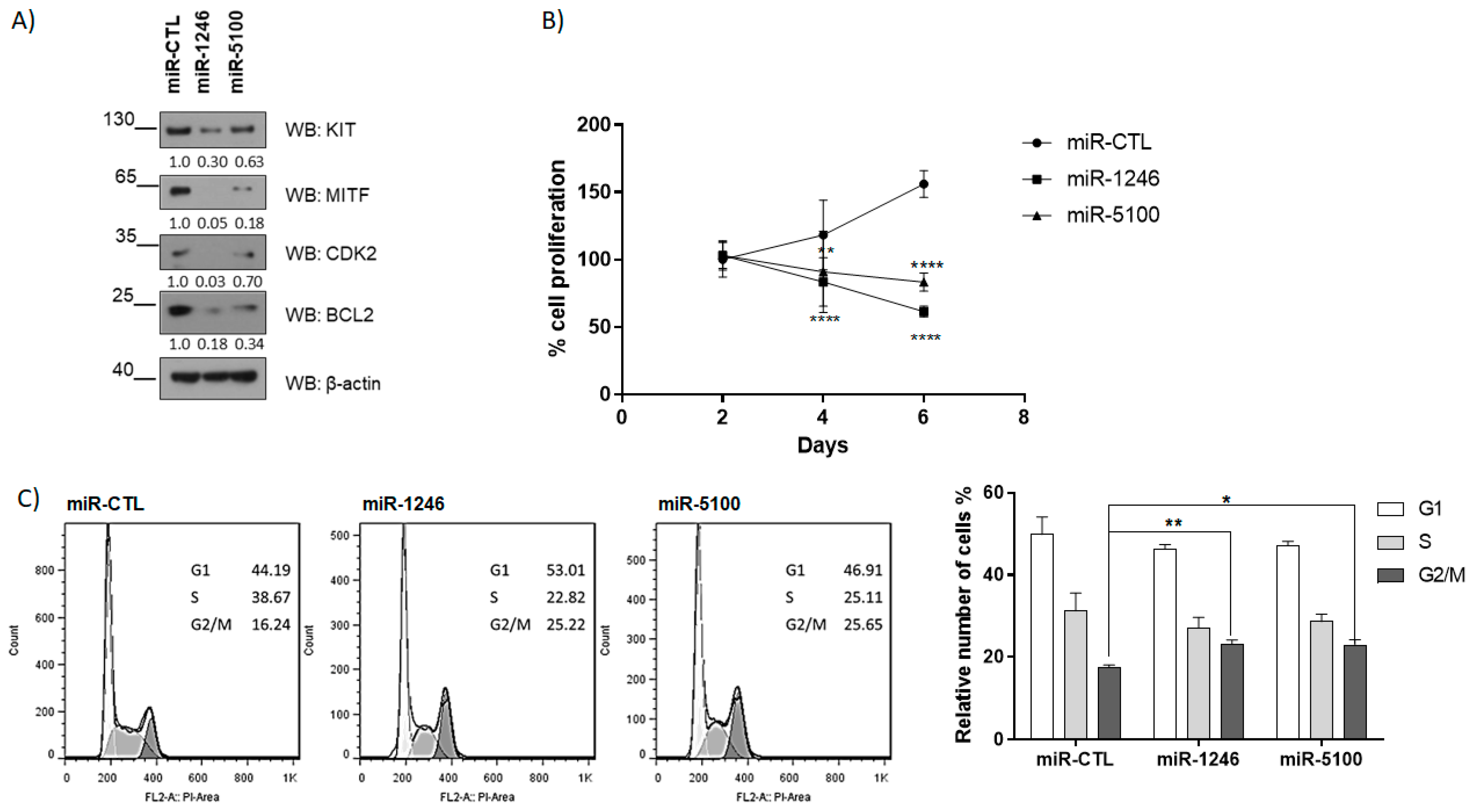
2.2. MiR1246 and miR5100 Target MITF, and Overexpression Significantly Affects Cell Proliferation and Cell Cycle Progression
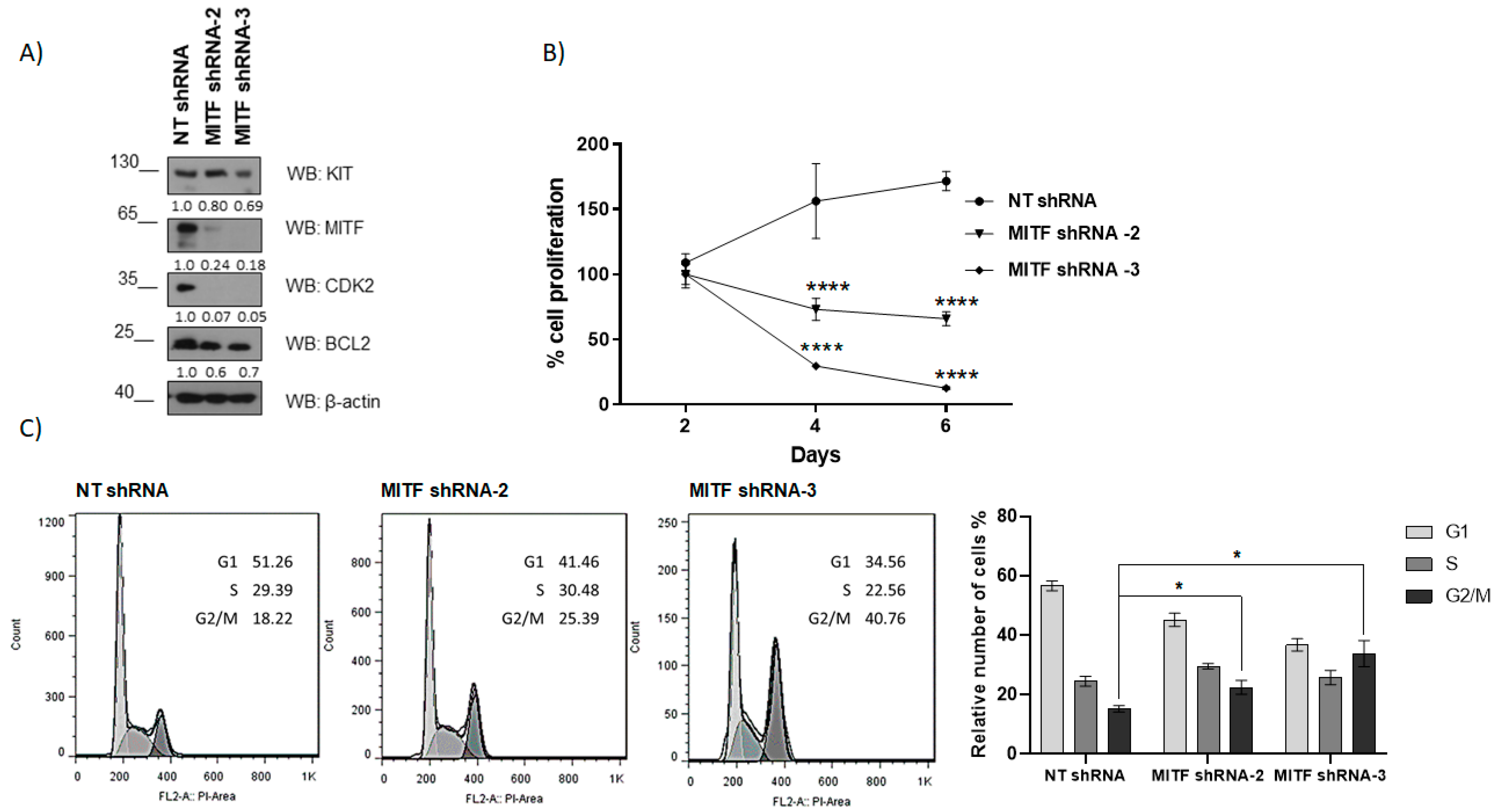
2.3. MITF Silencing Reduces KIT, BCL2, and CDK2 Expression in HMC-1 Cell Lines
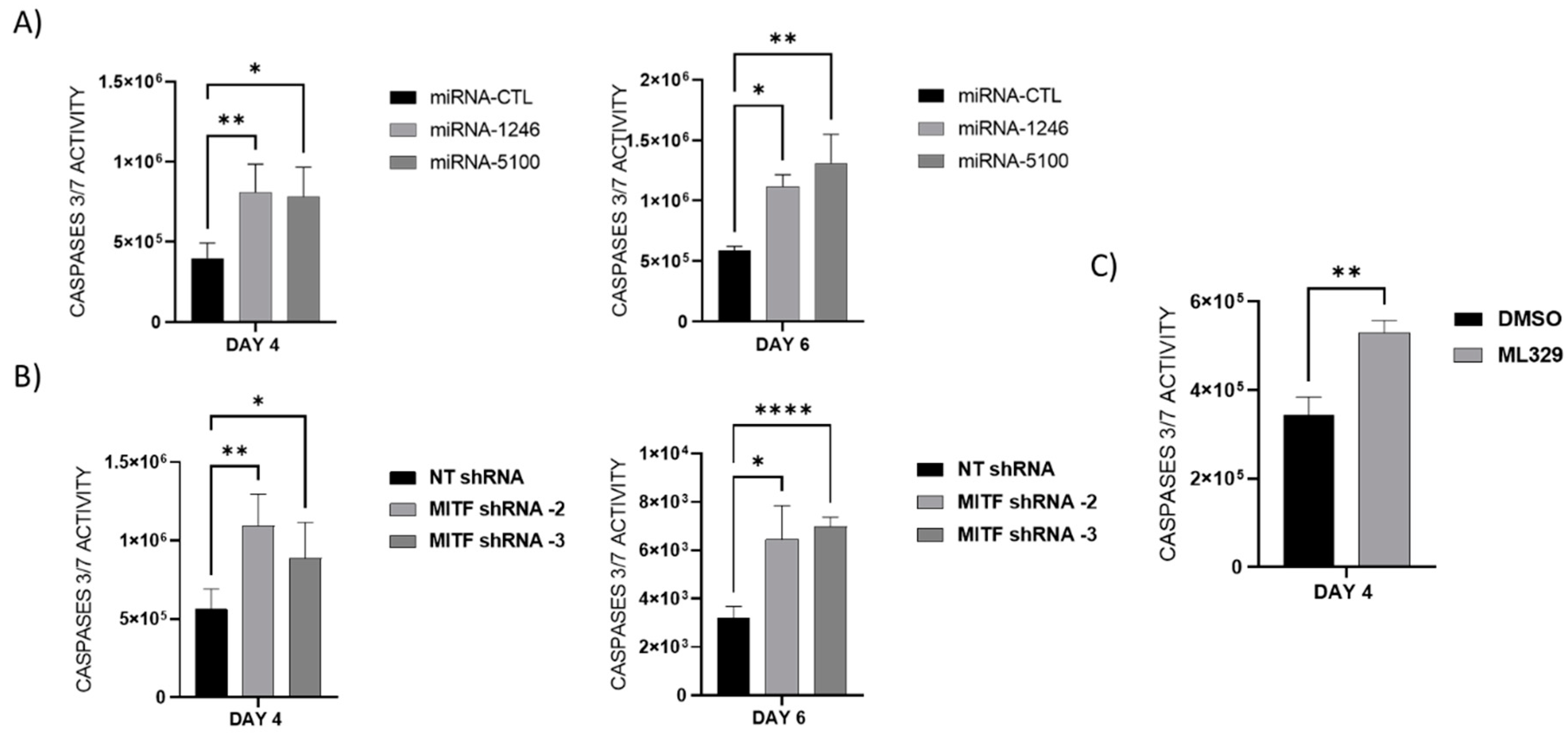
2.4. MITF Activity Inhibition by ML329 Reduces Cell Proliferation and Cycle Arrest
2.5. MITF Downregulation Promotes Apoptosis in HMC-1 Cells
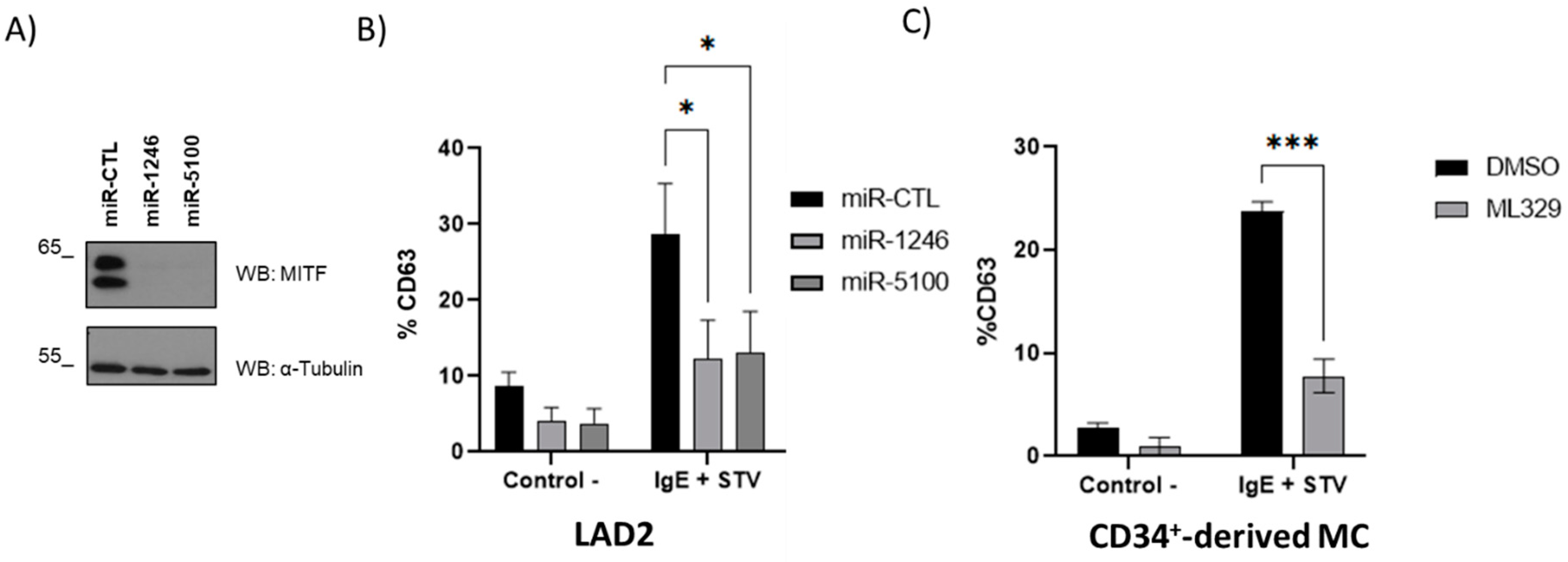
2.6. MITF Downregulation Reduces IgE-Dependent Mast Cell Degranulation
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Culture
4.3. RNA Extraction, Retrotranscription, and PCR Assays
4.4. Lentiviral Transduction
4.5. GFP-miRNA Overexpression
4.6. Cell Viability and Proliferation
4.7. Caspases 3/7 Activity Assay
4.8. Western Blotting
4.9. Cell Cycle Analysis by Flow Cytometry
4.10. Degranulation Assays
4.11. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valent, P.; Akin, C.; Metcalfe, D.D. Mastocytosis: 2016 updated WHO classification and novel emerging treatment concepts. Blood 2017, 129, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Horny, H.-P.; Sotlar, K.; Valent, P.; Hartmann, K. Mastocytosis—A Disease of the Hematopoietic Stem Cell. Dtsch. Arztebl. Int. 2008, 105, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.C.; Metcalfe, D.D.; Komarow, H.D. Mastocytosis. Immunol. Allergy Clin. N. Am. 2014, 34, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.S.; Ekoff, M.; Grootens, J.; Löf, L.; Amini, R.-M.; Hagberg, H.; Ungerstedt, J.S.; Olsson-Strömberg, U.; Nilsson, G. KIT signaling is dispensable for human mast cell progenitor development. Blood 2017, 130, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.M.; Maric, I.; Simakova, O.; Bai, Y.; Chan, E.C.; Olivares, N.; Carter, M.; Maric, D.; Robyn, J.; Metcalfe, D.D. Clonal analysis of NRAS activating mutations in KIT-D816V systemic mastocytosis. Haematologica 2011, 96, 459–463. [Google Scholar] [CrossRef]
- Sundström, M.; Vliagoftis, H.; Karlberg, P.; Butterfield, J.H.; Nilsson, K.; Metcalfe, D.D.; Nilsson, G. Functional and phenotypic studies of two variants of a human mast cell line with a distinct set of mutations in the c-kit proto-oncogene. Immunology 2003, 108, 89–97. [Google Scholar] [CrossRef]
