Abstract
The role of dysbiosis in the development and progression of oral potentially malignant disorders (OPMDs) remains largely unknown. Here, we aim to characterize and compare the oral microbiome of homogeneous leucoplakia (HL), proliferative verrucous leukoplakia (PVL), oral squamous cell carcinoma (OSCC), and OSCC preceded by PVL (PVL-OSCC). Fifty oral biopsies from HL (n = 9), PVL (n = 12), OSCC (n = 10), PVL-OSCC (n = 8), and healthy (n = 11) donors were obtained. The sequence of the V3–V4 region of the 16S rRNA gene was used to analyze the composition and diversity of bacterial populations. In the cancer patients, the number of observed amplicon sequence variants (ASVs) was lower and Fusobacteriota constituted more than 30% of the microbiome. PVL and PVL-OSCC patients had a higher abundance of Campilobacterota and lower Proteobacteria than any other group analyzed. A penalized regression was performed to determine which species were able to distinguish groups. HL is enriched in Streptococcus parasanguinis, Streptococcus salivarius, Fusobacterium periodonticum, Prevotella histicola, Porphyromonas pasteri, and Megasphaera micronuciformis; PVL is enriched in Prevotella salivae, Campylobacter concisus, Dialister pneumosintes, and Schaalia odontolytica; OSCC is enriched in Capnocytophaga leadbetteri, Capnocytophaga sputigena, Capnocytophaga gingivalis, Campylobacter showae, Metamycoplasma salivarium, and Prevotella nanceiensis; and PVL-OSCC is enriched in Lachnospiraceae bacterium, Selenomonas sputigena, and Prevotella shahii. There is differential dysbiosis in patients suffering from OPMDs and cancer. To the best of our knowledge, this is the first study comparing the oral microbiome alterations in these groups; thus, additional studies are needed.
Keywords:
oral leukoplakia; oral cancer; microbiota; head and neck cancer; pathogens; Campylobacter; 16S rRNA 1. Introduction
According to a recent international consensus, oral potentially malignant disorders (OPMDs) are defined as any oral mucosal abnormality that is associated with an increased risk of occurrence of oral cancer [1]. Among them, oral leucoplakias are the most frequent form of OPMD and consist of a predominantly white plaque of uncertain risk, having excluded other known diseases that carry no increased risk for cancer [2]. In that sense, a recent systematic review and meta-analysis concluded that their potential for malignant transformation is 9.7% [3]. Furthermore, their location on the tongue and floor of the mouth, the existence of non-homogeneous clinical forms [4], and the histological findings of high-grade epithelial dysplasia [3] contribute to a higher risk of cancer development. Among oral leucoplakias, the proliferative verrucous leukoplakia (PVL) is a low-frequency, progressive, persistent, and irreversible subtype with distinctive clinical and evolutionary characteristics [5,6]. PVL is frequently located on the gingiva, has a much higher potential for malignant transformation (ranging from 43.87 to 71.4%), and shows an increased tendency to develop second primary tumors, with a very high recurrence rate after various treatments [7].
The morphological and cytological changes of OPMDs are similar to those of early invasive oral squamous cell carcinomas (OSCCs) and some of the chromosomal, genomic, and molecular alterations detected in them are also found in OPMDs [8]. Unfortunately, the literature is sparse about the microbiome associated with OPMDs and the potential role of microbial dysbiosis in OPMDs remains largely unknown.
The mouth, along with the aerodigestive tract, harbors the second most abundant microbiota in the human body. As of December 2022, the expanded Human Oral Microbiome Database (eHOMD) includes information on 774 bacterial species, 74% of them cultivable and 26% belonging to uncultivated phylotypes [9]. These oral microorganisms have a direct impact on their hosts, ranging from metabolic reactions to immune responses. Indeed, oral microbial dysbiosis has been reported in many diseases, including diabetes, bacteremia, endocarditis, cancer, autoimmune disease, and atherosclerosis, making it important to recognize the diversity of the oral microbiota and how it changes under altered conditions to determine its potential role in disease [10,11,12,13,14]. Dysbiotic oral microflora has already been associated with chronic periodontal disease [15,16] and oral cancer [17] and for PVL, a controversy exists around the possible implication of some pathogens, such as human papillomavirus (HPV) 16 [18,19,20,21]. In this regard, a recent study published by our group comparing PVL patients with healthy controls revealed that oral dysbiosis is a common state in PVL patients, who exhibited a general loss of diversity and enrichment of some protumorigenic pathogens, such as Campylobacter jejuni [22]. The aim of our new study, following the same line of research as the previous one, is to analyze the microbiota differences between homogeneous leukoplakia (HL) and PVL, as well as with two groups of cancers, one of them after the evolution of PVL.
2. Results
2.1. Participant Characteristics and Sequencing Data Summary
The clinical profile and clinicopathological information of the healthy donors (group I), the patients with HL (group II), PVL (group III), OSCC (group IV), and PVL-OSCC (group V) are shown in Supplementary Data S1. The mean patient age was 70.31 years, 52% were females, and 32% were smokers. No significant differences in clinicopathological characteristics were found between groups. Five patients with non-neoplastic lesions (HL and PVL) did not have dysplasia, twelve showed mild dysplasia, three presented moderate dysplasia, and one PVL case had severe dysplasia. In addition, PVL-OSCC patients tended to show a lower frequency of poorly differentiated tumors (G0/1: 50% vs. 75%), perineural infiltrations (25% vs. 70%), presence of lymph node metastasis in the neck (12.5% vs. 30%), and stage IV tumors (37.5% vs. 60%) compared to other OSCC.
