IKAROS in Acute Leukemia: A Positive Influencer or a Mean Hater?
Abstract
1. Introduction
2. Genetic, Expression and Post-Translational Modification: A “Bio” of IKAROS
2.1. Genetic and Structure of IKZF1
2.2. Expression of IKZF1
2.3. Post-Translational Modification of IKAROS
3. IKAROS Is “LinkedIn” in Many Biological Pathways
3.1. Lymphoid Landscape
3.2. Myeloid Landscape
4. IKZF1 Dysregulation in ALL: A Mean “Hater”
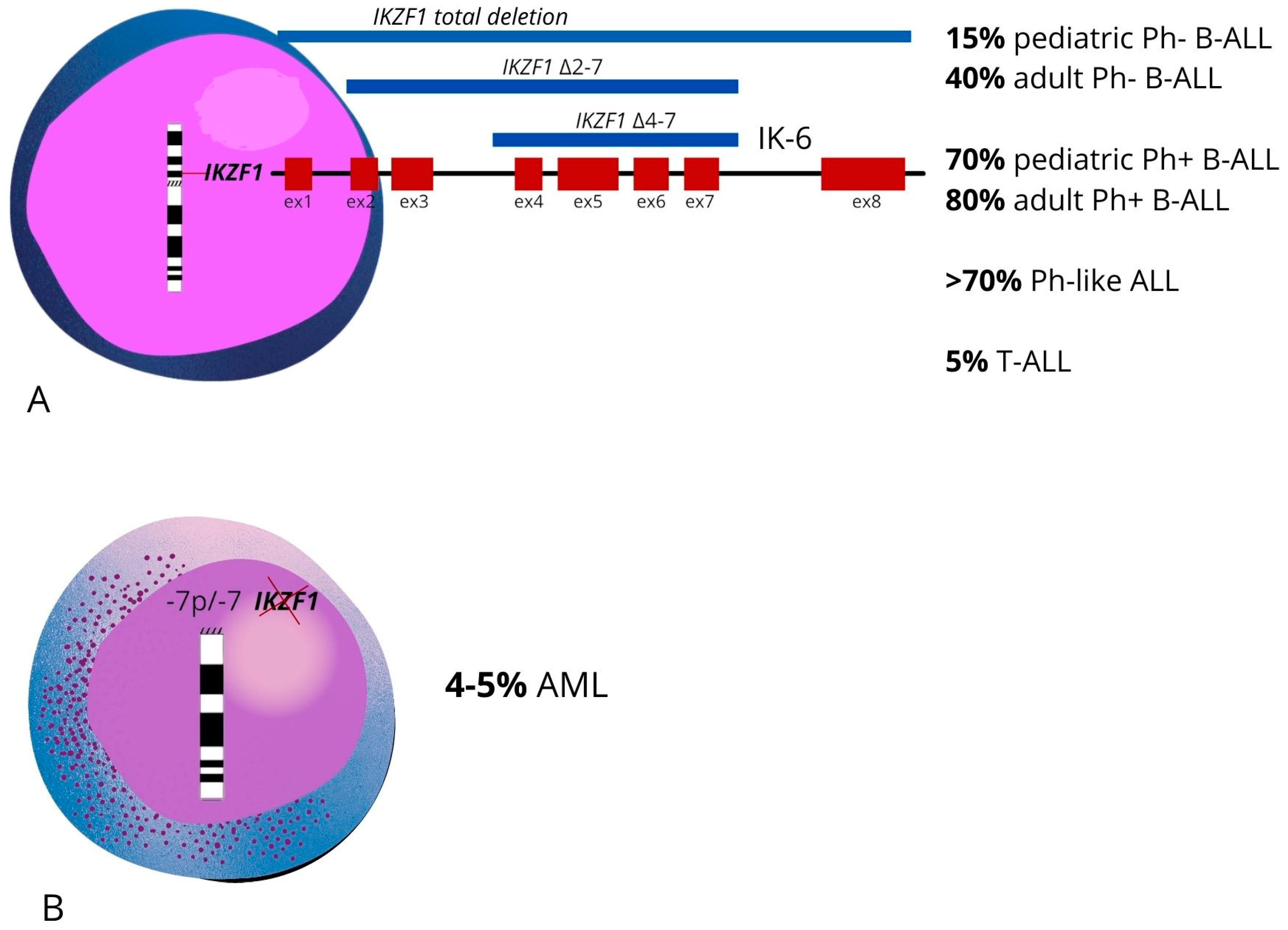
4.1. Deletions of IKZF1 Exons
4.2. Epistatic Effects of IKZF1 Deletion Impact All Patients’ Prognosis
4.3. Other IKZF1 Alterations
5. Loss of Function of IKAROS in ALL: What’s Up?
5.1. Cellular Proliferation Pathways
5.2. Cell Commitment: Pre-B-Cell Receptor Signaling and B/T-Cell Differentiation
5.3. Daily Cellular Occupations: Adhesion and Metabolism Activities
5.4. Cell Chatting: Signal Transducer and Cell Surface Receptors
5.5. DNA Makeover: Epigenetic Signaling
5.6. IKAROS Remodelling: IKAROS/CK2/PP1 Axis
6. IKAROS in AML: A “Tweet” of Interest
6.1. IKZF1: An Emerging Character in the Pediatric AML Scenario
6.2. IKZF1 Loss in AML: Is It Only a Consequence of Cytogenetic Alterations?
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winandy, S.; Wu, P.; Georgopoulos, K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 1995, 83, 289–299. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Downing, J.R. Global Genomic Characterization of Acute Lymphoblastic Leukemia. Semin. Hematol. 2009, 46, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Li, X.; Lv, Y.; Zhu, Y.; Wang, J.; Jin, J.; Yu, W. The specific distribution pattern of IKZF1 mutation in acute myeloid leukemia. J. Hematol. Oncol. 2020, 13, 140. [Google Scholar] [CrossRef]
- Lo, K.; Landau, N.R.; Smale, S.T. LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specific genes. Mol. Cell. Biol. 1991, 11, 5229–5243. [Google Scholar] [CrossRef] [PubMed]
- Georgopoulos, K.; Moore, D.D.; Derfler, B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 1992, 258, 808–812. [Google Scholar] [CrossRef]
- Hahm, K.; Ernst, P.; Lo, K.; Kim, G.S.; Turck, C.; Smalel, S.T. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol. 1994, 14, 7111. [Google Scholar] [CrossRef] [PubMed]
- John, L.B.; Ward, A.C. The Ikaros gene family: Transcriptional regulators of hematopoiesis and immunity. Mol. Immunol. 2011, 48, 1272–1278. [Google Scholar] [CrossRef]
- Georgopoulos, K.; Winandy, S.; Avitahl, N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997, 15, 155–176. [Google Scholar] [CrossRef]
- McCarty, A.S.; Kleiger, G.; Eisenberg, D.; Smale, S.T. Selective dimerization of a C2H2 zinc finger subfamily. Mol. Cell 2003, 11, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Miller, C.B.; Radtke, I.; Phillips, L.A.; Dalton, J.; Ma, J.; White, D.; Hughes, T.P.; Le Beau, M.M.; Pui, C.H.; et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008, 453, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Su, X.; Zhang, J.; Radtke, I.; Phillips, L.A.A.; Miller, C.B.; Ma, J.; Liu, W.; Cheng, C.; Schulman, B.A.; et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2009, 360, 470–480. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Phillips, L.A.; Su, X.; Ma, J.; Miller, C.B.; Shurtleff, S.A.; Downing, J.R. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008, 322, 1377–1380. [Google Scholar] [CrossRef]
- Caye, A.; Beldjord, K.; Mass-Malo, K.; Drunat, S.; Soulier, J.; Gandemer, V.; Baruchel, A.; Bertrand, Y.; Cavé, H.; Clappier, E. Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica 2013, 98, 597–601. [Google Scholar] [CrossRef]
- Schwab, C.J.; Chilton, L.; Morrison, H.; Jones, L.; Al-Shehhi, H.; Erhorn, A.; Russell, L.J.; Moorman, A.V.; Harrison, C.J. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: Association with cytogenetics and clinical features. Haematologica 2013, 98, 1081–1088. [Google Scholar] [CrossRef]
- Boer, J.M.; Van Der Veer, A.; Rizopoulos, D.; Fiocco, M.; Sonneveld, E.; De Groot-Kruseman, H.A.; Kuiper, R.P.; Hoogerbrugge, P.; Horstmann, M.; Zaliova, M.; et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: An international collaborative study. Leukemia 2016, 30, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Molnar, A.; Georgopoulos, K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 1994, 14, 8292–8303. [Google Scholar] [CrossRef]
- Sun, L.; Liu, A.; Georgopoulos, K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996, 15, 5358. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.M.; Chen, C.; Chunder, N.; Ma, L.; Taylor, J.; Pearce, E.J.; Wells, A.D. Ikaros Silences T-bet Expression and Interferon-γ Production during T Helper 2 Differentiation. J. Biol. Chem. 2010, 285, 2545–2553. [Google Scholar] [CrossRef]
- Heizmann, B.; Sellars, M.; Macias-Garcia, A.; Chan, S.; Kastner, P. Ikaros limits follicular B cell activation by regulating B cell receptor signaling pathways. Biochem. Biophys. Res. Commun. 2016, 470, 714–720. [Google Scholar] [CrossRef]
- Dumortier, A.; Kirstetter, P.; Kastner, P.; Chan, S. Ikaros regulates neutrophil differentiation. Blood 2003, 101, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.N.; Smuda, C.; Gregory, G.D.; Min, B.; Brown, M.A. Ikaros limits basophil development by suppressing C/EBP-α expression. Blood 2013, 122, 2572–2581. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Li, Z.; Erbe, A.K.; Savic, A.; Dovat, S. Regulation of Ikaros function by casein kinase 2 and protein phosphatase 1. World J. Biol. Chem. 2011, 2, 126. [Google Scholar] [CrossRef]