- Valent, P. Diagnostic evaluation and classification of mastocytosis. Immunol. Allergy Clin. N. Am. 2006, 26, 515–534. [Google Scholar] [CrossRef]
- Ainsua-Enrich, E.; Serrano-Candelas, E.; Álvarez-Errico, D.; Picado, C.; Sayós, J.; Rivera, J.; Martín, M. The Adaptor 3BP2 Is Required for KIT Receptor Expression and Human Mast Cell Survival. J. Immunol. 2015, 194, 4309–4318. [Google Scholar] [CrossRef]
- Serrano-Candelas, E.; Ainsua-Enrich, E.; Navinés-Ferrer, A.; Martín, M.; Serrano-Candelas, E.; Ainsua-Enrich, E.; Navinés-Ferrer, A.; Rodrigues, P.; García-Valverde, A.; Bazzocco, S.; et al. Silencing of adaptor protein SH3BP2 reduces KIT/PDGFRA receptors expression and impairs gastrointestinal stromal tumors growth. Mol. Oncol. 2018, 12, 1383–1397. [Google Scholar] [CrossRef]
- Hemesath, T.J.; Steingrímsson, E.; McGill, G.; Hansen, M.J.; Vaught, J.; Hodgkinson, C.A.; Arnheiter, H.; Copeland, N.G.; Jenkins, N.A.; Fisher, D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994, 8, 2770–2780. [Google Scholar] [CrossRef]
- Haq, R.; Yokoyama, S.; Hawryluk, E.B.; Jönsson, G.B.; Frederick, D.T.; McHenry, K.; Porter, D.; Tran, T.N.; Love, K.T.; Langer, R.; et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl. Acad. Sci. USA 2013, 110, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- McGill, G.G.; Horstmann, M.; Widlund, H.R.; Du, J.; Motyckova, G.; Nishimura, E.K.; Lin, Y.L.; Ramaswamy, S.; Avery, W.; Ding, H.F.; et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002, 109, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Widlund, H.R.; Horstmann, M.A.; Ramaswamy, S.; Ross, K.; Huber, W.E.; Nishimura, E.K.; Golub, T.R.; Fisher, D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004, 6, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R.; Arnheiter, H. MITF—The first 25 years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Morii, E.; Jippo, T.; Ito, A. Regulation of Mast Cell Phenotype by MITF. Int. Arch. Allergy Immunol. 2002, 127, 106–109. [Google Scholar] [CrossRef]