Sequencing of oral samples and filtering for sequence quality resulted in a total of 2,435,181 effective reads, with a median of 44,927 reads per sample (range 16,160–110,317). Rarefaction curves (number of reads vs. alpha diversity) began to level off for most of the samples, indicating that samples were sequenced to a sufficient depth such that a complete microbiome profile was likely captured for most samples (Figure 1a). A total of 14 phyla, 24 classes, 62 orders, 102 families, and 189 genera were identified for Bacteria. Phylum-level classification of the bacterial community was also identified (Figure 1b) using the feature prevalence, which is the number of samples in which an amplicon sequence variant (ASV) appears at least once. The bacterial community was heavily dominated by phylum Fusobacteriota (relative abundance >26.7%), Firmicutes (>21.0%), Bacteroidota (>19.3%), and Proteobacteria (>17.0%) (Figure 1b). Regarding Archea, low numbers of Woesearchaeales of Nanoarchaeota and Aenigmarchaeota were also identified.
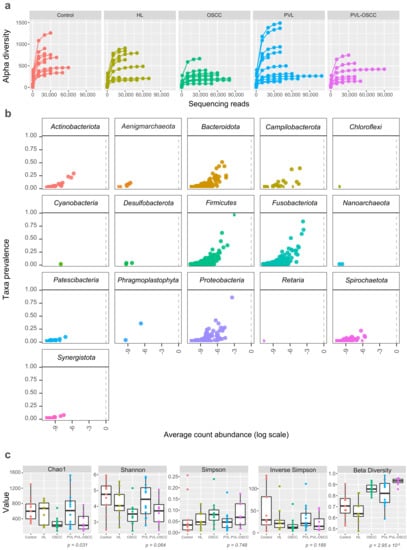
Figure 1.
Microbiome diversity for oral samples collected from healthy controls and HL, PVL, OSCC, and PVL-OSCC patients. (a) Rarefaction curves (alpha diversity versus sequencing library size). (b) Prevalence plot (taxa prevalence versus average count abundance). Each point corresponds to a different amplicon sequence variant. (c) Microbial richness (Chao1), diversity (Shannon), dominance (Simpson and Inverse Simpson), and similarity (beta divergence) of oral samples.
2.2. Differences in Microbiome Diversity
To study the complexity of the microbiota community structure within the samples, we determined alpha- and beta-diversity scores for all patients (Supplementary Data S2). We found that richness, as assessed by Chao1, was significantly higher for healthy donors (637.9 ± 306.8), PVL (652.7 ± 476.7), and HL (565.5 ± 295.8) than for OSCC (284.2 ± 166.9) and PVL-OSCC (320.5 ± 244.0) patients (p = 0.031) (Figure 1c). No significant differences were found in terms of diversity (Shannon–Wiener) or dominance (Simpson and Inverse Simpson). Regarding similarity, beta diversity revealed that the microbiome from OSCC and PVL-OSCC patients is more homogeneous than that from control, HL, and PVL patients (p = 2.95 × 10−5, Figure 1c).
2.3. Differences in Microbiome Composition
Krona plots representing an overview of the microbiome of all the samples included in the study are available in Supplementary Data S3. Additionally, a summarized description of the microbiome composition for each group of patients is provided in Table 1. The most abundant phyla found for all the groups were Fusobacteriota, Bacteroidota, Proteobacteria, and Firmicutes (Figure 2a). However, relative abundances significantly varied among groups. Thus, Fusobacteriota constituted 34% and 30% of the sequences detected in OSCC and PVL-OSCC patients, respectively, in contrast to the relative abundance of this phylum for healthy donors (24%), HL (25%), and PVL (19%). Of note, the relative abundance of Campilobacterota was higher in PVL (11%) and PVL-OSCC (14%) patients than in any other group analyzed. On the contrary, the relative abundance of Proteobacteria was lower in PVL (11%) and PVL-OSCC (11%) patients than in any other group (20% in controls, 25% in HL, and 19% in OSCC).

Table 1.
Phylogenetic composition of the most common taxa by group. Abundances at the phylum and genus levels are presented as a percentage of the total microbiome. Abundances < 1% are presented as empty cells. Asterisks indicate p < 0.05.
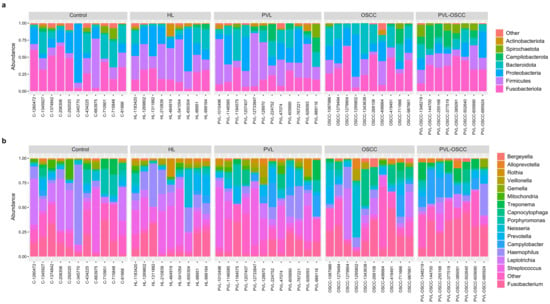
Figure 2.
Stacked bar plot of the phylogenetic composition of the most common taxa at the phylum (a) and genus (b) levels.
At the genus level, the most frequently detected genera in controls were Haemophilus (15%), Fusobacterium (14%), Leptotrichia (10%), and Streptococcus (10%) (Figure 2b). In contrast, Streptococcus (19%), Fusobacterium (19%), and Haemophilus (19%) were the most frequently registered genera in HL, and Fusobacterium (25%) and Capnocytophaga (13%) in OSCC. For PVL patients, Streptococcus (13%), Fusobacterium (14%), and Campylobacter (11%) were the genera with the highest relative abundance, whereas these were Fusobacterium (26%), Campylobacter (14%), and Treponema (10%) in the case of PVL-OSCC patients.
2.4. Community Structure Reveals Differently Abundant ASVs in Oral Disorders
In addition to the differences found in microbiome diversity and composition, specific ASVs were identified that exhibited differences in abundance between groups. Before further analysis, ASVs were filtered to remove microorganisms with very low counts (<100 reads) across all libraries since they provide little evidence for differential distribution analysis. After filtering, five genera were found to be exclusively present in healthy donors, whereas two were exclusive of PVL, four of OSCC, and four of PVL-OSCC (Figure 3a). Of note, no genera were found to be exclusive of HL, one was shared among OPMDs, and two were shared between OSCC and PVL-OSCC.
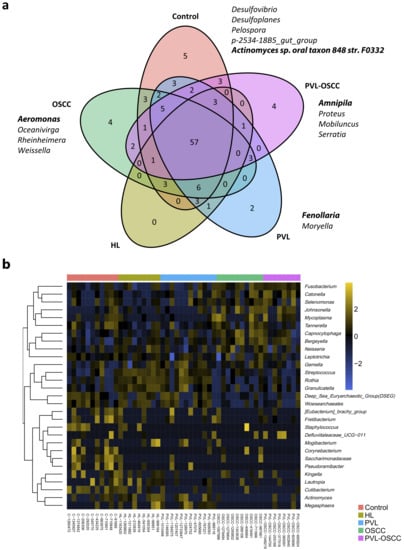
Figure 3.
Differently abundant genera between samples from healthy donors, HL, PVL, OSCC, and PVL-OSCC patients. (a) Venn diagram summarizing the distribution of genera along groups. (b) Clustering of the differentially distributed genera along samples as selected by elastic net analysis.
Afterward, a penalized regression was performed to determine which genera were able to distinguish groups, finding that 30 genera were differentially distributed among samples (Supplementary Data S4). Control samples were enriched in Fretibacterium, Tannerella, Kingella, Corynebacterium, Lautropia, Cutibacterium, Saccharimonadaceae, and Pseudoramibacter, whereas HL was enriched in Rothia, Streptococcus, Megasphaera, and Mogibacterium. PVL had a higher abundance of Granulicatella, Gemella, Eubacterium, Actinomyces and Deep Sea Euryarchaeotic Group (DSEG); OSCC were enriched in Capnocytophaga, Fusobacterium, Leptotrichia, Neisseria, Bergeyella, Mycoplasma, Johnsonella and Staphylococcus; and PVL-OSCC in Selenomonas, Catonella and Defluviitaleaceae UCG−011 (Figure 3b).
At the species level, seven microorganisms were found to be exclusively present in control samples, whereas three were exclusive of PVL, one of OSCC, and six of PVL-OSCC (Figure 4a). Again, no species were found to be exclusive of HL, three were shared between HL and PVL, and three were shared between OSCC and PVL-OSCC. In this case, the elastic net multinomial regression revealed that samples from healthy donors were enriched in Campylobacter gracilis, Filifactor alocis, Phocaeicola abscessus, and Treponema maltophilum; HL had more Streptococcus parasanguinis, Streptococcus salivarius, Fusobacterium periodonticum, Prevotella histicola, Porphyromonas pasteri, and Megasphaera micronuciformis; PVL were enriched in Prevotella salivae, Campylobacter concisus, Dialister pneumosintes, and Schaalia odontolytica; OSCC had a higher abundance of Capnocytophaga leadbetteri, Capnocytophaga sputigena, Capnocytophaga gingivalis, Campylobacter showae, Metamycoplasma salivarium, and Prevotella nanceiensis; and PVL-OSCC were enriched in Lachnospiraceae bacterium, Selenomonas sputigena and Prevotella shahii (Supplementary Data S5, Figure 4b).
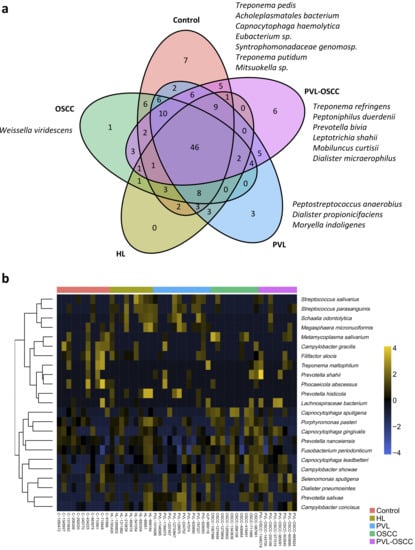
Figure 4.
Differently abundant species between samples from healthy donors, HL, PVL, OSCC, and PVL-OSCC patients. (a) Venn diagram summarizing the distribution of species along groups. (b) Clustering of the differentially distributed species along samples as selected by elastic net analysis.
3. Discussion
Microbiome studies, motivated by the availability of high-throughput technologies, have exhibited how the disturbance of the microbiota is associated with a great number of human diseases [23]. To date, the vast majority of studies have been performed on the gut, which constitutes the body niche where most of the commensal microorganisms reside. Although it has been less studied, oral microbiome dysbiosis seems to be linked to oral cancer and other oral diseases through several mechanisms, including the direct metabolism of chemical carcinogens and general inflammatory effects [24]. The oral cavity is home to more than 700 microbial species, including commensal and opportunistic bacterium, fungi, and viruses, however, there are still few studies addressing the influence of changes in the oral microbiome in the development and progression of OPMDs.
In this study, we characterized oral microbial communities of patients with HL, PVL, OSCC or OSCC preceded by PVL (PVL-OSCC). In terms of overall diversity, the average number of different species detected was lower in OSCC and PVL-OSCC than in disease-free, age-matched controls, but not in HL and PVL patients. This decrease in oral microbial diversity has been previously reported in several forms of head and neck cancers, including oral, esophageal, and nasopharyngeal carcinomas [25,26,27]. As with other studies comparing disease versus healthy microbiome, it is not possible to say whether the microbial alterations found are the cause or the consequence of the disease, however, the absence of this reduction in premalignant lesions could indicate that the decrease of the oral microbial diversity detected in cancer is a consequence of the disease. Nevertheless, reduction in oral microbial diversity has been reported in other oral diseases such as caries, recurrent aphthous stomatitis, or oral lichen planus, thus caution should be exerted when interpreting these findings.
At the phylum level, Firmicutes, Bacteroidota, Proteobacteria, Actinobacteriota, Spirochaetota, and Fusobacteriota have been reported to constitute 96% of the total oral bacteria [28], which aligns with the most abundant phyla found for all patients in our study. However, the abundance of Campylobacterota was >10% in PVL and PVL-OSCC in our study. Previous reports have shown the association between oral Campylobacter infections and increased risk of inflammatory bowel disease [29], esophageal [30], and oral [31] cancers. In addition, increased levels of Campylobacterota have been reported in oral leukoplakia (OL) [32] and a previous study performed by our group concluded that the microbiome of PVL patients is significantly enriched in Campylobacter jejuni [22]. Importantly, it is known that a Campylobacter-derived genotoxin, called cytolethal distending toxin (CDT), induces DNA double-strand breaks and facilitates colorectal tumorigenesis [33]. These findings, together with our data, suggest that Campylobacter species might be associated with PVL development and oral carcinogenesis via the induction of DNA damage.
At the genus level, the main components of the oral microbiome according to Bik et al. are Actinomyces, Atopobium, Corynebacterium, Rothia of Actinobacteria; Bergeyella, Capnocytophaga, Prevotella of Bacteroidetes, Granulicatella, Streptococcus, Veillonella of Firmicutes, Fusobacterium, Campylobacter, Cardiobacterium, Haemophilus, Neisseria of Proteobacteria, and TM7 [34]. However, differences can be found between studies, since many factors, including smoking habits, diet, and varying geographical and climatic conditions alter the oral microbiota, making comparisons difficult [35]. In our study, these bacteria were the most frequently detected genera in all the patients included, except for Cardiobacterium, Actinomyces, Atopobium, Corynebacterium, and TM7, which were less frequently detected than Porphyromonas, Leptotrichia or Treponema. Specifically, the presence of Rothia, Streptococcus, Megasphaera, and Mogibacterium was remarkable in HL, whereas PVL had a higher abundance of Granulicatella, Gemella, Eubacterium, Actinomyces and Deep Sea Euryarchaeotic Group (DSEG). Amer and colleagues compared swabs from OL to contralateral healthy sites and control samples and concluded that the microbiome of OL exhibits enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia species [36]. Moreover, another study comparing salivary samples from OL, OSCC and healthy controls reported that healthy controls could be distinguished from the former two disorders based on the abundance of Megaspheara, unclassified enterobacteria, Prevotella, Porphyromonas, Rothia, Salmonella, Streptococcus, and Fusobacterium [36]. Regarding oral cancers, in this study, OSCC were enriched in Capnocytophaga, Fusobacterium, Leptotrichia, Neisseria, Bergeyella, Mycoplasma, Johnsonella, and Staphylococcus; and PVL-OSCC in Selenomonas, Catonella, and Defluviitaleaceae UCG−011. Previous studies have reported the increased abundance of Capnocytophaga, Fusobacterium, Leptotrichia, Staphylococcus, Selenomonas, and Catonella in oral cancers [37,38,39,40]. In particular, Weiwen and collaborators reported that Capnocytophaga species are potential tumor promotors in oral cancer [37] and Saxena et al. reported that Capnocytophaga and Fusobacterium are differentially abundant in OSCC-associated microbiomes and can be considered as potential microbiome marker genera for oral cancer [39]. In addition, Catonella species are considered periodontitis-associated bacteria that may be related to primary endodontic infections [41,42].
In addition to the differences exposed in overall microbiome profiles, we also identified several species that were specifically over-represented in every oral disease. HL had a higher abundance of the protumorigenic pathogens Prevotella histicola and Streptococcus parasanguinis. P. histicola has been reported to produce acetaldehyde [43], whereas a study comparing the oral microbiota in tumor and non-tumor tissues of OSCC patients concluded that S. parasanguinis was highly associated with the tumor site [44]. The relative abundance of S. parasanguinis was also significantly higher in tongue/pharyngeal cancer patients [45]. Importantly, the relative abundance of the oral probiotic Streptococcus salivarius and Porphyromonas pasteri were also higher in HL. S. salivarius has been extensively reported as a probiotic microbe [46], whereas P. pasteri has been inversely associated with OSCC progression [47]. In contrast to HL, the oral microbiome of PVL was enriched in Campylobacter concisus, Prevotella salivae, and Dialister pneumosintes. Elevated levels of C. concisus have been associated with severe dysplasia in patients with OL, and a case-control study involving 25 OSCC cases and 27 fibroepithelial polyps (FEP) concluded that C. concisus was more abundant in OSCC than in FEP [48]. Additionally, this case-control study reported the enrichment of P. salivae in OSCC samples compared to FEP ones [48], whereas the study conducted by Coker and col. concluded that D. pneumosintes had significant centralities in the gastric cancer ecological network [49].
In OSCC, Capnocytophaga leadbetteri, Capnocytophaga sputigena, and Capnocytophaga gingivalis had a higher abundance. These three have been previously reported to be increased in OSCC patients, suggesting a potential association between these bacteria and OSCC [42,50,51]. Indeed, C. sputigena was suggested to cause invasive gingival disease with hyperplasia in immunocompromised patients [52] and C. sputigena bacteremia, most likely induced by an oral pathology, occurred in several patients with acute leukemia [53]. C. gingivalis is also considered a potential tumor promotor in oral cancer. A high salivary count of C. gingivalis has been suggested as a diagnostic indicator of OSCC [51], and its supernatant was found to induce epithelial to mesenchymal transition (EMT), causing OSCC cells to acquire highly invasive and metastatic properties [37]. Metamycoplasma salivarium and Prevotella nanceiensis were also elevated in OSCC. M. salivarium was considered a non-pathogenic commensal at first, however, its detection in the epithelial cells of OL and oral lichen planus and as a dominant colonizer of Fanconi anemia-associated oral carcinoma has cast doubt on this [54,55]. On the other hand, the higher abundance of P. nanceiensis has been suggested to influence OSCC through inflammation [56]. In contrast to OSCC, PVL-OSCC were enriched in Selenomonas sputigena, Lachnospiraceae bacterium, and Prevotella shahii, and, to the best of our knowledge, it is the first time that these bacteria have been linked to oral cancers.
4. Materials and Methods
4.1. Patients and Tissue Samples
This study included 50 individuals who visited and were treated at the Stomatology and Maxillofacial Surgery Department of the General University Hospital of Valencia. Participants were distributed into five groups according to their oral disorder: group I consisted of nine patients with HL, group II included twelve patients with PVL, group III comprised ten OSCC patients, group IV was composed of eight patients with OSCC preceded by the evolution of PVL (PVL-OSCC), and group V included eleven healthy donors. For groups I to IV, two representative biopsies were taken from the same area of the lesions, including epithelium and the underlying connective tissue between 2017 and 2021. One of each pair of specimens was analyzed with the routine histopathological methods to ensure that the observed lesions met the histopathological criteria to establish the diagnosis together with the clinical data of each patient. PVL diagnosis was determined following the criteria provided by Villa et al. [6]. The other sample was used for the 16S sequencing. For the control group (group V), samples were obtained from healthy mucosa areas adjacent to the teeth (vestibular fundus). All tissue samples were frozen at −80 °C until their analysis.
4.2. DNA Extraction and 16S rDNA Gene Sequencing
Total DNA from clinical samples was extracted using the DNAeasy kit (QiaGen, Barcelona, Spain). Variable V3 and V4 regions of the 16S rDNA gene were amplified following the 16S Metagenomic Sequencing Library Preparation Illumina protocol (Cod. 15044223 Rev. A, Illumina, Inc., San Diego, CA, USA). The full-length primer sequences including Illumina adapter overhang nucleotide sequences were selected according to Klindworth et al. [57] as follows:
Forward primer: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG-GGNGGCWGCAG-3′.
Reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTA-CHVGGGTATCTAATCC-3′.
After 16S rDNA gene amplification, the multiplexing step was performed using Nextera XT Index Kit (FC-131-2001). A volume of 1 μL of the PCR product was run on a Bioanalyzer DNA 1000 chip to verify the size (~550 bp expected) on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). After size verification, libraries were sequenced using a 2 × 300 bp paired-end run (MiSeq Reagent kit v3, MS-102-3003) on an Illumina MiSeq Sequencer according to the manufacturer’s instructions.
4.3. Bioinformatic Analysis and Taxonomic Annotation
Sequencing data were demultiplexed using the Illumina bcl2fastq© program. Demultiplexed paired FASTQ sequences were processed using QIIME2 v2021.4. Quality control was carried out using the DADA2 pipeline incorporated into QIIME2 [58]. The DADA2 pipeline filtered out phiX reads, removed chimeric sequences, and assigned reads into Amplicon Sequence Variants (ASVs) [59]. Taxonomic affiliations were assigned using the Naive Bayesian classifier integrated into QIIME2 plugins. The SILVA v138 database was used for taxonomic annotation [60]. The taxonomic composition of the oral microbiota was generated by different levels: kingdom, phylum, class, order, family, genus, and species.
4.4. Statistical Analysis
Data were summarized using mean (standard deviation) and median (first and third quartiles) in the case of continuous variables, and absolute (relative) frequencies in the case of categorical variables. Unsupervised analyses were performed using principal coordinates analysis and hierarchical clustering. Chi-squared test and nonparametric tests (Mann–Whitney U or Kruskal–Wallis) were applied to evaluate associations between patient clinicopathological characteristics and microbiota composition. Compositional differences among groups were assessed by elastic net multinomial regression. The penalization factor for the elastic net analysis was selected using three-fold cross-validation. Statistical analyses were performed using R software v.4.2.2 (R Foundation for Statistical Computing, Vienna, Austria), and the R packages microbiome (version 1.18.0), pgirmess (version 2.0.0), glmnet (version 4.1-4) and clickR (version 0.9.27).
