Abstract
Prostate cancer (PC) is one of the most common types of cancer in males. Although early stages of PC are generally associated with favorable outcomes, advanced phases of the disease present a significantly poorer prognosis. Moreover, currently available therapeutic options for the treatment of PC are still limited, being mainly focused on androgen deprivation therapies and being characterized by low efficacy in patients. As a consequence, there is a pressing need to identify alternative and more effective therapeutics. In this study, we performed large-scale 2D and 3D similarity analyses between compounds reported in the DrugBank database and ChEMBL molecules with reported anti-proliferative activity on various PC cell lines. The analyses included also the identification of biological targets of ligands with potent activity on PC cells, as well as investigations on the activity annotations and clinical data associated with the more relevant compounds emerging from the ligand-based similarity results. The results led to the prioritization of a set of drugs and/or clinically tested candidates potentially useful in drug repurposing against PC.
1. Introduction
Prostate cancer (PC) is one of the most common types of tumors in men, with the number of diagnosed cases being significantly increased in recent years [1]. PC is particularly diffused in countries where advanced screening protocols (e.g., prostate-specific antigen (PSA) detection, MRI, and prostate biopsy) are available [2], accounting for more than 350,000 deaths per year [3]. According to recent studies, the prognosis and survival rates of PC patients heavily depend on tumor grade and the stage encountered at the primary diagnosis [4]. In particular, localized PC (~80% of the cases) presents a 5-year survival rate more than 90% [5], while more aggressive metastatic castration-sensitive (mCSPC) and castration-resistant PC (mCRPC) present a significantly lower survival rate (i.e., around 60–80% for locoregional metastases and around 30–40% for distant metastases) [6].
Standard early-stage PC therapy includes interventions, such as radiotherapy (RT) or radical prostatectomy, in combination with androgen deprivation therapy (ADT) or pelvic lymph node dissection (PLND) for patients with high-risk localized PC [7,8]. More aggressive late-stage prostate cancers are generally treated with ADT, albeit very often obtaining low therapeutic efficacy. Indeed, ADT is generally administered to PC patients on a lifelong basis in combination with Docetaxel (or its second-generation derivative Cabazitaxel) [9], drugs as androgen pathway inhibitors [10], or poly(ADP-ribose) polymerase inhibitors (PARPi) [11], considering the role of the androgen receptor (AR) for the tumor proliferation in mCRPC [12]. Additional androgen pathway inhibitors, such as abiraterone (ABI), enzalutamide (ENZ), or apalutamide (APA), have also recently been approved [11]. Moreover, new PARPi such as Olaparib and Rucaparib recently entered clinical use for mCRPC forms with germline alterations (i.e., BRCA2 mutation) [13]. Despite these recent developments, there still is a pressing need for the development of novel therapeutic agents, especially for the treatment of advanced stages of PC [14].
Considering that de novo drug discovery usually: (i) presents low success rates (~2%) [15], which are typically due to efficacy or safety issues [16,17]; (ii) requires a significant amount of resources and investments [18]; (iii) requires several years to deliver a new drug to the market, other strategies have been proposed to more efficiently identify effective therapeutics. Among them, drug repurposing (also known as drug repositioning) emerged as a valuable alternative or complementary approach [19], being able to potentially circumvent issues typically associated with de novo drug discovery [20,21,22]. The integration of different in silico approaches and workflows has been demonstrated to provide significant advantages for the identification of new drug repurposing possibilities [23]. Such data-driven approaches have been fueled by the constant increase in structural, biological, and chemical information reported into public repositories, such as DrugBank [24], ChEMBL [25], PubChem [26], UniProt [27], Therapeutic Target Database (TTD) [28], and clinicaltrials.gov [29,30,31,32,33]. Although many attempts to integrate such data have been proposed with success in this context [34,35,36], such information remains often scattered among various sources, complicating drug repurposing approaches.