- Lee, Y.-N.; Brandal, S.; Noel, P.; Wentzel, E.; Mendell, J.T.; McDevitt, M.A.; Kapur, R.; Carter, M.; Metcalfe, D.D.; Takemoto, C.M. KIT signaling regulates MITF expression through miRNAs in normal and malignant mast cell proliferation. Blood 2011, 117, 3629–3640. [Google Scholar] [CrossRef]
- Lee, Y.-N.N.; Nechushtan, H.; Figov, N.; Razin, E. The Function of Lysyl-tRNA Synthetase and Ap4A as Signaling Regulators of MITF Activity in FcϵRI-Activated Mast Cells. Immunity 2004, 20, 145–151. [Google Scholar] [CrossRef]
- Ribó, P.; Guo, Y.; Aranda, J.; Ainsua-Enrich, E.; Navinés-Ferrer, A.; Guerrero, M.; Pascal, M.; de la Cruz, C.; Orozco, M.; Muñoz-Cano, R.; et al. Mutation in KARS: A novel mechanism for severe anaphylaxis. J. Allergy Clin. Immunol. 2020, 147, 1855–1864. [Google Scholar] [CrossRef]
- Proaño-Pérez, E.; Serrano-Candelas, E.; Mancia, C.; Navinés-Ferrer, A.; Guerrero, M.; Martin, M. SH3BP2 Silencing Increases miRNAs Targeting ETV1 and Microphthalmia-Associated Transcription Factor, Decreasing the Proliferation of Gastrointestinal Stromal Tumors. Cancers 2022, 14, 6198. [Google Scholar] [CrossRef]
- Faloon, P.W.; Bennion, M.; Weiner, W.S.; Smith, R.A.; Wurst, J.; Weiwer, M.; Hartland, C.; Mosher, C.M.; Johnston, S.; Porubsky, P.; et al. A Small Molecule Inhibitor of the MITF Molecular Pathway; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Hsu, J.; Chiu, C.-M.; Hsu, S.-D.; Huang, W.-Y.; Chien, C.-H.; Lee, T.-Y.; Huang, H.-D. miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011, 12, 300. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk—Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—Integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Mitra, J.; van den Heuvel, S.; Enders, G.H. S and G2 Phase Roles for Cdk2 Revealed by Inducible Expression of a Dominant-Negative Mutant in Human Cells. Mol. Cell. Biol. 2001, 21, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Sundaramurthi, H.; García-Mulero, S.; Tonelotto, V.; Slater, K.; Marcone, S.; Piulats, J.M.; Watson, R.W.; Tobin, D.J.; Jensen, L.D.; Kennedy, B.N. Uveal Melanoma Cell Line Proliferation Is Inhibited by Ricolinostat, a Histone Deacetylase Inhibitor. Cancers 2022, 14, 782. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, G.; Blom, T.; Kusche-Gullberg, M.; Kjellen, L.; Butterfield, J.H.; Sundström, C.; Nilsson, K.; Hellman, L. Phenotypic Characterization of the Human Mast-Cell Line HMC-1. Scand. J. Immunol. 1994, 39, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, D.D.; Akin, C. Mastocytosis: Molecular mechanisms and clinical disease heterogeneity. Leuk. Res. 2001, 25, 577–582. [Google Scholar] [CrossRef]
- Valent, P.; Akin, C.; Hartmann, K.; George, T.I.; Sotlar, K.; Peter, B.; Gleixner, K.V.; Blatt, K.; Sperr, W.R.; Manley, P.W.; et al. Midostaurin: A magic bullet that blocks mast cell expansion and activation. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 2367–2376. [Google Scholar] [CrossRef]
- Metcalfe, D.D.; Mekori, Y.A. Pathogenesis and Pathology of Mastocytosis. Annu. Rev. Pathol. 2017, 12, 487–514. [Google Scholar] [CrossRef] [PubMed]
- DeAngelo, D.J.; Radia, D.H.; George, T.I.; Robinson, W.A.; Quiery, A.T.; Drummond, M.W.; Bose, P.; Hexner, E.O.; Winton, E.F.; Horny, H.P.; et al. Safety and efficacy of avapritinib in advanced systemic mastocytosis: The phase 1 EXPLORER trial. Nat. Med. 2021, 27, 2183–2191. [Google Scholar] [CrossRef] [PubMed]
- Gotlib, J.; Reiter, A.; DeAngelo, D.J. Avapritinib for advanced systemic mastocytosis. Blood 2022, 140, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Landolina, N.; Zaffran, I.; Smiljkovic, D.; Serrano-Candelas, E.; Schmiedel, D.; Friedman, S.; Arock, M.; Hartmann, K.; Pikarsky, E.; Mandelboim, O.; et al. Activation of Siglec-7 results in inhibition of in vitro and in vivo growth of human mast cell leukemia cells. Pharmacol. Res. 2020, 158, 104682. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-N.; Noel, P.; Shahlaee, A.; Carter, M.; Kapur, R.; Wayne, A.; Metcalfe, D.D.; Takemoto, C. Kit Signaling Regulates Mitf Expression in Mastocytosis. Blood 2006, 108, 3601. [Google Scholar] [CrossRef]
- Tsujimura, T.; Morii, E.; Nozaki, M.; Hashimoto, K.; Moriyama, Y.; Takebayashi, K.; Kondo, T.; Kanakura, Y.; Kitamura, Y. Involvement of Transcription Factor Encoded by the mi Locus in the Expression of c-kit Receptor Tyrosine Kinase in Cultured Mast Cells of Mice. Blood 1996, 88, 1225–1233. [Google Scholar] [CrossRef]