5. Conclusions
Using a 16S rRNA gene sequencing-based approach, oral microbial dysbiosis was found in patients with HL, PVL, OSCC, and PVL-OSCC. Loss of diversity was found in oral cancers but not in premalignant lesions, which could indicate that the decrease of the oral microbial diversity detected in cancer is a consequence of the disease. In addition, the relative abundance of Campylobacterota was higher in PVL and PVL-OSCC, which may constitute an important risk factor for PVL development and progression. Oral diseases could be distinguished by the abundance of certain species. A better understanding of the role of the oral microbiome in OPMDs and cancer could direct to novel non-invasive diagnostic and prognostic options, as well as to more personalized treatments and microbiome-targeted therapeutic interventions.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043466/s1.
Author Contributions
Methodology, A.H.-P., D.H. and J.B.; Software, D.H.; Validation, A.H.-P.; Formal analysis, A.H.-P. and D.H.; Investigation, A.H.-P., D.H. and J.B.; Data curation, D.H.; Writing—original draft, A.H.-P., D.H. and J.B.; Writing—review & editing, A.H.-P., D.H. and J.B.; Supervision, E.J.-L., L.B.-D., C.G.-C. and J.B.; Funding acquisition, J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by PI19/00790 from Fondo de Investigación Sanitaria, ISCIII (PI: Jose Bagan).
Institutional Review Board Statement
This study was approved by the Ethics Committee for Human Research of the University of Valencia (Ref. H1523722754549). Informed written consent was obtained from all participants after an explanation of the nature of the study, as approved by the Ethics Committee for Human Research of the University General Hospital, Valencia, Spain.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Raw data have been deposited at the NCBI SRA archive with BioProject record PRJNA916491 and BioSample records from SAMN32471763 to SAMN32471812.The samples were managed and processed by the Biobank of the Hospital Universitario y Politécnico, authorised biobank (B.0000723) following the requirements of RD1716/2011.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
ASV: Amplicon Sequence Variant; CDT, Cytolethal Distending Toxin; DSEG, Deep Sea Euryarchaeotic Group; eHOMD, expanded Human Oral Microbiome Database; FEP, Fibroepithelial polyp; EMT, Epithelial–Mesenchymal Transition; HL, Homogeneous Leukoplakia; HPV, Human Papillomavirus; OL, Oral Leukoplakia; OPMD, Oral Potentially Malignant Disorder; PVL, Proliferative Verrucous Leukoplakia, OSCC, Oral Squamous Cell Carcinoma; PVL-OSCC, Oral Squamous Cell Carcinoma preceded by Proliferative Verrucous Leukoplakia.
References
- Warnakulasuriya, S.; Kujan, O.; Aguirre-Urizar, J.M.; Bagan, J.V.; González-Moles, M.Á.; Kerr, A.R.; Lodi, G.; Mello, F.W.; Monteiro, L.; Ogden, G.R.; et al. Oral potentially malignant disorders: A consensus report from an international seminar on nomenclature and classification, convened by the WHO Collaborating Centre for Oral Cancer. Oral Dis. 2021, 27, 1862–1880. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S.; Johnson, N.W.; Van Der Waal, I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J. Oral Pathol. Med. 2007, 36, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Urizar, J.M.; Lafuente-Ibáñez de Mendoza, I.; Warnakulasuriya, S. Malignant transformation of oral leukoplakia: Systematic review and meta-analysis of the last 5 years. Oral Dis. 2021, 27, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Bagan, J.; Martorell, M.; Cebrián, J.L.; Rubert, A.; Bagán, L.; Mezquida, C.; Hervás, D. Effect of clinical and histologic features on time to malignancy in 224 cases of oral leukoplakia treated by surgery. Clin. Oral Investig. 2022, 26, 5181–5188. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á.; Mello, F.W.; Bagan, J.V.; Warnakulasuriya, S. Malignant transformation of oral proliferative verrucous leukoplakia: A systematic review and meta-analysis. Oral Dis. 2021, 27, 1896–1907. [Google Scholar] [CrossRef]
- Villa, A.; Menon, R.S.; Kerr, A.R.; De Abreu Alves, F.; Guollo, A.; Ojeda, D.; Woo, S.B. Proliferative leukoplakia: Proposed new clinical diagnostic criteria. Oral Dis. 2018, 24, 749–760. [Google Scholar] [CrossRef]
- Bagan, J.; Murillo-Cortes, J.; Poveda-Roda, R.; Leopoldo-Rodado, M.; Bagan, L. Second primary tumors in proliferative verrucous leukoplakia: A series of 33 cases. Clin. Oral Investig. 2020, 24, 1963–1969. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.-H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, 2010, baq013. [Google Scholar] [CrossRef]
- Ganesan, S.M.; Joshi, V.; Fellows, M.; Dabdoub, S.M.; Nagaraja, H.N.; O’Donnell, B.; Deshpande, N.R.; Kumar, P.S. A tale of two risks: Smoking, diabetes and the subgingival microbiome. ISME J. 2017, 11, 2075–2089. [Google Scholar] [CrossRef]