On these premises, in this work, we aimed to identify potential drug repurposing candidates via an integrated, ad hoc in silico approach. In particular, we devised a repurposing workflow based on extensive ligand-based similarity estimations between DrugBank [24] compounds and a set of molecules with potent activity on PC cells retrieved from ChEMBL [25] ligands, these databases being widely employed in in silico drug discovery repurposing research [34,37,38].
We selected compounds from DrugBank for the analyses as it contains freely available data related to FDA-approved drugs, and pre-clinical and clinical candidates, including information on their properties, mechanisms of action, interactions with different molecules, pathways, and information on clinical trials [24]. Compounds with potent activity on PC cells were collected from ChEMBL, which is one among the most large, open-access resources of molecules with manually curated bioactivity data on targets and cell lines currently available, reporting also information about properties, pharmacokinetics, and safety profiles of ligands.
The adopted approach allowed us to identify a set of DrugBank compounds significantly similar to molecules with known potent PC anti-proliferative activity. Moreover, the analyses highlighted biological targets of ligands with reported potent activity on PC cells, which could be potentially relevant for their antiproliferative activity. Overall, the analyses on activity annotations and clinical data allowed us to confirm five top-scoring drugs that are already in clinical trials against PC, and to identify ten drugs, already assessed at various preclinical and clinical stages for different pathologies, which could be considered as valuable candidates for fast repurposing against PC.
2. Results and Discussion
Therapeutic options for advanced-stage prostate cancer are still limited and very often not effective [12]. Considering the extended timelines usually required for the development and approval of novel drugs, drug repurposing approaches, especially when driven by in silico methods, can represent faster yet valuable alternatives or complement methods for the identification of novel therapeutic treatments against PC, similarly to what is already observed for other diseases [21,22,30,31,32,33]. On these premises, we devised and applied a computationally driven drug repurposing workflow (Figure 1) in the search for novel drug candidates potentially suitable for PC therapy.
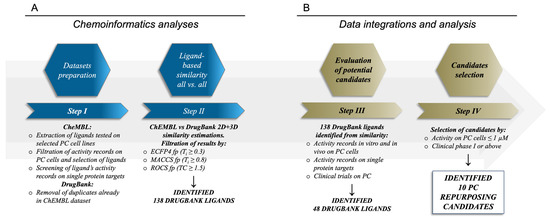
Figure 1.
Workflow adopted for the identification of potential drug repurposing candidates for PC. Panel (A) describes the ligand-based procedure, while panel (B) contains the data integration and analysis procedure for the selection of the repurposing candidates.
Moreover, a series of analyses were also conducted on the annotations of ChEMBL compounds with reported PC activity, in the search for molecular targets that are more likely to be involved in the observed cell-based antiproliferative effects (see below).
2.1. Analyses of the Compounds in the ChEMBL Dataset and of Their Target Activity Data
A set of ChEMBL ligands tested also on PC cells was first curated as detailed in the Methods section. The majority of these ligands derive from experiments on the PC-3, DU-145, and LNCaP cells (Table S1). Of note, most of the reported activity annotations of these compounds were below 1 µM (i.e., “highly active” records) in at least one PC cell line, while the remaining annotations followed a decreasing trend toward higher values (Figure S1 and Table S2). Moreover, the compounds showed a good degree of variability in terms of LogP, PSA, HBD, HBA, MW, volume, and ovality molecular descriptors, with distributions similar to those evaluated for the DrugBank ligands under investigation (Figure S2). In particular, most of the compounds in the ChEMBL dataset presented acceptable solubility and good membrane permeability, with LogP in the range of 2–4, and PSA values in the range of 50–100 Å2 [39]. Moreover, most of the investigated molecules presented 1 or 2 HBD and 3 to 6 HBA, which is in line with “Lipinski’s rule of five” [40]. The distribution of molecular weight, volume, and ovality, which are three indicators of small and spherical-shaped molecules, resulted in the 200–450 Da, 200–400 Å3, and 0.65–0.75 ranges, respectively. Moreover, comparison of these physicochemical properties highlighted a significant overlap between the properties of compounds in the two curated datasets (i.e., ChEMBL and DrugBank datasets, Figure S2), which is interesting in the view of repositioning molecules employing in silico similarity estimations.