- Proaño-Pérez, E.; Serrano-Candelas, E.; García-Valverde, A.; Rosell, J.; Gómez-Peregrina, D.; Navinés-Ferrer, A.; Guerrero, M.; Serrano, C.; Martín, M. The microphthalmia-associated transcription factor is involved in gastrointestinal stromal tumor growth. Cancer Gene Ther. 2022, 115–117. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, J.M.; Zeng, S.X.; Lu, H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011, 12, 811–817. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Hu, J.; Wan, X.; Huang, W.; Zhang, H.; Jiang, N. MiR-1246 regulates the PI3K/AKT signaling pathway by targeting PIK3AP1 and inhibits thyroid cancer cell proliferation and tumor growth. Mol. Cell. Biochem. 2022, 477, 649–661. [Google Scholar] [CrossRef]
- Bhagirath, D.; Yang, T.L.; Bucay, N.; Sekhon, K.; Majid, S.; Shahryari, V.; Dahiya, R.; Tanaka, Y.; Saini, S. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018, 78, 1833–1844. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, H.; Wang, G.X.; Wang, J.; Xiang, Y.; Huang, Y.; Shen, C.; Dai, Z.T.; Li, J.P.; Zhang, T.C.; et al. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia 2020, 22, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Iwama, H.; Sakamoto, T.; Okura, R.; Kobayashi, K.; Takano, J.; Katsura, A.; Tatsuta, M.; Maeda, E.; Mimura, S.; et al. Galectin-9 suppresses the growth of hepatocellular carcinoma via apoptosis in vitro and in vivo. Int. J. Oncol. 2015, 46, 2419–2430. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jiang, Y.; Wang, Y.; Chen, T.; Yang, L.; He, H.; Lin, Z.; Liu, T.; Yang, T.; Kamp, D.W.; et al. miR-5100 promotes tumor growth in lung cancer by targeting Rab6. Cancer Lett. 2015, 362, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiong, H.; Duan, L.; Li, Q.; Li, X.; Zhou, Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3β–mediated Wnt/β-Catenin Pathway. Cancer Res. Treat. 2019, 51, 1420–1429. [Google Scholar] [CrossRef]
- Kim, J.H.; Ahn, J.H.; Lee, M. Upregulation of MicroRNA-1246 Is Associated with BRAF Inhibitor Resistance in Melanoma Cells with Mutant BRAF. Cancer Res. Treat. 2017, 49, 947–959. [Google Scholar] [CrossRef]
- Sun, Z.; Meng, C.; Wang, S.; Zhou, N.; Guan, M.; Bai, C.; Lu, S.; Han, Q.; Zhao, R.C. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer 2014, 14, 616. [Google Scholar] [CrossRef]
- Witte, O.N. Steel locus defines new multipotent growth factor. Cell 1990, 63, 5–6. [Google Scholar] [CrossRef]
- Oppezzo, A.; Rosselli, F. The underestimated role of the microphthalmia-associated transcription factor (MiTF) in normal and pathological haematopoiesis. Cell Biosci. 2021, 11, 18. [Google Scholar] [CrossRef]
- Qi, X.; Hong, J.; Chaves, L.; Zhuang, Y.; Chen, Y.; Wang, D.; Chabon, J.; Graham, B.; Ohmori, K.; Li, Y.; et al. Antagonistic Regulation by the Transcription Factors C/EBPα and MITF Specifies Basophil and Mast Cell Fates. Immunity 2013, 39, 97–110. [Google Scholar] [CrossRef]
- Lee, S.-H.H.; Lee, J.-H.H.; Lee, Y.-M.M.; Kim, D.-K.K. Involvement of MITF-A, an alternative isoform of mi transcription factor, on the expression of tryptase gene in human mast cells. Exp. Mol. Med. 2010, 42, 366. [Google Scholar] [CrossRef]
- Li, Y.; Liu, B.; Harmacek, L.; Long, Z.; Liang, J.; Lukin, K.; Leach, S.M.; O’Connor, B.; Gerber, A.N.; Hagman, J.; et al. The transcription factors GATA2 and microphthalmia-associated transcription factor regulate Hdc gene expression in mast cells and are required for IgE/mast cell–mediated anaphylaxis. J. Allergy Clin. Immunol. 2018, 142, 1173–1184. [Google Scholar] [CrossRef]
- Morii, E.; Oboki, K. MITF Is Necessary for Generation of Prostaglandin D 2 in Mouse Mast Cells. J. Biol. Chem. 2004, 279, 48923–48929. [Google Scholar] [CrossRef]
- Shahlaee, A.H.; Brandal, S.; Lee, Y.-N.; Jie, C.; Takemoto, C.M. Distinct and Shared Transcriptomes Are Regulated by Microphthalmia-Associated Transcription Factor Isoforms in Mast Cells. J. Immunol. 2007, 178, 378–388. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Z.; Liang, Y.; Luo, F.; Zhang, J.; Tian, C.; Motzik, A.; Zheng, M.; Kang, J.; Zhong, G.; et al. Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in FcεRI-activated mast cells. Nat. Commun. 2019, 10, 4664. [Google Scholar] [CrossRef]
- King, R.; Weilbaecher, K.N.; McGill, G.; Cooley, E.; Mihm, M.; Fisher, D.E. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for MelanomaDiagnosis. Am. J. Pathol. 1999, 155, 731–738. [Google Scholar] [CrossRef]