- Long, J.; Cai, Q.; Steinwandel, M.; Hargreaves, M.K.; Bordenstein, S.R.; Blot, W.J.; Zheng, W.; Shu, X.O. Association of oral microbiome with type 2 diabetes risk. J. Periodontal Res. 2017, 52, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Kong, C.; Yang, Y.; Cai, S.; Li, X.; Cai, G.; Ma, Y. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics 2020, 10, 11595–11606. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-R.; Xu, Q.; Xiao, J.; Deng, Y.-M.; Tang, Z.-H.; Tang, Y.-L.; Liu, L.S. Role of oral microbiota in atherosclerosis. Clin. Chim. Acta 2020, 506, 191–195. [Google Scholar] [CrossRef]
- Ruff, W.E.; Greiling, T.M.; Kriegel, M.A. Host–microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 2020, 18, 521–538. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, A.; Susin, C.; Gustafsson, A. Smoking and inflammation: Evidence for a synergistic role in chronic disease. Periodontol. 2000 2014, 64, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Bullon, P.; Newman, H.N.; Battino, M. Obesity, diabetes mellitus, atherosclerosis and chronic periodontitis: A shared pathology via oxidative stress and mitochondrial dysfunction? Periodontol. 2000 2014, 64, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Tezal, M.; Sullivan, M.A.; Hyland, A.; Marshall, J.R.; Stoler, D.; Reid, M.E.; Loree, T.R.; Rigual, N.R.; Merzianu, M.; Hauck, L.; et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2406–2412. [Google Scholar] [CrossRef]
- Palefsky, J.M.; Silverman, S.J.; Abdel-Salaam, M.; Daniels, T.E.; Greenspan, J.S. Association between proliferative verrucous leukoplakia and infection with human papillomavirus type 16. J. Oral Pathol. Med. 1995, 24, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Campisi, G.; Giovannelli, L.; Ammatuna, P.; Capra, G.; Colella, G.; Di Liberto, C.; Gandolfo, S.; Pentenero, M.; Carrozzo, M.; Serpico, R.; et al. Proliferative verrucous vs conventional leukoplakia: No significantly increased risk of HPV infection. Oral Oncol. 2004, 40, 835–840. [Google Scholar] [CrossRef]
- García-López, R.; Moya, A.; Bagan, J.V.; Pérez-Brocal, V. Retrospective case-control study of viral pathogen screening in proliferative verrucous leukoplakia lesions. Clin. Otolaryngol. 2014, 39, 272–280. [Google Scholar] [CrossRef]
- Upadhyaya, J.D.; Fitzpatrick, S.G.; Islam, M.N.; Bhattacharyya, I.; Cohen, D.M. A Retrospective 20-Year Analysis of Proliferative Verrucous Leukoplakia and Its Progression to Malignancy and Association with High-risk Human Papillomavirus. Head Neck Pathol. 2018, 12, 500–510. [Google Scholar] [CrossRef]
- Herreros-Pomares, A.; Llorens, C.; Soriano, B.; Zhang, F.; Gallach, S.; Bagan, L.; Murillo, J.; Jantus-Lewintre, E.; Bagan, J. Oral microbiome in Proliferative Verrucous Leukoplakia exhibits loss of diversity and enrichment of pathogens. Oral Oncol. 2021, 120, 105404. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Eslami, H.; Yousefi, M.; Asgharzadeh, M.; Aghazadeh, M.; Kafil, H.S. Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 2016, 84, 552–558. [Google Scholar] [CrossRef]
- Michikawa, C.; Gopalakrishnan, V.; Harrandah, A.M.; Karpinets, T.V.; Garg, R.R.; Chu, R.A.; Park, Y.P.; Chukkapallia, S.S.; Yadlapalli, N.; Erikson-Carter, K.C.; et al. Fusobacterium is enriched in oral cancer and promotes induction of programmed death-ligand 1 (PD-L1). Neoplasia 2022, 31, 100813. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Sun, S.; Yue, J. Decrease of oral microbial diversity might correlate with radiation esophagitis in patients with esophageal cancer undergoing chemoradiation: A pilot study. Precis. Radiat. Oncol. 2020, 4, 81–88. [Google Scholar] [CrossRef]
- Debelius, J.W.; Huang, T.; Cai, Y.; Ploner, A.; Barrett, D.; Zhou, X.; Xiao, X.; Li, Y.; Liao, J.; Zheng, Y.; et al. Subspecies Niche Specialization in the Oral Microbiome Is Associated with Nasopharyngeal Carcinoma Risk. mSystems 2020, 5, e00065-20. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef]
- Ismail, Y.; Mahendran, V.; Octavia, S.; Day, A.S.; Riordan, S.M.; Grimm, M.C.; Lan, R.; Lemberg, D.; Tran, T.A.; Zhang, L. Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS ONE 2012, 7, e38217. [Google Scholar] [CrossRef]
- Poosari, A.; Nutravong, T.; Sa-ngiamwibool, P.; Namwat, W.; Chatrchaiwiwatana, S.; Ungareewittaya, P. Association between infection with Campylobacter species, poor oral health and environmental risk factors on esophageal cancer: A hospital-based case–control study in Thailand. Eur. J. Med. Res. 2021, 26, 82. [Google Scholar] [CrossRef]
- Su, S.-C.; Chang, L.-C.; Huang, H.-D.; Peng, C.-Y.; Chuang, C.-Y.; Chen, Y.-T.; Lu, M.Y.; Chiu, Y.W.; Chen, P.Y.; Yang, S.F. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis 2021, 42, 127–135. [Google Scholar] [CrossRef]
- Amer, A.; Galvin, S.; Healy, C.M.; Moran, G.P. The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front. Microbiol. 2017, 8, 2391. [Google Scholar] [CrossRef]
- He, Z.; Gharaibeh, R.Z.; Newsome, R.C.; Pope, J.L.; Dougherty, M.W.; Tomkovich, S.; Pons, B.; Mirey, G.; Vignard, J.; Hendrixson, D.R.; et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 2019, 68, 289–300. [Google Scholar] [CrossRef]