Besides the investigations on key molecular descriptors, analyses of the target activity annotations of the ChEMBL datasets were also performed. These analyses aimed at identifying correlations between activity on biological targets and antiproliferative effects on different PC cell lines. To this aim, target annotations were first filtered and processed as described in the Methods section. Targets with fewer than ten activity records were not considered in the analysis for statistical robustness. Moreover, the analyses were restricted to compounds with reported antiproliferative activity on PC-3, DU-145, and LNCaP cell lines, which are the most widely studied in prostate cancer research (Tables S1 and S2; Figure S3). Indeed, a broad spectrum of PC cell lines has been studied over the past decades to characterize cellular mechanisms and aberrations at a molecular level responsible for development and progression of different subtypes of prostate cancer [41]. However, major interest has been placed on PC-3, DU-145, and LNCaP cell lines due to their ability to represent prostate cancer with different levels of aggressiveness, metastatic potential and hormonal dependency. In particular, PC-3 are cells collected from a lumbar vertebral metastasis of advanced-staged prostate cancer, showing high metastatic potential and aggressiveness, and low androgen dependency [42]. DU-145 are androgen-refractory cells collected from central nervous system (CNS) metastases of prostate cancer, with moderated metastatic potential [43]. LNCaP are androgen-dependent cells with low metastatic potential, which were collected from lymph node metastases of PC [44]. Compounds tested on PC-3 and DU-145 cells revealed target activities with a good degree of correlation with antiproliferative cellular activity (Spearman’s Rho ρs ≥ 0.4, p-value < 0.001), whereas targets with a statistically significant correlation to cellular activity against LNCaP cells were not found, most likely because of the limited amount of data available for this cell line compared to PC-3 and DU-145 cells. The performed analyses allowed us to highlight a total of 9 biological targets with significant correlation (see Table 1 and Figure 2) with PC antiproliferative activity. The complete list of the targets investigated in this study is reported in Tables S3 and S4 and Figure S4.

Table 1.
Targets identified from the analyses performed in the CHEMBL dataset, whose inhibitory activity data correlate with the activity on PC cells. The table reports only targets whose lower confidence interval of Spearman’s Rho (ρs) resulted as equal or higher than 0.4 (ρs ≥ 0.4, p-value < 0.001).
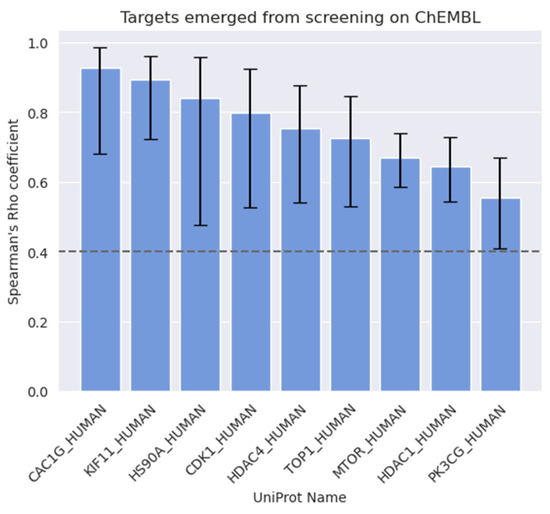
Figure 2.
Targets whose activity data correlate with antiproliferative activity on PC cells (ρs ≥ 0.4, p-value < 0.001).
Interestingly, all the targets reported in Figure 2 have been previously studied for their involvement in PC development and progression. For example, mTOR and PI3Kγ emerged as two of the most relevant targets for PC-3 treatment, regulating the PI3K/AKT/mTOR signaling pathway, and both have been found to be significantly overexpressed in several types of cancers, including PC [45,46]. