Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of Physicochemical Properties of MADS-Box Proteins
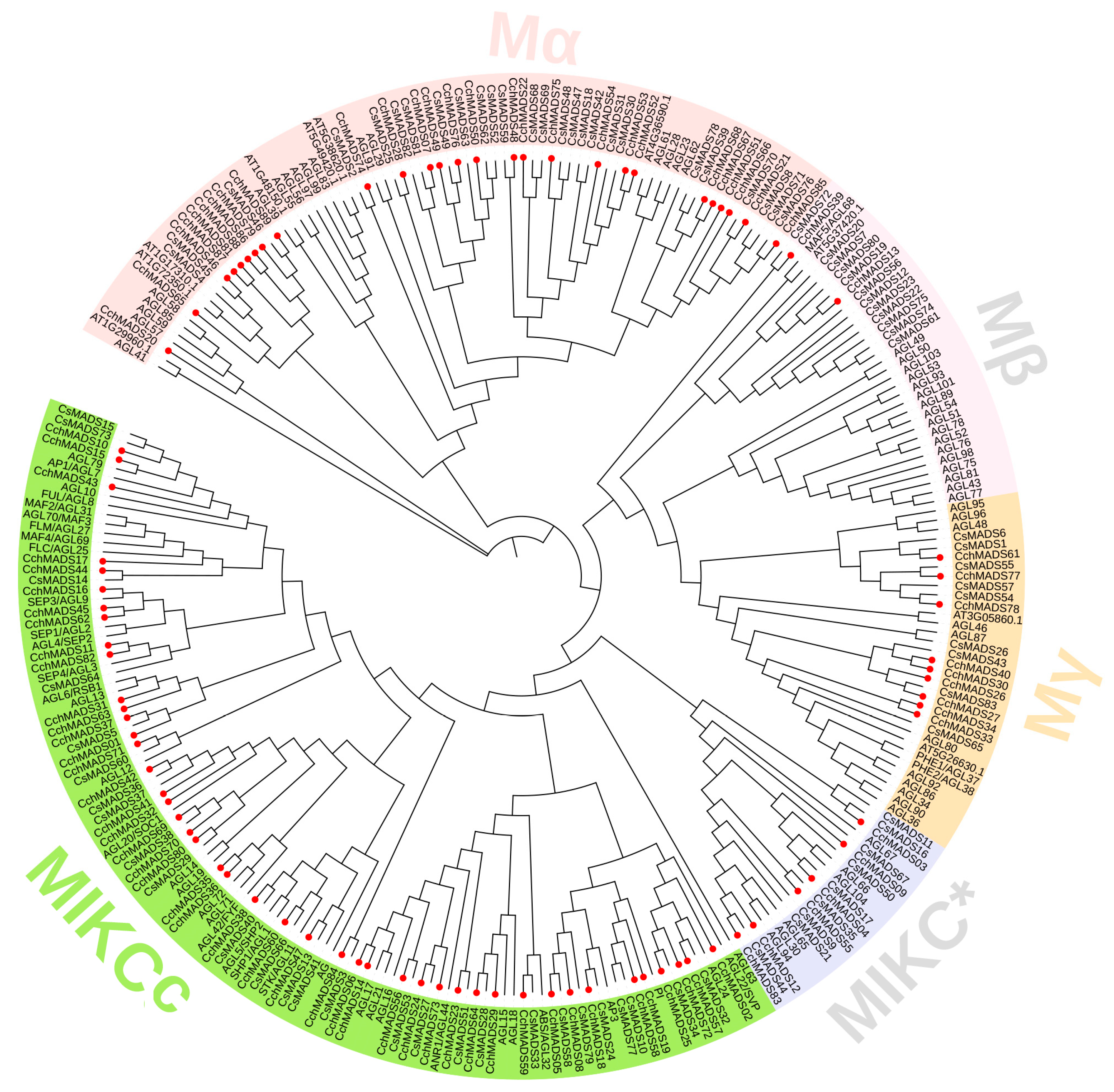
2.2. Phylogenetic Analysis of CchMADS Proteins
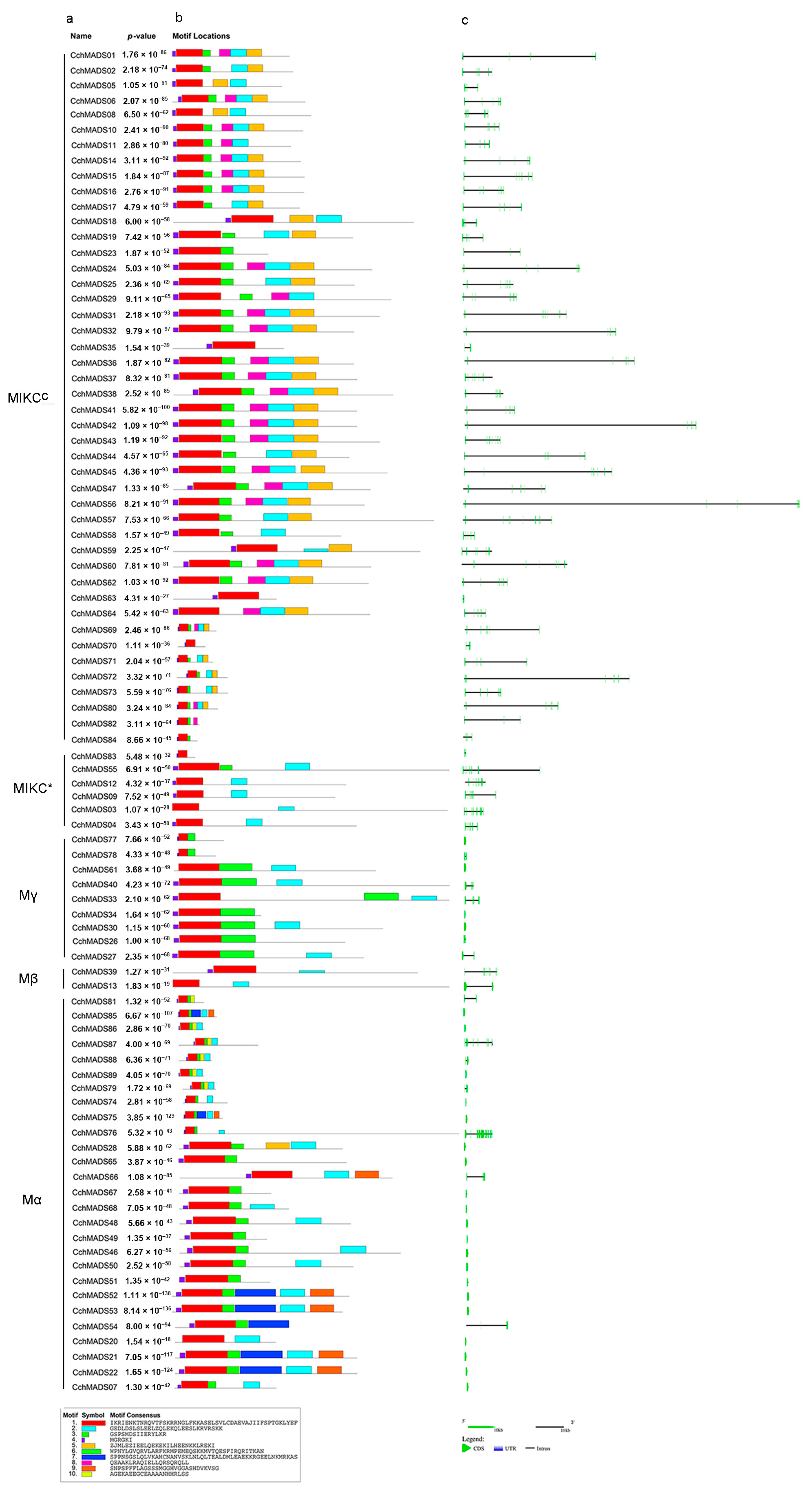
2.3. Gene Structure and Motif Analysis
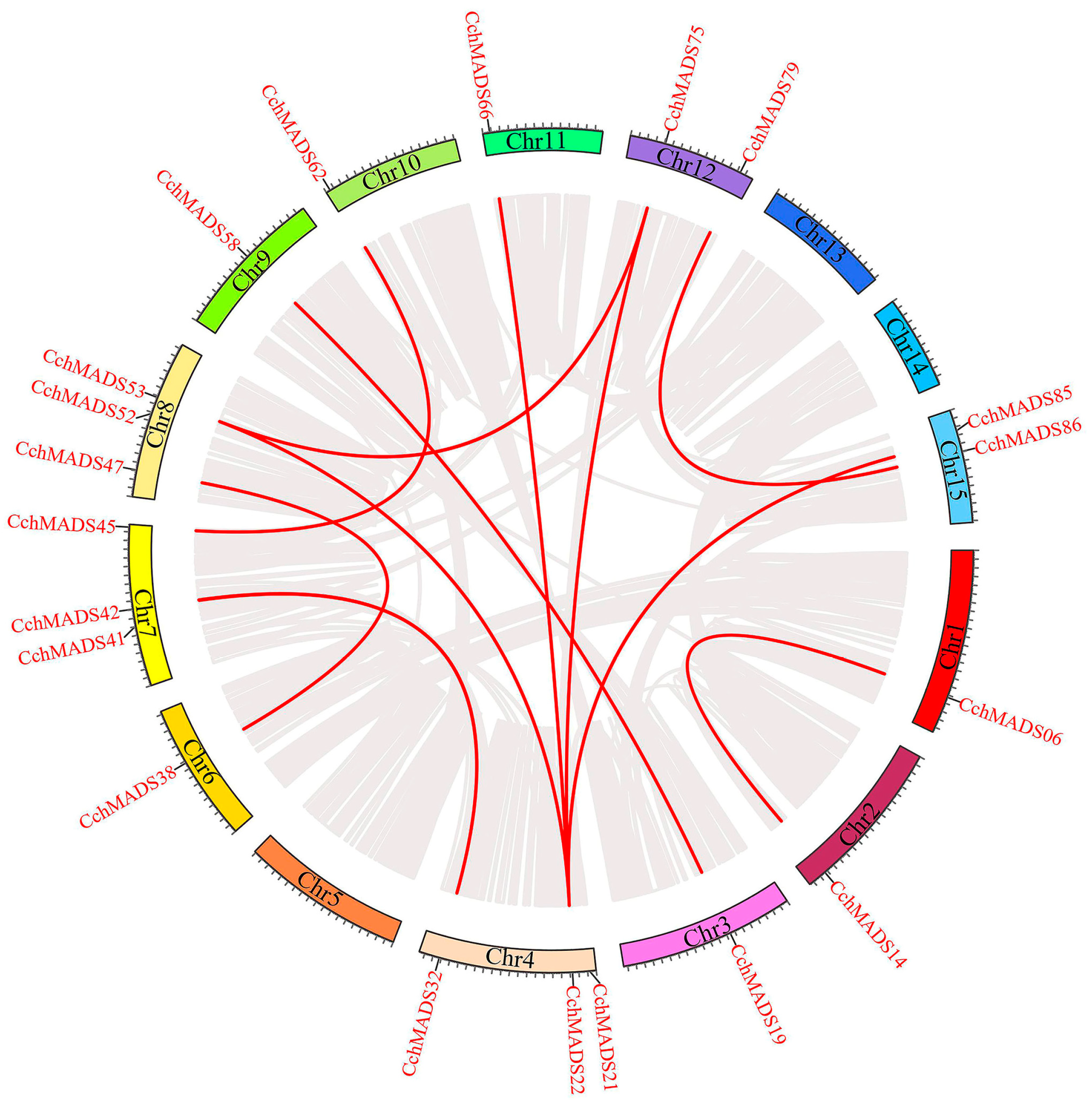
2.4. Chromosomal Localization and Duplication of CchMADS Genes
2.5. Cis-Acting Elements of MADS-Box Gene Family-Associated Promoters
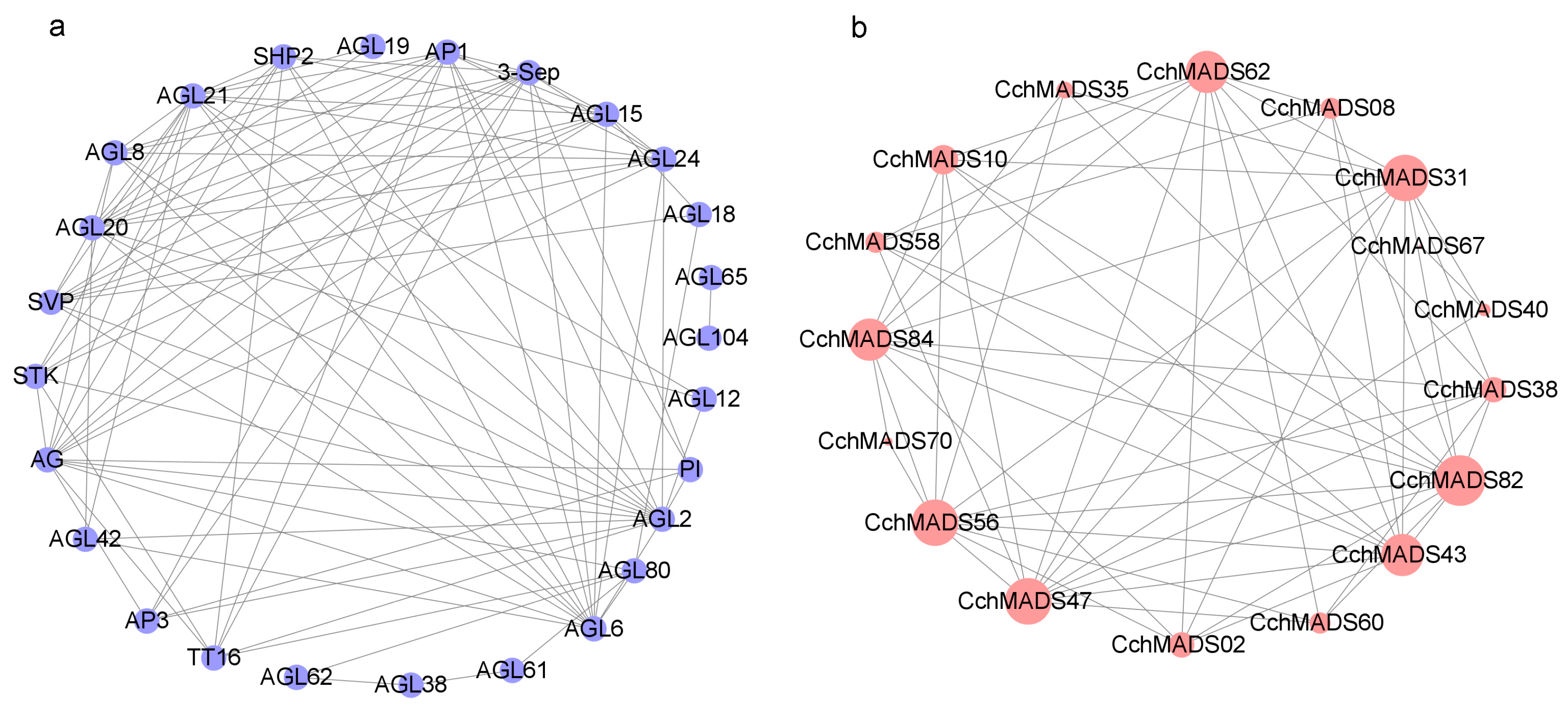
2.6. Protein–Protein Interaction Network of CchMADSs
2.7. Expression of CchMADS Genes in Different Tissues
3. Discussion
3.1. Number and Characteristics of CchMADS Genes
3.2. Expansion of the CchMADS Gene Family
3.3. Intron Specificity of CchMADS Genes
3.4. Gene Expression and Potential Function
4. Materials and Methods
4.1. Identification of C. chekiangoleosa MADS-Box Genes
4.2. Multiple Alignment and Phylogenetic Analysis
4.3. Chromosomal Localization and Gene Structure Analysis
4.4. Gene Duplication and Promoter Cis-Acting Regulatory Element Analysis
4.5. Protein–Protein Interaction Network Analysis
4.6. Plant Materials and Expression Analysis in Different Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, W.; Xiao, X.; Shen, J.; Wen, Q.; Li, T.; Zhu, H.; Yang, J.; Xun, C. Review and Breeding Strategy of Camellia chekiangoleosa. Germplasm Resour. 2019, 47, 20–24. [Google Scholar] [CrossRef]
- Wen, Q.; Li, T.; Yang, J.; Li, X.; Zhu, H.; Zhu, W.; Ye, J.; Xu, L. Discovery and Molecular Research on White Flower Variation of Camellia chekiangoleosa in Jiangxi Province. South For. Sci. 2018, 46, 1–6. [Google Scholar] [CrossRef]
- Tian, Q.; Huang, J.; Wen, Q.; Zhou, W.; Huang, B.; Gong, C.; Xu, C. Current Situation and Trend of Molecular Breeding of Camellia. South For. Sci. 2021, 49, 53–58. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Z.; Dong, L.; Wen, Q.; Huang, W.; Li, T.; Ye, J.; Xu, L. Analysis on the character diversity of fruit and seed of Camellia chekiangoleosa. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2021, 45, 51–59. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.; Huijser, P.; Pape, H.; Lönnig, W.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Boil. 2010, 11, 214. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Gramzow, L.; Theißen, G. Phylogenomics reveals surprising sets of essential and dispensable clades of MIKCc-group MADS-box genes in flowering plants. J. Exp. Zool. Part B 2015, 324, 353–362. [Google Scholar] [CrossRef]
- Grimplet, J.; Martinez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef]
- Weigel, D.; Meyerowitz, E.M. The ABCs of floral homeotic genes. Cell 1994, 78, 203–209. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.-L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.B.; Ryder, C.D.; Cevik, V.; Hammond, J.P.; Popovich, A.; King, G.J.; Vrebalov, J.; Giovannoni, J.J.; Manning, K. SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria× ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 2011, 62, 1179–1188. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef]
- Huo, S.; Li, Y.; Li, R.; Chen, R.; Xing, H.; Wang, J.; Zhao, Y.; Song, X. Genome-wide analysis of the MADS-box gene family in Rhododendron hainanense Merr. and expression analysis under heat and waterlogging stresses. Ind. Crops Prod. 2021, 172, 114007. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, L.-L.; Xu, Z.-S.; Fu, L.; Pang, H.-X.; Ma, Y.-Z.; Min, D.-H. Genome-Wide Analysis of MADS-Box Genes in Foxtail Millet (Setaria italica L.) and Functional Assessment of the Role of SiMADS51 in the Drought Stress Response. Front. Plant Sci. 2021, 12, 659474. [Google Scholar] [CrossRef]
- Zhang, Z.-B.; Jin, Y.-J.; Wan, H.-H.; Cheng, L.; Feng, Z.-G. Genome-wide identification and expression analysis of the MADS-box transcription factor family in Camellia sinensis. J. Appl. Genet. 2021, 62, 249–264. [Google Scholar] [CrossRef]
- Qu, Y.; Bi, C.; He, B.; Ye, N.; Yin, T.; Xu, L.-A. Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis. PeerJ 2019, 7, e8019. [Google Scholar] [CrossRef]
- Wei, X.; Wang, L.; Yu, J.; Zhang, Y.; Li, D.; Zhang, X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 2015, 569, 66–76. [Google Scholar] [CrossRef]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings tothe MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Leseberg, C.H.; Li, A.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef]
- Puig, J.; Meynard, D.; Khong, G.N.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns 2013, 13, 160–170. [Google Scholar] [CrossRef]
- Moon, J.; Suh, S.-S.; Lee, H.; Choi, K.-R.; Hong, C.B.; Paek, N.-C.; Kim, S.-G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef]
- de Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.H.; Francisca, A.; Soraya, P.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-wide analysis of the MADS-box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Airoldi, C.A.; Davies, B. Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genom. 2012, 39, 157–165. [Google Scholar] [CrossRef]
- Lee, S.; Woo, Y.M.; Ryu, S.; Shin, Y.D.; Woo, T.K.; Ky, Y.P.; Lee, I.J.; An, G. Further characterization of a rice AGL12 group MADS-box gene, OsMADS26. Plant Physiol. 2008, 147, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Bioinf. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Mehan, M.R.; Freimer, N.B.; Ophoff, R.A. A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum. Genom. 2004, 1, 335–344. [Google Scholar] [CrossRef]
- Wu, J.; Xiao, J.; Wang, L.; Zhong, J.; Yin, H.; Wu, S.; Zhang, Z.; Yu, J. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci. China Life Sci. 2013, 56, 968–974. [Google Scholar] [PubMed]
- Yang, Y.F.; Zhu, T.; Niu, D.K. Association of intron loss with high mutation rate in Arabidopsis: Implications for genome size evolution. Genome Biol. Evol. 2013, 5, 723–733. [Google Scholar] [CrossRef]
- Hong, X.; Scofield, D.G.; Lynch, M. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006, 23, 2392–2404. [Google Scholar] [CrossRef]
- Korb, M.; Ke, Y.; Johnson, L.F. Stimulation of gene expression by introns: Conversion of an inhibitory intron to a stimulatory intron by alteration of the splice donor sequence. Nucleic Acids Res. 1993, 21, 5901–5908. [Google Scholar] [CrossRef]
- Vain, P.; Finer, K.R.; Engler, D.E.; Pratt, R.C.; Finer, J.J. Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep. 1996, 15, 489–494. [Google Scholar] [CrossRef]
- Wang, L.; Fan, S.L.; Song, M.Z. Advances in the Research of MADS-box Gene in Plant. Biotechnol. Bull. 2010, 8, 12–19. [Google Scholar]
- Dreni, L.; Zhang, D. Flower development: The evolutionary history and functions of the AGL6 subfamily MADS-box genes. J. Exp. Bot. 2016, 67, 1625–1638. [Google Scholar] [CrossRef]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef]
- Adamczyk, B.J.; Fernandez, D.E. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009, 149, 1713–1723. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Schuster-Böckler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.P.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 1, gkz1020. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


| Gene Name | Sequence ID | Location | Length (aa) | MW (kDa) | pI | Subcellular Localization | Type | Intron |
|---|---|---|---|---|---|---|---|---|
| CchMADS01 | Cole01G001968.1 | Chr1 | 222 | 25.823 | 6.47 | Nucleus | MIKCC | 6 |
| CchMADS02 | Cole01G003080.1 | Chr1 | 229 | 25.997 | 5.75 | Nucleus | MIKCC | 7 |
| CchMADS03 | Cole01G003276.1 | Chr1 | 522 | 58.723 | 5.46 | Nucleus | MIKC* | 11 |
| CchMADS04 | Cole01G003279.1 | Chr1 | 349 | 39.503 | 4.96 | Nucleus | MIKC* | 10 |
| CchMADS05 | Cole01G003924.1 | Chr1 | 208 | 24.266 | 9.48 | Nucleus | MIKCC | 6 |
| CchMADS06 | Cole01G004072.1 | Chr1 | 252 | 29.436 | 8.76 | Nucleus | MIKCC | 6 |
| CchMADS07 | Cole01G004142.1 | Chr1 | 193 | 21.333 | 9.37 | Nucleus | Mα | 0 |
| CchMADS08 | Cole02G001598.1 | Chr2 | 263 | 29.794 | 9.01 | Nucleus | MIKCC | 5 |
| CchMADS09 | Cole02G002131.1 | Chr2 | 307 | 34.614 | 5.54 | Nucleus | MIKC* | 8 |
| CchMADS10 | Cole02G002392.1 | Chr2 | 246 | 28.233 | 8.93 | Nucleus | MIKCC | 7 |
| CchMADS11 | Cole02G002395.1 | Chr2 | 223 | 25.637 | 9 | Nucleus | MIKCC | 4 |
| CchMADS12 | Cole02G002503.1 | Chr2 | 329 | 37.934 | 6.64 | Nucleus | MIKC* | 8 |
| CchMADS13 | Cole02G003948.1 | Chr2 | 524 | 59.964 | 6.19 | Nucleus | Mβ | 1 |
| CchMADS14 | Cole02G004095.1 | Chr2 | 242 | 28.203 | 8.44 | Nucleus | MIKCC | 6 |
| CchMADS15 | Cole02G004285.1 | Chr2 | 249 | 28.652 | 9.02 | Nucleus | MIKCC | 7 |
| CchMADS16 | Cole02G004286.1 | Chr2 | 248 | 29.003 | 7.08 | Nucleus | MIKCC | 7 |
| CchMADS17 | Cole02G004715.1 | Chr2 | 240 | 27.737 | 7.75 | Nucleus | MIKCC | 6 |
| CchMADS18 | Cole03G000406.1 | Chr3 | 289 | 33.388 | 8.89 | Nucleus | MIKCC | 6 |
| CchMADS19 | Cole03G001914.1 | Chr3 | 216 | 25.379 | 6.77 | Nucleus | MIKCC | 6 |
| CchMADS20 | Cole04G000929.1 | Chr4 | 121 | 13.591 | 9.83 | Nucleus | Mα | 0 |
| CchMADS21 | Cole04G000931.1 | Chr4 | 219 | 24.905 | 9.49 | Nucleus | Mα | 0 |
| CchMADS22 | Cole04G000947.1 | Chr4 | 219 | 24.247 | 9.08 | Nucleus | Mα | 0 |
| CchMADS23 | Cole04G001061.1 | Chr4 | 114 | 12.833 | 9.61 | Nucleus | MIKCC | 2 |
| CchMADS24 | Cole04G001062.1 | Chr4 | 239 | 27.475 | 6.41 | Nucleus | MIKCC | 7 |
| CchMADS25 | Cole04G001125.1 | Chr4 | 218 | 25.004 | 8.61 | Nucleus | MIKCC | 7 |
| CchMADS26 | Cole04G001870.1 | Chr4 | 206 | 24.217 | 9.2 | Nucleus | Mγ | 1 |
| CchMADS27 | Cole04G001878.1 | Chr4 | 230 | 26.646 | 9.55 | Nucleus | Mγ | 1 |
| CchMADS28 | Cole04G002096.1 | Chr4 | 197 | 22.197 | 7.69 | Nucleus | Mα | 0 |
| CchMADS29 | Cole04G003100.1 | Chr4 | 262 | 30.047 | 6.13 | Nucleus | MIKCC | 7 |
| CchMADS30 | Cole04G003823.1 | Chr4 | 252 | 28.918 | 9.77 | Nucleus | Mγ | 0 |
| CchMADS31 | Cole04G004469.1 | Chr4 | 248 | 28.181 | 8.83 | Nucleus | MIKCC | 7 |
| CchMADS32 | Cole04G004470.1 | Chr4 | 217 | 24.824 | 6.47 | Nucleus | MIKCC | 6 |
| CchMADS33 | Cole05G000003.1 | Chr5 | 332 | 38.973 | 10.14 | Nucleus | Mγ | 4 |
| CchMADS34 | Cole05G000013.1 | Chr5 | 106 | 12.359 | 10.12 | Nucleus | Mγ | 0 |
| CchMADS35 | Cole06G000488.1 | Chr6 | 131 | 14.624 | 9.16 | Nucleus | MIKCC | 2 |
| CchMADS36 | Cole06G000572.1 | Chr6 | 213 | 24.628 | 8.8 | Nucleus | MIKCC | 6 |
| CchMADS37 | Cole06G000576.1 | Chr6 | 217 | 24.795 | 7.05 | Nucleus | MIKCC | 7 |
| CchMADS38 | Cole06G002198.1 | Chr6 | 259 | 29.666 | 9.2 | Nucleus | MIKCC | 8 |
| CchMADS39 | Cole06G002899.1 | Chr6 | 289 | 32.705 | 8.65 | Nucleus | Mβ | 10 |
| CchMADS40 | Cole07G000640.1 | Chr7 | 326 | 37.273 | 9.82 | Nucleus | Mγ | 2 |
| CchMADS41 | Cole07G001913.1 | Chr7 | 217 | 25.053 | 6.39 | Nucleus | MIKCC | 6 |
| CchMADS42 | Cole07G001920.1 | Chr7 | 217 | 25.042 | 6.18 | Nucleus | MIKCC | 6 |
| CchMADS43 | Cole07G004098.1 | Chr7 | 244 | 28.547 | 8.44 | Nucleus | MIKCC | 7 |
| CchMADS44 | Cole07G004743.1 | Chr7 | 208 | 23.709 | 9.49 | Nucleus | MIKCC | 6 |
| CchMADS45 | Cole07G004746.1 | Chr7 | 253 | 28.615 | 8.74 | Nucleus | MIKCC | 7 |
| CchMADS46 | Cole08G000724.1 | Chr8 | 261 | 28.432 | 6.76 | Nucleus | Mα | 0 |
| CchMADS47 | Cole08G000781.1 | Chr8 | 233 | 26.711 | 9.78 | Nucleus | MIKCC | 8 |
| CchMADS48 | Cole08G002218.1 | Chr8 | 202 | 22.749 | 8.45 | Nucleus | Mα | 0 |
| CchMADS49 | Cole08G002233.1 | Chr8 | 103 | 11.521 | 9.56 | Nucleus | Mα | 0 |
| CchMADS50 | Cole08G002238.1 | Chr8 | 205 | 23.551 | 6.72 | Nucleus | Mα | 0 |
| CchMADS51 | Cole08G002611.1 | Chr8 | 107 | 11.853 | 9.96 | Nucleus | Mα | 1 |
| CchMADS52 | Cole08G002612.1 | Chr8 | 219 | 24.053 | 9.32 | Nucleus | Mα | 0 |
| CchMADS53 | Cole08G002613.1 | Chr8 | 211 | 23.132 | 9.12 | Nucleus | Mα | 0 |
| CchMADS54 | Cole08G002614.1 | Chr8 | 141 | 15.725 | 9.94 | Nucleus | Mα | 1 |
| CchMADS55 | Cole09G000962.1 | Chr9 | 342 | 38.905 | 5.65 | Nucleus | MIKC* | 10 |
| CchMADS56 | Cole09G001241.1 | Chr9 | 237 | 27.304 | 9.38 | Nucleus | MIKCC | 6 |
| CchMADS57 | Cole09G001278.1 | Chr9 | 323 | 35.838 | 6.85 | Nucleus | MIKCC | 9 |
| CchMADS58 | Cole09G001941.1 | Chr9 | 208 | 24.372 | 9.11 | Nucleus | MIKCC | 6 |
| CchMADS59 | Cole09G001965.1 | Chr9 | 306 | 35.993 | 9.35 | Nucleus | MIKCC | 9 |
| CchMADS60 | Cole09G002445.1 | Chr9 | 245 | 28.275 | 9.22 | Nucleus | MIKCC | 9 |
| CchMADS61 | Cole09G004317.1 | Chr9 | 250 | 28.592 | 4.92 | Nucleus | Mγ | 0 |
| CchMADS62 | Cole10G000146.1 | Chr10 | 242 | 27.683 | 8.96 | Nucleus | MIKCC | 7 |
| CchMADS63 | Cole10G001610.1 | Chr10 | 128 | 14.444 | 9.9 | Nucleus | MIKCC | 1 |
| CchMADS64 | Cole10G003622.1 | Chr10 | 244 | 27.614 | 8.24 | Nucleus | MIKCC | 7 |
| CchMADS65 | Cole11G000155.1 | Chr11 | 208 | 23.141 | 5.22 | Nucleus | Mα | 0 |
| CchMADS66 | Cole11G000294.1 | Chr11 | 263 | 30.297 | 9.35 | Nucleus | Mα | 2 |
| CchMADS67 | Cole11G000296.1 | Chr11 | 114 | 12.381 | 10.48 | Nucleus | Mα | 1 |
| CchMADS68 | Cole11G000299.1 | Chr11 | 136 | 15.199 | 7.92 | Nucleus | Mα | 1 |
| CchMADS69 | Cole11G000477.1 | Chr11 | 213 | 24.686 | 7.66 | Nucleus | MIKCC | 6 |
| CchMADS70 | Cole11G003071.1 | Chr11 | 152 | 17.325 | 9.32 | Nucleus | MIKCC | 2 |
| CchMADS71 | Cole12G000125.1 | Chr12 | 199 | 22.838 | 7.02 | Nucleus | MIKCC | 6 |
| CchMADS72 | Cole12G000456.1 | Chr12 | 279 | 31.438 | 9 | Nucleus | MIKCC | 8 |
| CchMADS73 | Cole12G000684.1 | Chr12 | 282 | 32.291 | 8.63 | Nucleus | MIKCC | 7 |
| CchMADS74 | Cole12G000760.2 | Chr12 | 246 | 26.972 | 6.54 | Nucleus | Mα | 0 |
| CchMADS75 | Cole12G001299.1 | Chr12 | 219 | 24.101 | 9.25 | Nucleus | Mα | 0 |
| CchMADS76 | Cole12G001525.1 | Chr12 | 1528 | 176.716 | 9.12 | Nucleus | Mα | 12 |
| CchMADS77 | Cole12G002412.1 | Chr12 | 257 | 29.344 | 5.7 | Nucleus | Mγ | 0 |