- Popescu, M.; Gurel, Z.; Ronni, T.; Song, C.; Hung, K.Y.; Payne, K.J.; Dovat, S. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J. Biol. Chem. 2009, 284, 13869–13880. [Google Scholar] [CrossRef]
- Lindner, S.; Krönke, J. The molecular mechanism of thalidomide analogs in hematologic malignancies. J. Mol. Med. 2016, 94, 1327–1334. [Google Scholar] [CrossRef]
- Arco, P.G.; Maki, K.; Georgopoulos, K. Phosphorylation Controls Ikaros’s Ability To Negatively Regulate the G1-S Transition. Mol. Cell. Biol. 2004, 24, 2797. [Google Scholar] [CrossRef] [PubMed]
- Apostolov, A.; Litim-Mecheri, I.; Oravecz, A.; Goepp, M.; Kirstetter, P.; Marcha, P.; Ittel, A.; Mauvieux, L.; Chan, S.; Kastner, P. Sumoylation Inhibits the Growth Suppressive Properties of Ikaros. PLoS ONE 2016, 11, e0157767. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Qazi, S.; Ozer, Z.; Zhang, J.; Ishkhanian, R.; Uckun, F.M. Regulatory Phosphorylation of Ikaros by Bruton’s Tyrosine Kinase. PLoS ONE 2013, 8, 71302. [Google Scholar] [CrossRef]
- Georgopoulos, K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2002, 2, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, I.; Piechoki, M.; Liu, G.; Ratner, S.; Galy, A. Regulation of L-Selectin Expression by a Dominant Negative Ikaros Protein. Available online: https://pubmed.ncbi.nlm.nih.gov/11310856/ (accessed on 25 November 2022).
- Lopez, R.A.; Schoetz, S.; DeAngelis, K.; O’Neill, D.; Bank, A. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc. Natl. Acad. Sci. USA 2002, 99, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Dhanyamraju, P.K.; Iyer, S.; Smink, G.; Bamme, Y.; Bhadauria, P.; Payne, J.L.; Dovat, E.; Klink, M.; Ding, Y. Transcriptional regulation of genes by ikaros tumor suppressor in acute lymphoblastic leukemia. Int. J. Mol. Sci. 2020, 21, 1377. [Google Scholar] [CrossRef]
- Payne, J.L.; Song, C.; Ding, Y.; Dhanyamraju, P.K.; Bamme, Y.; Schramm, J.W.; Desai, D.; Sharma, A.; Gowda, C.; Dovat, S. Regulation of Small GTPase Rab20 by Ikaros in B-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1718. [Google Scholar] [CrossRef] [PubMed]
- Koipally, J.; Heller, E.J.; Seavitt, J.R.; Georgopoulos, K. Unconventional potentiation of gene expression by Ikaros. J. Biol. Chem. 2002, 277, 13007–13015. [Google Scholar] [CrossRef]
- Song, C.; Pan, X.; Ge, Z.; Gowda, C.; Ding, Y.; Li, H.; Li, Z.; Yochum, G.; Muschen, M.; Li, Q.; et al. Epigenetic regulation of gene expression by Ikaros, HDAC1 and Casein Kinase II in leukemia. Leukemia 2016, 30, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Koipally, J.; Georgopoulos, K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 2000, 275, 19594–19602. [Google Scholar] [CrossRef]
- Bottardi, S.; Zmiri, F.A.; Bourgoin, V.; Ross, J.; Mavoungou, L.; Milot, E. Ikaros interacts with P-TEFb and cooperates with GATA-1 to enhance transcription elongation. Nucleic Acids Res. 2011, 39, 3505–3519. [Google Scholar] [CrossRef]
- Heizmann, B.; Kastner, P.; Chan, S. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 2018, 51, 14–23. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, D. The Ikaros family of zinc-finger proteins. Acta Pharm. Sin. B 2016, 6, 513–521. [Google Scholar] [CrossRef]
- Powell, M.D.; Read, K.A.; Sreekumar, B.K.; Oestreich, K.J. Ikaros zinc finger transcription factors: Regulators of cytokine signaling pathways and CD4+ T helper cell differentiation. Front. Immunol. 2019, 10, 1299. [Google Scholar] [CrossRef]
- Cippitelli, M.; Stabile, H.; Kosta, A.; Petillo, S.; Gismondi, A.; Santoni, A.; Fionda, C. Role of Aiolos and Ikaros in the Antitumor and Immunomodulatory Activity of IMiDs in Multiple Myeloma: Better to Lose Than to Find Them. Int. J. Mol. Sci. 2021, 22, 1103. [Google Scholar] [CrossRef] [PubMed]
- Schwickert, T.A.; Tagoh, H.; Gültekin, S.; Dakic, A.; Axelsson, E.; Minnich, M.; Ebert, A.; Werner, B.; Roth, M.; Cimmino, L.; et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 2014, 15, 283–293. [Google Scholar] [CrossRef]
- Alkhatib, A.; Werner, M.; Hug, E.; Herzog, S.; Eschbach, C.; Faraidun, H.; Köhler, F.; Wossning, T.; Jumaa, H. FoxO1 induces Ikaros splicing to promote immunoglobulin gene recombination. J. Exp. Med. 2012, 209, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Maeda, A.; Kurosaki, M.; Tezuka, T.; Hironaka, K.; Yamamoto, T.; Kurosaki, T. Cbl Suppresses B Cell Receptor–Mediated Phospholipase C (Plc)-γ2 Activation by Regulating B Cell Linker Protein–Plc-γ2 Binding. J. Exp. Med. 2000, 191, 641–650. [Google Scholar] [CrossRef]
- Nera, K.P.; Alinikula, J.; Terho, P.; Narvi, E.; Törnquist, K.; Kurosaki, T.; Buerstedde, J.M.; Lassila, O. Ikaros has a crucial role in regulation of B cell receptor signaling. Eur. J. Immunol. 2006, 36, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Mecarozzi, M.; Campese, A.F.; Grazioli, P.; Talora, C.; Frati, L.; Gulino, A.; Screpanti, I. Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 2007, 26, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Dumortier, A.; Jeannet, R.; Kirstetter, P.; Kleinmann, E.; Sellars, M.; dos Santos, N.R.; Thibault, C.; Barths, J.; Ghysdael, J.; Punt, J.A.; et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 2006, 26, 209–220. [Google Scholar] [CrossRef]
- Joshi, I.; Yoshida, T.; Jena, N.; Qi, X.; Zhang, J.; Van Etten, R.A.; Georgopoulos, K. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat. Immunol. 2014, 15, 294–304. [Google Scholar] [CrossRef]
- Ma, S.; Pathak, S.; Trinh, L.; Lu, R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood 2008, 111, 1396–1403. [Google Scholar] [CrossRef]
- Fang, C.M.; Roy, S.; Nielsen, E.; Paul, M.; Maul, R.; Paun, A.; Koentgen, F.; Raval, F.M.; Szomolanyi-Tsuda, E.; Pitha, P.M. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012, 13, 421–430. [Google Scholar] [CrossRef]
- Ferreirós-Vidal, I.; Carroll, T.; Taylor, B.; Terry, A.; Liang, Z.; Bruno, L.; Dharmalingam, G.; Khadayate, S.; Cobb, B.S.; Smale, S.T.; et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood 2013, 121, 1769–1782. [Google Scholar] [CrossRef]
- Trinh, L.A.; Ferrini, R.; Cobb, B.S.; Weinmann, A.S.; Hahm, K.; Ernst, P.; Garraway, I.P.; Merkenschlager, M.; Smale, S.T. Down-regulation of TDT transcription in CD4+CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 2001, 15, 1817–1832. [Google Scholar] [CrossRef]
- Reynaud, D.; Demarco, I.A.; Reddy, K.L.; Schjerven, H.; Bertolino, E.; Chen, Z.; Smale, S.T.; Winandy, S.; Singh, H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 2008, 9, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Harker, N.; Naito, T.; Cortes, M.; Hostert, A.; Hirschberg, S.; Tolaini, M.; Roderick, K.; Georgopoulos, K.; Kioussis, D. The CD8α Gene Locus Is Regulated by the Ikaros Family of Proteins. Mol. Cell 2002, 10, 1403–1415. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Duré, M.; Paroder, M.; Soto-Nieves, N.; Puga, I.; Macián, F. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells. Blood 2007, 109, 2878–2886. [Google Scholar] [CrossRef]
- Wong, L.Y.; Hatfield, J.K.; Brown, M.A. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development. J. Biol. Chem. 2013, 288, 35170–35179. [Google Scholar] [CrossRef] [PubMed]
- Yap, W.H.; Yeoh, E.; Tay, A.; Brenner, S.; Venkatesh, B. STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. FEBS Lett. 2005, 579, 4470–4478. [Google Scholar] [CrossRef]
- Ma, S.; Pathak, S.; Mandal, M.; Trinh, L.; Clark, M.R.; Lu, R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell. Biol. 2010, 30, 4149–4158. [Google Scholar] [CrossRef]
- Ochiai, K.; Yamaoka, M.; Swaminathan, A.; Bouvet, P.; Kundu, T.K.; Igarashi, K. Chromatin Protein PC4 Orchestrates B Cell Differentiation by Collaborating with IKAROS and IRF4. CellReports 2020, 33, 108517. [Google Scholar] [CrossRef]
- Heizmann, B.; Kastner, P.; Chan, S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med. 2013, 210, 2823. [Google Scholar] [CrossRef] [PubMed]
- Macias-Garcia, A.; Heizmann, B.; Sellars, M.; Marchal, P.; Dali, H.; Pasquali, J.L.; Muller, S.; Kastner, P.; Chan, S. Ikaros Is a Negative Regulator of B1 Cell Development and Function. J. Biol. Chem. 2016, 291, 9073–9086. [Google Scholar] [CrossRef]
- Mazzurana, L.; Forkel, M.; Rao, A.; Van Acker, A.; Kokkinou, E.; Ichiya, T.; Almer, S.; Höög, C.; Friberg, D.; Mjösberg, J. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3−ILC1/NK cell transdifferentiation. Eur. J. Immunol. 2019, 49, 1344–1355. [Google Scholar] [CrossRef]
- Naik, A.K.; Byrd, A.T.; Lucander, A.C.K.; Krangel, M.S. Hierarchical assembly and disassembly of a transcriptionally active RAG locus in CD4+CD8+ thymocytes. J. Exp. Med. 2019, 216, 231–243. [Google Scholar] [CrossRef]
- Winandy, S.; Wu, L.; Wang, J.H.; Georgopoulos, K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J. Exp. Med. 1999, 190, 1039–1048. [Google Scholar] [CrossRef]
- Schwickert, T.A.; Tagoh, H.; Schindler, K.; Fischer, M.; Jaritz, M.; Busslinger, M. Ikaros prevents autoimmunity by controlling anergy and Toll-like receptor signaling in B cells. Nat. Immunol. 2019, 20, 1517. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Huh, J.E.; Song, J.; Rhee, D.K.; Pyo, S. Ikaros negatively regulates inducible nitric oxide synthase expression in macrophages: Involvement of Ikaros phosphorylation by casein kinase 2. Cell. Mol. Life Sci. 2008, 65, 3290–3303. [Google Scholar] [CrossRef] [PubMed]
- Malinge, S.; Thiollier, C.; Chlon, T.M.; Doré, L.C.; Diebold, L.; Bluteau, O.; Mabialah, V.; Vainchenker, W.; Dessen, P.; Winandy, S.; et al. Ikaros inhibits megakaryopoiesis through functional interaction with GATA-1 and NOTCH signaling. Blood 2013, 121, 2440–2451. [Google Scholar] [CrossRef]
- Cytlak, U.; Resteu, A.; Pagan, S.; Green, K.; Milne, P.; Maisuria, S.; McDonald, D.; Hulme, G.; Filby, A.; Carpenter, B.; et al. Differential IRF8 Transcription Factor Requirement Defines Two Pathways of Dendritic Cell Development in Humans. Immunity 2020, 53, 353–370.e8. [Google Scholar] [CrossRef] [PubMed]
- Mastio, J.; Simand, C.; Cova, G.; Kastner, P.; Chan, S.; Kirstetter, P. Ikaros cooperates with Notch activation and antagonizes TGFβ signaling to promote pDC development. PLoS Genet. 2018, 14, e1007485. [Google Scholar] [CrossRef]
- Churchman, M.L.; Mullighan, C.G. Ikaros: Exploiting and targeting the hematopoietic stem cell niche in B-progenitor acute lymphoblastic leukemia. Exp. Hematol. 2017, 46, 1–8. [Google Scholar] [CrossRef]
- Iacobucci, I.; Storlazzi, C.T.; Cilloni, D.; Lonetti, A.; Ottaviani, E.; Soverini, S.; Astolfi, A.; Chiaretti, S.; Vitale, A.; Messa, F.; et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: On behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leuke. Blood 2009, 114, 2159–2167. [Google Scholar] [CrossRef]
- Iacobucci, I.; Mullighan, C.G. Genetic basis of acute lymphoblastic leukemia. J. Clin. Oncol. 2017, 35, 975–983. [Google Scholar] [CrossRef]
- Kuiper, R.P.; Waanders, E.; Van Der Velden, V.H.J.; Van Reijmersdal, S.V.; Venkatachalam, R.; Scheijen, B.; Sonneveld, E.; Van Dongen, J.J.M.; Veerman, A.J.P.; Van Leeuwen, F.N.; et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 2010, 24, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Mullighan, C.G.; Goorha, S.; Radtke, I.; Miller, C.B.; Coustan-Smith, E.; Dalton, J.D.; Girtman, K.; Mathew, S.; Ma, J.; Pounds, S.B.; et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007, 446, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, K.; Yamaguchi, S.; Iwanaga, E.; Nanri, T.; Shimomura, T.; Suzushima, H.; Mitsuya, H.; Asou, N. High frequency of IKZF1 genetic alterations in adult patients with B-cell acute lymphoblastic leukemia. Eur. J. Haematol. 2013, 91, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Clappier, E.; Grardel, N.; Bakkus, M.; Rapion, J.; De Moerloose, B.; Kastner, P.; Caye, A.; Vivent, J.; Costa, V.; Ferster, A.; et al. IKZF1 deletion is an independent prognostic marker in childhood B-cell precursor acute lymphoblastic leukemia, and distinguishes patients benefiting from pulses during maintenance therapy: Results of the EORTC Children’s Leukemia Group study 58,951. Leukemia 2015, 29, 2154–2161. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Gu, Z.; Payne-Turner, D.; McCastlain, K.; Harvey, R.C.; Chen, I.M.; Pei, D.; Iacobucci, I.; Valentine, M.; Pounds, S.B.; et al. High Frequency and Poor Outcome of Philadelphia Chromosome-Like Acute Lymphoblastic Leukemia in Adults. J. Clin. Oncol. 2017, 35, 394–401. [Google Scholar] [CrossRef]
- Churchman, M.L.; Qian, M.; te Kronnie, G.; Zhang, R.; Yang, W.; Zhang, H.; Lana, T.; Tedrick, P.; Baskin, R.; Verbist, K.; et al. Germline Genetic IKZF1 Variation and Predisposition to Childhood Acute Lymphoblastic Leukemia. Cancer Cell 2018, 33, 937–948.e8. [Google Scholar] [CrossRef]
- Gurel, Z.; Ronni, T.; Ho, S.; Kuchar, J.; Payne, K.J.; Turk, C.W.; Dovat, S. Recruitment of Ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J. Biol. Chem. 2008, 283, 8291–8300. [Google Scholar] [CrossRef]
- Dupuis, A.; Gaub, M.P.; Legrain, M.; Drenou, B.; Mauvieux, L.; Lutz, P.; Herbrecht, R.; Chan, S.; Kastner, P. Biclonal and biallelic deletions occur in 20% of B-ALL cases with IKZF1 mutations. Leukemia 2013, 27, 503–507. [Google Scholar] [CrossRef]
- Martinelli, G.; Iacobucci, I.; Storlazzi, C.T.; Vignetti, M.; Paoloni, F.; Cilloni, D.; Soverini, S.; Vitale, A.; Chiaretti, S.; Cimino, G.; et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: A GIMEMA AL WP report. J. Clin. Oncol. 2009, 27, 5202–5207. [Google Scholar] [CrossRef]
- Klein, F.; Feldhahn, N.; Herzog, S.; Sprangers, M.; Mooster, J.L.; Jumaa, H.; Müschen, M. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene 2006, 25, 1118–1124. [Google Scholar] [CrossRef]
- Ruiz, A.; Jiang, J.; Kempski, H.; Brady, H.J.M. Overexpression of the Ikaros 6 isoform is restricted to t(4;11) acute lymphoblastic leukaemia in children and infants and has a role in B-cell survival. Br. J. Haematol. 2004, 125, 31–37. [Google Scholar] [CrossRef]
- Steeghs, E.M.P.; Boer, J.M.; Hoogkamer, A.Q.; Boeree, A.; de Haas, V.; de Groot-Kruseman, H.A.; Horstmann, M.A.; Escherich, G.; Pieters, R.; den Boer, M.L. Copy number alterations in B-cell development genes, drug resistance, and clinical outcome in pediatric B-cell precursor acute lymphoblastic leukemia. Sci. Rep. 2019, 9, 4634. [Google Scholar] [CrossRef] [PubMed]
- Scheijen, B.; Boer, J.M.; Marke, R.; Tijchon, E.; van Ingen Schenau, D.; Waanders, E.; van Emst, L.; van der Meer, L.T.; Pieters, R.; Escherich, G.; et al. Tumor suppressors BTG1 and IKZF1 cooperate during mouse leukemia development and increase relapse risk in B-cell precursor acute lymphoblastic leukemia patients. Haematologica 2017, 102, 541–551. [Google Scholar] [CrossRef]
- Clappier, E.; Auclerc, M.F.; Rapion, J.; Bakkus, M.; Caye, A.; Khemiri, A.; Giroux, C.; Hernandez, L.; Kabongo, E.; Savola, S.; et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia 2014, 28, 70–77. [Google Scholar] [CrossRef]
- Vrooman, L.M.; Blonquist, T.M.; Harris, M.H.; Stevenson, K.E.; Place, A.E.; Hunt, S.K.; O’Brien, J.E.; Asselin, B.L.; Athale, U.H.; Clavell, L.A.; et al. Refining risk classification in childhood b acute lymphoblastic leukemia: Results of DFCI ALL consortium protocol 05-001. Blood Adv. 2018, 2, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Stanulla, M.; Dagdan, E.; Zaliova, M.; Möricke, A.; Palmi, C.; Cazzaniga, G.; Eckert, C.; Te Kronnie, G.; Bourquin, J.P.; Bornhauser, B.; et al. IKZF1 plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric b-cell precursor acute lymphoblastic leukemia. J. Clin. Oncol. 2018, 36, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Zaliova, M.; Stuchly, J.; Winkowska, L.; Musilova, A.; Fiser, K.; Slamova, M.; Starkova, J.; Vaskova, M.; Hrusak, O.; Sramkova, L.; et al. Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematologica 2019, 104, 1396–1406. [Google Scholar] [CrossRef]
- Burmeister, T.; Bartels, G.; Gröger, D.; Trautmann, H.; Schwartz, S.; Lenz, K.; Tietze-Bürger, C.; Viardot, A.; Wäsch, R.; Horst, H.A.; et al. Germline variants in IKZF1, ARID5B, and CEBPE as risk factors for adult-onset acute lymphoblastic leukemia: An analysis from the GMALL study group. Haematologica 2014, 99. [Google Scholar] [CrossRef]
- Olsson, L.; Castor, A.; Behrendtz, M.; Biloglav, A.; Forestier, E.; Paulsson, K.; Johansson, B. Deletions of IKZF1 and SPRED1 are associated with poor prognosis in a population-based series of pediatric B-cell precursor acute lymphoblastic leukemia diagnosed between 1992 and 2011. Leukemia 2014, 28, 302–310. [Google Scholar] [CrossRef]
- Churchman, M.L.; Low, J.; Qu, C.; Paietta, E.M.; Kasper, L.H.; Chang, Y.; Payne-Turner, D.; Althoff, M.J.; Song, G.; Chen, S.C.; et al. Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell 2015, 28, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Vairy, S.; Tran, T.H. IKZF1 alterations in acute lymphoblastic leukemia: The good, the bad and the ugly. Blood Rev. 2020, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Dai, Y.T.; Lilljebjörn, H.; Shen, S.H.; Cui, B.W.; Bai, L.; Liu, Y.F.; Qian, M.X.; Kubota, Y.; Kiyoi, H.; et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1223 cases. Proc. Natl. Acad. Sci. USA 2018, 115, E11711–E11720. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Churchman, M.L.; Roberts, K.G.; Moore, I.; Zhou, X.; Nakitandwe, J.; Hagiwara, K.; Pelletier, S.; Gingras, S.; Berns, H.; et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019, 51, 296–307. [Google Scholar] [CrossRef]
- Lilljebjörn, H.; Henningsson, R.; Hyrenius-Wittsten, A.; Olsson, L.; Orsmark-Pietras, C.; Von Palffy, S.; Askmyr, M.; Rissler, M.; Schrappe, M.; Cario, G.; et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat. Commun. 2016, 7, 11790. [Google Scholar] [CrossRef]
- Rahmani, M.; Fardi, M.; Hagh, M.F.; Feizi, A.A.H.; Talebi, M.; Solali, S. An investigation of methylation pattern changes in the IKZF1 promoter in patients with childhood B-cell acute lymphoblastic leukemia. Blood Res. 2019, 54, 144–148. [Google Scholar] [CrossRef]
- Gowda, C.; Song, C.; Ding, Y.; Iyer, S.; Dhanyamraju, P.K.; McGrath, M.; Bamme, Y.; Soliman, M.; Kane, S.; Payne, J.L.; et al. Cellular signaling and epigenetic regulation of gene expression in leukemia. Adv. Biol. Regul. 2020, 75. [Google Scholar] [CrossRef]
- Durchdewald, M.; Angel, P.; Hess, J. The transcription factor Fos: A Janus-type regulator in health and disease. Histol. Histopathol. 2009, 24, 1451–1461. [Google Scholar] [CrossRef]
- Ge, Z.; Gu, Y.; Zhao, G.; Li, J.; Chen, B.; Han, Q.; Guo, X.; Liu, J.; Li, H.; Yu, M.D.; et al. High CRLF2 expression associates with IKZF1 dysfunction in adult acute lymphoblastic leukemia without CRLF2 rearrangement. Oncotarget 2016, 7, 49722–49732. [Google Scholar] [CrossRef]
- Xhabija, B.; Kidder, B.L. KDM5B is a master regulator of the H3K4-methylome in stem cells, development and cancer. Semin. Cancer Biol. 2019, 57, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Qiu, Y.; Li, G.; Liu, C.; She, L.; Zhang, D.; Chen, X.; Zhu, G.; Zhang, X.; Tian, Y.; et al. KDM5B overexpression predicts a poor prognosis in patients with squamous cell carcinoma of the head and neck. J. Cancer 2018, 9, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Gu, Y.; Han, Q.; Sloane, J.; Ge, Q.; Gao, G.; Ma, J.; Song, H.; Hu, J.; Chen, B.; et al. Plant homeodomain finger protein 2 as a novel IKAROS target in acute lymphoblastic leukemia. Epigenomics 2018, 10, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Han, Q.; Gu, Y.; Ge, Q.; Ma, J.; Sloane, J.; Gao, G.; Payne, K.J.; Szekely, L.; Song, C.; et al. Aberrant ARID5B expression and its association with Ikaros dysfunction in acute lymphoblastic leukemia. Oncogenesis 2018, 7, 84. [Google Scholar] [CrossRef]
- Chan, L.N.; Chen, Z.; Braas, D.; Lee, J.W.; Xiao, G.; Geng, H.; Cosgun, K.N.; Hurtz, C.; Shojaee, S.; Cazzaniga, V.; et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 2017, 542, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Vitanza, N.A.; Zaky, W.; Blum, R.; Meyer, J.A.; Wang, J.; Bhatla, T.; Morrison, D.J.; Raetz, E.A.; Carroll, W.L. Ikaros deletions in BCR-ABL-negative childhood acute lymphoblastic leukemia are associated with a distinct gene expression signature but do not result in intrinsic chemoresistance. Pediatr. Blood Cancer 2014, 61, 1779–1785. [Google Scholar] [CrossRef]
- Churchman, M.L.; Evans, K.; Richmond, J.; Robbins, A.; Jones, L.; Shapiro, I.M.; Pachter, J.A.; Weaver, D.T.; Houghton, P.J.; Smith, M.A.; et al. Synergism of FAK and tyrosine kinase inhibition in Ph+ B-ALL. JCI Insight 2016, 1, e86082. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sørensen, A.B.; Wang, B.; Wabl, M.; Nielsen, A.L.; Pedersen, F.S. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol. Biol. 2009, 10, 2. [Google Scholar] [CrossRef]
- Trageser, D.; Iacobucci, I.; Nahar, R.; Duy, C.; Von Levetzow, G.; Klemm, L.; Park, E.; Schuh, W.; Gruber, T.; Herzog, S.; et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 2009, 206, 1739–1753. [Google Scholar] [CrossRef]
- Schjerven, H.; Ayongaba, E.F.; Aghajanirefah, A.; McLaughlin, J.; Cheng, D.; Geng, H.; Boyd, J.R.; Eggesbø, L.M.; Lindeman, I.; Heath, J.L.; et al. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J. Exp. Med. 2017, 214, 793–814. [Google Scholar] [CrossRef]
- Ge, Z.; Guo, X.; Li, J.; Hartman, M.; Kawasawa, Y.I.; Dovat, S.; Song, C. Clinical significance of high c-MYC and low MYCBP2 expression and their association with Ikaros dysfunction in adult acute lymphoblastic leukemia. Oncotarget 2015, 6, 42300–42311. [Google Scholar] [CrossRef]
- Ge, Z.; Zhou, X.; Gu, Y.; Han, Q.; Li, J.; Chen, B.; Ge, Q.; Dovat, E.; Payne, J.L.; Sun, T.; et al. Ikaros regulation of the BCL6/BACH2 axis and its clinical relevance in acute lymphoblastic leukemia. Oncotarget 2017, 8, 8022–8034. [Google Scholar] [CrossRef]
- Mascle, X.; Albagli, O.; Lemercier, C. Point mutations in BCL6 DNA-binding domain reveal distinct roles for the six zinc fingers. Biochem. Biophys. Res. Commun. 2003, 300, 391–396. [Google Scholar] [CrossRef]
- Hurtz, C.; Hatzi, K.; Cerchietti, L.; Braig, M.; Park, E.; Kim, Y.M.; Herzog, S.; Ramezani-Rad, P.; Jumaa, H.; Müller, M.C.; et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. J. Exp. Med. 2011, 208, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Ma, J.; Gu, Y.; Song, H.; Kapadia, M.; Kawasawa, Y.I.; Dovat, S.; Song, C.; Ge, Z. RAG1 high expression associated with IKZF1 dysfunction in adult B-cell acute lymphoblastic leukemia. J. Cancer 2019, 10, 3842–3850. [Google Scholar] [CrossRef] [PubMed]
- Schjerven, H.; Mclaughlin, J.; Arenzana, T.L.; Frietze, S.; Cheng, D.; Wadsworth, S.E.; Lawson, G.W.; Bensinger, S.J.; Farnham, P.J.; Witte, O.N.; et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 2013, 14, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Katerndahl, C.D.S.; Heltemes-Harris, L.M.; Willette, M.J.L.; Henzler, C.M.; Frietze, S.; Yang, R.; Schjerven, H.; Silverstein, K.A.T.; Ramsey, L.B.; Hubbard, G.; et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol. 2017, 18, 694–704. [Google Scholar] [CrossRef]
- Shochat, C.; Tal, N.; Gryshkova, V.; Birger, Y.; Bandapalli, O.R.; Cazzaniga, G.; Gershman, N.; Kulozik, A.E.; Biondi, A.; Mansour, M.R.; et al. Novel activating mutations lacking cysteine in type I cytokine receptors in acute lymphoblastic leukemia. Blood 2014, 124, 106–110. [Google Scholar] [CrossRef]
- Ge, Z.; Gu, Y.; Xiao, L.; Han, Q.; Li, J.; Chen, B.; Yu, J.; Kawasawa, Y.I.; Payne, K.J.; Dovat, S.; et al. Co-existence of IL7R high and SH2B3 low expression distinguishes a novel high-risk acute lymphoblastic leukemia with Ikaros dysfunction. Oncotarget 2016, 7, 46014–46027. [Google Scholar] [CrossRef]
- Liang, Z.; Brown, K.E.; Carroll, T.; Taylor, B.; Vidal, I.F.; Hendrich, B.; Rueda, D.; Fisher, A.G.; Merkenschlager, M. A high-resolution map of transcriptional repression. Elife 2017, 6, e22767. [Google Scholar] [CrossRef] [PubMed]
- Findley, H.W., Jr.; Cooper, M.D.; Kim, T.H.; Alvarado, C.; Ragab, A.H. Two new acute lymphoblastic leukemia cell lines with early B-cell phenotypes-PubMed. Blood 1982, 60, 1305–1309. [Google Scholar] [CrossRef]
- Song, C.; Gowda, C.; Pan, X.; Ding, Y.; Tong, Y.; Tan, B.H.; Wang, H.; Muthusami, S.; Ge, Z.; Sachdev, M.; et al. Targeting casein kinase II restores Ikaros tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood 2015, 126, 1813–1822. [Google Scholar] [CrossRef]
- Wu, H.B.; Lv, W.F.; Wang, Y.X.; Li, Y.Y.; Guo, W. BCL6 promotes the methotrexate-resistance by upregulating ZEB1 expression in children with acute B lymphocytic leukemia. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5240–5247. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Duy, C.; Müschen, M. BACH2-BCL6 balance regulates selection at the pre-B cell receptor checkpoint. Trends Immunol. 2014, 35, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Tashiro, S.; Nakajima, O.; Hoshino, H.; Takahashi, S.; Sakoda, E.; Ikebe, D.; Yamamoto, M.; Igarashi, K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 2004, 429, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.H.; Mai, Y. A Bach2 link between pre-B cell receptor checkpoint and pre-B cell ALL. Cancer Cell 2013, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Dave, S.S. The polyphony of BACH2. Blood 2014, 123, 950. [Google Scholar] [CrossRef]
- Muto, A.; Hoshino, H.; Madisen, L.; Yanai, N.; Obinata, M.; Karasuyama, H.; Hayashi, N.; Nakauchi, H.; Yamamoto, M.; Groudine, M.; et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3’ enhancer. EMBO J. 1998, 17, 5734–5743. [Google Scholar] [CrossRef]
- Casolari, D.A.; Makri, M.; Yoshida, C.; Muto, A.; Igarashi, K.; Melo, J. V Transcriptional suppression of BACH2 by the Bcr-Abl oncoprotein is mediated by PAX5. Leukemia 2013, 27, 409–415. [Google Scholar] [CrossRef]
- Swaminathan, S.; Huang, C.; Geng, H.; Chen, Z.; Harvey, R.; Kang, H.; Ng, C.; Titz, B.; Hurtz, C.; Sadiyah, M.F.; et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat. Med. 2013, 19, 1014–1022. [Google Scholar] [CrossRef]
- Witkowski, M.T.; Hu, Y.; Roberts, K.G.; Boer, J.M.; McKenzie, M.D.; Liu, G.J.; Le Grice, O.D.; Tremblay, C.S.; Ghisi, M.; Willson, T.A.; et al. Conserved IKAROS-regulated genes associated with B-progenitor acute lymphoblastic leukemia outcome. J. Exp. Med. 2017, 214, 773–791. [Google Scholar] [CrossRef]
- Nakada, D.; Saunders, T.L.; Morrison, S.J. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 2010, 468, 653–658. [Google Scholar] [CrossRef]
- Gan, B.; Hu, J.; Jiang, S.; Liu, Y.; Sahin, E.; Zhuang, L.; Fletcher-Sananikone, E.; Colla, S.; Wang, Y.A.; Chin, L.; et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 2010, 468, 701–704. [Google Scholar] [CrossRef]
- Gurumurthy, S.; Xie, S.Z.; Alagesan, B.; Kim, J.; Yusuf, R.Z.; Saez, B.; Tzatsos, A.; Ozsolak, F.; Milos, P.; Ferrari, F.; et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 2010, 468, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Bazdar, D.A.; Kalinowska, M.; Panigrahi, S.; Sieg, S.F. Recycled IL-7 Can Be Delivered to Neighboring T Cells. J. Immunol. 2015, 194, 4698–4704. [Google Scholar] [CrossRef] [PubMed]
- Noronha, E.P.; Marques, L.V.C.; Andrade, F.G.; Sardou-Cezar, I.; Dos Santos-Bueno, F.V.; Da Paz Zampier, C.; Terra-Granado, E.; Pombo-de-Oliveira, M.S. T-lymphoid/myeloid mixed phenotype acute leukemia and early T-cell precursor lymphoblastic leukemia similarities with NOTCH1 mutation as a good prognostic factor. Cancer Manag. Res. 2019, 11, 3933–3943. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.F.; Morella, K.K.; Anderson, D.; Kumaki, N.; Leonard, W.J.; Cosman, D.; Baumann, H. Reconstitution of a functional interleukin (IL)-7 receptor demonstrates that the IL-2 receptor gamma chain is required for IL-7 signal transduction. Eur. J. Immunol. 1995, 25, 399–404. [Google Scholar] [CrossRef]