- King, R.; Googe, P.B.; Weilbaecher, K.N.; Mihm, M.C.J.; Fisher, D.E. Microphthalmia transcription factor expression in cutaneous benign, malignant melanocytic, and nonmelanocytic tumors. Am. J. Surg. Pathol. 2001, 25, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Ennas, M.G.; Vacca, A.; Ferreli, F.; Nico, B.; Orru, S.; Sirigu, P. Tumor vascularity and tryptase-positive mast cells correlate with a poor prognosis in melanoma. Eur. J. Clin. Invest. 2003, 33, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Willmore-Payne, C.; Holden, J.A.; Tripp, S.; Layfield, L.J. Human malignant melanoma: Detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum. Pathol. 2005, 36, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Capo, A.; Goteri, G.; Mozzicafreddo, G.; Serresi, S.; Giacchetti, A. Melanoma and mastocytosis: Is really only a coincidence? Clin. Exp. Dermatol. 2019, 44, 76–77. [Google Scholar] [CrossRef] [PubMed]
- Nooron, N.; Ohba, K.; Takeda, K.; Shibahara, S.; Chiabchalard, A. Dysregulated Expression of MITF in Subsets of Hepatocellular Carcinoma and Cholangiocarcinoma. Tohoku J. Exp. Med. 2017, 242, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Leachman, S.A.; Lucero, O.M.; Sampson, J.E.; Cassidy, P.; Bruno, W.; Queirolo, P.; Ghiorzo, P. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017, 36, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qin, X.; Wang, B.; Xu, G.; Zhang, J.; Jiang, X.; Chen, C.; Qiu, F.; Zou, Z. MiTF is Associated with Chemoresistance to Cisplatin in A549 Lung Cancer Cells via Modulating Lysosomal Biogenesis and Autophagy. Cancer Manag. Res. 2020, 12, 6563–6573. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, S.; Lee, M.-W.; Jeon, H.-J.; Ryu, H.; Kim, J.-M.; Lee, H.-J. MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway. Cancers 2021, 13, 2920. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.; Vocke, C.D.; Ricketts, C.J.; Metwalli, A.R.; Ball, M.W.; Schmidt, L.S.; Linehan, W.M. Clinical and Molecular Characterization of Microphthalmia-associated Transcription Factor (MITF)-related Renal Cell Carcinoma. Urology 2021, 149, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kirshenbaum, A.S.; Akin, C.; Wu, Y.; Rottem, M.; Goff, J.P.; Beaven, M.A.; Rao, V.K.; Metcalfe, D.D. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcεRI or FcγRI. Leuk. Res. 2003, 27, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Rådinger, M.; Jensen, B.M.; Kuehn, H.S.; Kirshenbaum, A.; Gilfillan, A.M. Generation, Isolation, and Maintenance of Human Mast Cells and Mast Cell Lines Derived from Peripheral Blood or Cord Blood. In Current Protocols in Immunology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Álvarez-Errico, D.; Oliver-Vila, I.; Ainsua-Enrich, E.; Gilfillan, A.M.; Picado, C.; Sayós, J.; Martín, M. CD84 Negatively Regulates IgE High-Affinity Receptor Signaling in Human Mast Cells. J. Immunol. 2011, 187, 5577–5586. [Google Scholar] [CrossRef] [PubMed]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of Cell Cycle by Flow Cytometry. In Checkpoint Controls and Cancer; Humana Press: Totowa, NJ, USA, 2004; Volume 281, pp. 301–312. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proaño-Pérez, E.; Ollé, L.; Guo, Y.; Aparicio, C.; Guerrero, M.; Muñoz-Cano, R.; Martin, M. MITF Downregulation Induces Death in Human Mast Cell Leukemia Cells and Impairs IgE-Dependent Degranulation. Int. J. Mol. Sci. 2023, 24, 3515. https://doi.org/10.3390/ijms24043515
Proaño-Pérez E, Ollé L, Guo Y, Aparicio C, Guerrero M, Muñoz-Cano R, Martin M. MITF Downregulation Induces Death in Human Mast Cell Leukemia Cells and Impairs IgE-Dependent Degranulation. International Journal of Molecular Sciences. 2023; 24(4):3515. https://doi.org/10.3390/ijms24043515
Chicago/Turabian StyleProaño-Pérez, Elizabeth, Laia Ollé, Yanru Guo, Cristina Aparicio, Mario Guerrero, Rosa Muñoz-Cano, and Margarita Martin. 2023. "MITF Downregulation Induces Death in Human Mast Cell Leukemia Cells and Impairs IgE-Dependent Degranulation" International Journal of Molecular Sciences 24, no. 4: 3515. https://doi.org/10.3390/ijms24043515
APA StyleProaño-Pérez, E., Ollé, L., Guo, Y., Aparicio, C., Guerrero, M., Muñoz-Cano, R., & Martin, M. (2023). MITF Downregulation Induces Death in Human Mast Cell Leukemia Cells and Impairs IgE-Dependent Degranulation. International Journal of Molecular Sciences, 24(4), 3515. https://doi.org/10.3390/ijms24043515