- Bik, E.M.; Long, C.D.; Armitage, G.C.; Loomer, P.; Emerson, J.; Mongodin, E.F.; Nelson, K.E.; Gill, S.R.; Fraser-Liggett, C.M.; Relman, D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010, 4, 962–974. [Google Scholar] [CrossRef]
- Li, J.; Quinque, D.; Horz, H.-P.; Li, M.; Rzhetskaya, M.; Raff, J.A.; Hayes, M.G.; Stoneking, M. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014, 14, 316. [Google Scholar] [CrossRef]
- Gopinath, D.; Kunnath Menon, R.; Chun Wie, C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; George Botelho, M.; et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome. J. Oral Microbiol. 2020, 13, 1857998. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shen, W.; Wang, J.; Xu, Y.; Zhai, R.; Zhang, J.; Wang, M.; Wang, M.; Liu, L. Capnocytophaga gingivalis is a potential tumor promotor in oral cancer. Oral Dis. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; He, P.; Zhou, M.; Li, S.; Zhang, J.; Tao, X.; Wang, A.; Wu, X. Variations in oral microbiome and its predictive functions between tumorous and healthy individuals. J. Med. Microbiol. 2022, 71, 001568. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; PK, V.P.; Gupta, S.V.; Gupta, S.; Waiker, P.; Samaiya, A.; Sharma, A.K. Assessing the Effect of Smokeless Tobacco Consumption on Oral Microbiome in Healthy and Oral Cancer Patients. Front. Cell. Infect. Microbiol. 2022, 12, 841465. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kwon, E.J.; Yu, Y.; Kim, J.; Woo, S.-Y.; Choi, H.-S.; Kwon, M.; Jung, K.; Kim, H.S.; Park, H.R.; et al. Microbial and molecular differences according to the location of head and neck cancers. Cancer Cell Int. 2022, 22, 135. [Google Scholar] [CrossRef]
- Siqueira, J.F.J.; Rôças, I.N. Catonella morbi and Granulicatella adiacens: New species in endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2006, 102, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zheng, H.J.; Zhang, C.P. The Oral Microbiota May Have Influence on Oral Cancer. Front. Cell. Infect. Microbiol. 2019, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Moritani, K.; Takeshita, T.; Shibata, Y.; Ninomiya, T.; Kiyohara, Y.; Yamashita, Y. Acetaldehyde production by major oral microbes. Oral Dis. 2015, 21, 748–754. [Google Scholar] [CrossRef]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, S.; Takeshita, T.; Takeuchi, K.; Asakawa, M.; Matsumi, R.; Furuta, M.; Shibata, Y.; Nagai, K.; Ikebe, M.; Morita, M.; et al. Characteristics of the Salivary Microbiota in Patients With Various Digestive Tract Cancers. Front. Microbiol. 2019, 10, 1780. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Zhang, H.; Zheng, X.; Wang, J.; Jia, X.; Peng, X.; Xie, Q.; Zou, J.; Zheng, L.; et al. Probiotic Streptococcus salivarius K12 Alleviates Radiation-Induced Oral Mucositis in Mice. Front. Immunol. 2021, 12, 684824. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yeh, Y.-M.; Yu, H.-Y.; Chin, C.-Y.; Hsu, C.-W.; Liu, H.; Huang, P.J.; Hu, S.N.; Liao, C.T.; Chang, K.P.; et al. Oral Microbiota Community Dynamics Associated With Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef]
- Perera, M.; Al-Hebshi, N.N.; Perera, I.; Ipe, D.; Ulett, G.C.; Speicher, D.J.; Chen, T.; Johnson, N.W. Inflammatory Bacteriome and Oral Squamous Cell Carcinoma. J. Dent. Res. 2018, 97, 725–732. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.; Qi, Y.; Wen, X.; Zhang, L. Metagenomic Analysis Reveals a Changing Microbiome Associated With the Depth of Invasion of Oral Squamous Cell Carcinoma. Front. Microbiol. 2022, 13, 795777. [Google Scholar] [CrossRef]
- Mager, D.L.; Haffajee, A.D.; Devlin, P.M.; Norris, C.M.; Posner, M.R.; Goodson, J.M. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Fredricks, D.N.; Schubert, M.M.; Myerson, D. Molecular identification of an invasive gingival bacterial community. Clin. Infect. Dis. 2005, 41, e1–e4. [Google Scholar] [CrossRef] [PubMed]
- Funada, H.; Machi, T.; Yoneyama, H.; Matsuda, T.; Miura, H.; Ezaki, T.; Yokota, Y. Capnocytophaga sputigena bacteremia associated with acute leukemia. Kansenshogaku zasshi. J. Jpn. Assoc. Infect. Dis. 1993, 67, 622–628. [Google Scholar]
- Mizuki, H.; Abe, R.; Mikami, T. Ultrastructural Changes during the Life Cycle of Mycoplasma salivarium in Oral Biopsies from Patients with Oral Leukoplakia. Front. Cell. Infect. Microbiol. 2017, 7, 403. [Google Scholar] [CrossRef]
- Henrich, B.; Rumming, M.; Sczyrba, A.; Velleuer, E.; Dietrich, R.; Gerlach, W.; Gombert, M.; Rahn, S.; Stoye, J.; Borkhardt, A.; et al. Mycoplasma salivarium as a dominant coloniser of Fanconi anaemia associated oral carcinoma. PLoS ONE 2014, 9, e92297. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Panda, M.; Das, A.K.; Rahman, T.; Das, R.; Das, K.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch. Microbiol. 2021, 203, 137–152. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).