- Raetz, E.A.; Bhatla, T. Where do we stand in the treatment of relapsed acute lymphoblastic leukemia? Hematol. Am. Soc. Hematol. Educ. Progr. 2012, 2012, 129–136. [Google Scholar] [CrossRef]
- Roberts, K.G.; Yang, Y.L.; Payne-Turner, D.; Lin, W.; Files, J.K.; Dickerson, K.; Gu, Z.; Taunton, J.; Janke, L.J.; Chen, T.; et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017, 1, 1657–1671. [Google Scholar] [CrossRef]
- Gianfelici, V.; Messina, M.; Paoloni, F.; Peragine, N.; Lauretti, A.; Fedullo, A.L.; Di Giacomo, F.; Vignetti, M.; Vitale, A.; Guarini, A.; et al. IL7R overexpression in adult acute lymphoblastic leukemia is associated to JAK/STAT pathway mutations and identifies patients who could benefit from targeted therapies. Leuk. Lymphoma 2019, 60, 829–832. [Google Scholar] [CrossRef]
- Baran-Marszak, F.; Magdoud, H.; Desterke, C.; Alvarado, A.; Roger, C.; Harel, S.; Mazoyer, E.; Cassinat, B.; Chevret, S.; Tonetti, C.; et al. Expression level and differential JAK2-V617F-binding of the adaptor protein Lnk regulates JAK2-mediated signals in myeloproliferative neoplasms. Blood 2010, 116, 5961–5971. [Google Scholar] [CrossRef]
- Jang, W.; Park, J.; Kwon, A.; Choi, H.; Kim, J.; Lee, G.D.; Han, E.; Jekarl, D.W.; Chae, H.; Han, K.; et al. CDKN2B downregulation and other genetic characteristics in T-acute lymphoblastic leukemia. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.B.; Gu, Z.; Iacobucci, I.; Dickerson, K.; Choi, J.K.; Xu, B.; Payne-Turner, D.; Yoshihara, H.; Loh, M.L.; Horan, J.; et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018, 562, 373–406. [Google Scholar] [CrossRef]
- Maslah, N.; Cassinat, B.; Verger, E.; Kiladjian, J.J.; Velazquez, L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia 2017, 31, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Song, C.; Ding, Y.; Pan, X.; Ge, Z.; Tan, B.H.; Gowda, C.; Sachdev, M.; Muthusami, S.; Ouyang, H.; et al. Transcriptional Regulation of JARID1B/KDM5B Histone Demethylase by Ikaros, Histone Deacetylase 1 (HDAC1), and Casein Kinase 2 (CK2) in B-cell Acute Lymphoblastic Leukemia. J. Biol. Chem. 2016, 291, 4004–4018. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.C.; Chang, J.; Wang, L.C.; Ren, H.M.; Pang, J.R.; Liu, H.M. Lysine demethylase 5B (KDM5B): A potential anti-cancer drug target. Eur. J. Med. Chem. 2019, 161, 131–140. [Google Scholar] [CrossRef]
- Lin, C.; Song, W.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Zhao, H. Recent advances in the ARID family: Focusing on roles in human cancer. Onco. Targets. Ther. 2014, 7, 315–324. [Google Scholar] [CrossRef]
- Leong, W.Z.; Tan, S.H.; Ngoc, P.C.T.; Amanda, S.; Yam, A.W.Y.; Liau, W.S.; Gong, Z.; Lawton, L.N.; Tenen, D.G.; Sanda, T. ARID5B as a critical downstream target of the TAL1 complex that activates the oncogenic transcriptional program and promotes T-cell leukemogenesis. Genes Dev. 2017, 31, 2343–2360. [Google Scholar] [CrossRef]
- Lahoud, M.H.; Ristevski, S.; Venter, D.J.; Jermiin, L.S.; Bertoncello, I.; Zavarsek, S.; Hasthorpe, S.; Drago, J.; De Kretser, D.; Hertzog, P.J.; et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001, 11, 1327–1334. [Google Scholar] [CrossRef]
- Baba, A.; Ohtake, F.; Okuno, Y.; Yokota, K.; Okada, M.; Imai, Y.; Ni, M.; Meyer, C.A.; Igarashi, K.; Kanno, J.; et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 2011, 13, 668–675. [Google Scholar] [CrossRef]
- Joshi, P.; Greco, T.M.; Guise, A.J.; Luo, Y.; Yu, F.; Nesvizhskii, A.I.; Cristea, I.M. The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 2013, 9, 672. [Google Scholar] [CrossRef]
- Trevĩo, L.R.; Yang, W.; French, D.; Hunger, S.P.; Carroll, W.L.; Devidas, M.; Willman, C.; Neale, G.; Downing, J.; Raimondi, S.C.; et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1001–1005. [Google Scholar] [CrossRef]
- Rudant, J.; Orsi, L.; Bonaventure, A.; Goujon-Bellec, S.; Corda, E.; Baruchel, A.; Bertrand, Y.; Nelken, B.; Robert, A.; Michel, G.; et al. Are ARID5B and IKZF1 polymorphisms also associated with childhood acute myeloblastic leukemia: The ESCALE study (SFCE)? Leukemia 2013, 27, 746–748. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, H.; Ben Hassine, I.; Soltani, I.; Safra, I.; Ouerhani, S.; Bel Haj Othmen, H.; Teber, M.; Farah, A.; Amouri, H.; Toumi, N.H.; et al. Association of genetic variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the risk of acute lymphoblastic leukemia in Tunisian children and their contribution to racial differences in leukemia incidence. Pediatr. Hematol. Oncol. 2016, 33, 157–167. [Google Scholar] [CrossRef]
- Bartram, T.; Burkhardt, B.; Wössmann, W.; Seidemann, K.; Zimmermann, M.; Cario, G.; Lisfeld, J.; Ellinghaus, E.; Franke, A.; Houlston, R.S.; et al. Childhood acute lymphoblastic leukemia-associated risk-loci IKZF1, ARID5B and CEBPE and risk of pediatric non-Hodgkin lymphoma: A report from the Berlin-Frankfurt-Münster Study Group. Leuk. Lymphoma 2015, 56, 814–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nolan, M.; Yamada, H.; Watanabe, M.; Nasu, Y.; Takei, K.; Takeda, T. Dynamin2 GTPase contributes to invadopodia formation in invasive bladder cancer cells. Biochem. Biophys. Res. Commun. 2016, 480, 409–414. [Google Scholar] [CrossRef]
- Le Borgne, R.; Bardin, A.; Schweisguth, F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 2005, 132, 1751–1762. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Gu, Y.; Han, Q.; Zhao, G.; Li, M.; Li, J.; Chen, B.; Sun, T.; Dovat, S.; Gale, R.P.; et al. Targeting High Dynamin-2 (DNM2) Expression by Restoring Ikaros Function in Acute Lymphoblastic Leukemia. Sci. Rep. 2016, 6, 38004. [Google Scholar] [CrossRef] [PubMed]