| CchMADS78 | Cole12G002414.1 | Chr12 | 213 | 24.174 | 5.3 | Nucleus | Mγ | 1 |
| CchMADS79 | Cole12G003350.1 | Chr12 | 184 | 20.516 | 6.45 | Nucleus | Mα | 1 |
| CchMADS80 | Cole13G001797.1 | Chr13 | 226 | 25.852 | 6.09 | Nucleus | MIKCC | 6 |
| CchMADS81 | Cole13G003210.1 | Chr13 | 149 | 17.171 | 10.44 | Nucleus | Mα | 1 |
| CchMADS82 | Cole14G000617.1 | Chr14 | 124 | 14.314 | 9.67 | Nucleus | MIKCC | 3 |
| CchMADS83 | Cole14G000851.1 | Chr14 | 102 | 12.018 | 9.78 | Nucleus | MIKC* | 1 |
| CchMADS84 | Cole14G001528.1 | Chr14 | 113 | 13.234 | 9.84 | Nucleus | MIKCC | 2 |
| CchMADS85 | Cole15G000774.1 | Chr15 | 219 | 24.682 | 9.72 | Nucleus | Mα | 0 |
| CchMADS86 | Cole15G000995.1 | Chr15 | 141 | 15.648 | 6.32 | Nucleus | Mα | 0 |
| CchMADS87 | Cole00G064455.1 | ChrN | 439 | 48.72 | 5.9 | Nucleus | Mα | 7 |
| CchMADS88 | Cole00G064458.1 | ChrN | 184 | 20.505 | 5.75 | Nucleus | Mα | 1 |
| CchMADS89 | Cole00G064466.1 | ChrN | 141 | 15.536 | 6.32 | Nucleus | Mα | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, P.; Qu, Y.; Wang, Z.; Huang, B.; Wen, Q.; Xin, Y.; Ni, Z.; Xu, L. Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa. Int. J. Mol. Sci. 2023, 24, 3434. https://doi.org/10.3390/ijms24043434
Zhou P, Qu Y, Wang Z, Huang B, Wen Q, Xin Y, Ni Z, Xu L. Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa. International Journal of Molecular Sciences. 2023; 24(4):3434. https://doi.org/10.3390/ijms24043434
Chicago/Turabian StyleZhou, Pengyan, Yanshu Qu, Zhongwei Wang, Bin Huang, Qiang Wen, Yue Xin, Zhouxian Ni, and Li’an Xu. 2023. "Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa" International Journal of Molecular Sciences 24, no. 4: 3434. https://doi.org/10.3390/ijms24043434
APA StyleZhou, P., Qu, Y., Wang, Z., Huang, B., Wen, Q., Xin, Y., Ni, Z., & Xu, L. (2023). Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa. International Journal of Molecular Sciences, 24(4), 3434. https://doi.org/10.3390/ijms24043434