- Dovat, S.; Song, C.; Payne, K.J.; Li, Z. Ikaros, CK2 kinase, and the road to leukemia. Mol. Cell. Biochem. 2011, 356, 201–207. [Google Scholar] [CrossRef]
- Chen, L.; Niu, Q.; Huang, Z.; Yang, B.; Wu, Y.; Zhang, J. IKZF1 polymorphisms are associated with susceptibility, cytokine levels, and clinical features in systemic lupus erythematosus. Medicine 2020, 99, e22607. [Google Scholar] [CrossRef]
- Schott, C.A.; Ascoli, C.; Huang, Y.; Perkins, D.L.; Finn, P.W. Declining Pulmonary Function in Interstitial Lung Disease Linked to Lymphocyte Dysfunction. Am. J. Respir. Crit. Care Med. 2020, 201, 610–613. [Google Scholar] [CrossRef]
- Gowda, C.; Soliman, M.; Kapadia, M.; Ding, Y.; Payne, K.; Dovat, S. Casein Kinase II (CK2), Glycogen Synthase Kinase-3 (GSK-3) and Ikaros mediated regulation of leukemia. Adv. Biol. Regul. 2017, 65, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Linabery, A.M.; Blommer, C.N.; Langer, E.K.; Spector, L.G.; Hilden, J.M.; Heerema, N.A.; Radloff, G.A.; Tower, R.L.; Davies, S.M. Genetic variants modify susceptibility to leukemia in infants: A Children’s Oncology Group report. Pediatr. Blood Cancer 2013, 60, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Dijon, M.; Bardin, F.; Murati, A.; Batoz, M.; Chabannon, C.; Tonnelle, C. The role of Ikaros in human erythroid differentiation. Blood 2008, 111, 1138–1146. [Google Scholar] [CrossRef]
- Ruiz, A.; Williams, O.; Brady, H.J.M. The Ikaros splice isoform, Ikaros 6, immortalizes murine haematopoietic progenitor cells. Int. J. Cancer 2008, 123, 1240–1245. [Google Scholar] [CrossRef]
- Jin, S.K.; Ju, I.E.; Cheong, J.W.; Ae, J.C.; Jin, K.L.; Woo, I.Y.; Yoo, H.M. Protein kinase CK2alpha as an unfavorable prognostic marker and novel therapeutic target in acute myeloid leukemia. Clin. Cancer Res. 2007, 13, 1019–1028. [Google Scholar] [CrossRef]
- Goldman, F.D.; Gurel, Z.; Al-Zubeidi, D.; Fried, A.J.; Icardi, M.; Song, C.; Dovat, S. Congenital pancytopenia and absence of B lymphocytes in a neonate with a mutation in the Ikaros gene. Pediatr. Blood Cancer 2012, 58, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Hasle, H.; Alonzo, T.A.; Auvrignon, A.; Behar, C.; Chang, M.; Creutzig, U.; Fischer, A.; Forestier, E.; Fynn, A.; Haas, O.A.; et al. Monosomy 7 and deletion 7q in children and adolescents with acute myeloid leukemia: An international retrospective study. Blood 2007, 109, 4641–4647. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, J.D.E.; Beuling, E.; van den Heuvel-Eibrink, M.M.; Obulkasim, A.; Baruche, A.; Trka, J.; Reinhardt, D.; Sonneveld, E.; Gibson, B.E.S.; Pieters, R.; et al. Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia. Haematologica 2015, 100, 1151–1159. [Google Scholar] [CrossRef]
- Breems, D.A.; Van Putten, W.L.J.; De Greef, G.E.; Van Zelderen-Bhola, S.L.; Gerssen-Schoorl, K.B.J.; Mellink, C.H.M.; Nieuwint, A.; Jotterand, M.; Hagemeijer, A.; Beverloo, H.B.; et al. Monosomal karyotype in acute myeloid leukemia: A better indicator of poor prognosis than a complex karyotype. J. Clin. Oncol. 2008, 26, 4791–4797. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, T.; Harutyunyan, A.; Berg, T.; Gisslinger, B.; Schalling, M.; Bagienski, K.; Olcaydu, D.; Passamonti, F.; Rumi, E.; Pietra, D.; et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood 2011, 118, 167–176. [Google Scholar] [CrossRef]
- Milosevic, J.D.; Puda, A.; Malcovati, L.; Berg, T.; Hofbauer, M.; Stukalov, A.; Klampfl, T.; Harutyunyan, A.S.; Gisslinger, H.; Gisslinger, B.; et al. Clinical significance of genetic aberrations in secondary acute myeloid leukemia. Am. J. Hematol. 2012, 87, 1010–1016. [Google Scholar] [CrossRef]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018, 562, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, A.; Liu, L.; Qin, J.; Wang, C.; Yang, M.; Wang, L.; Ni, X.; Hu, X.; Tang, G.; et al. The clinical impact of IKZF1 mutation in acute myeloid leukemia. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Westman, B.J.; Mackay, J.P.; Gell, D. Ikaros: A key regulator of haematopoiesis. Int. J. Biochem. Cell Biol. 2002, 34, 1304–1307. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conserva, M.R.; Redavid, I.; Anelli, L.; Zagaria, A.; Tarantini, F.; Cumbo, C.; Tota, G.; Parciante, E.; Coccaro, N.; Minervini, C.F.; et al. IKAROS in Acute Leukemia: A Positive Influencer or a Mean Hater? Int. J. Mol. Sci. 2023, 24, 3282. https://doi.org/10.3390/ijms24043282
Conserva MR, Redavid I, Anelli L, Zagaria A, Tarantini F, Cumbo C, Tota G, Parciante E, Coccaro N, Minervini CF, et al. IKAROS in Acute Leukemia: A Positive Influencer or a Mean Hater? International Journal of Molecular Sciences. 2023; 24(4):3282. https://doi.org/10.3390/ijms24043282
Chicago/Turabian StyleConserva, Maria Rosa, Immacolata Redavid, Luisa Anelli, Antonella Zagaria, Francesco Tarantini, Cosimo Cumbo, Giuseppina Tota, Elisa Parciante, Nicoletta Coccaro, Crescenzio Francesco Minervini, and et al. 2023. "IKAROS in Acute Leukemia: A Positive Influencer or a Mean Hater?" International Journal of Molecular Sciences 24, no. 4: 3282. https://doi.org/10.3390/ijms24043282
APA StyleConserva, M. R., Redavid, I., Anelli, L., Zagaria, A., Tarantini, F., Cumbo, C., Tota, G., Parciante, E., Coccaro, N., Minervini, C. F., Minervini, A., Specchia, G., Musto, P., & Albano, F. (2023). IKAROS in Acute Leukemia: A Positive Influencer or a Mean Hater? International Journal of Molecular Sciences, 24(4), 3282. https://doi.org/10.3390/ijms24043282










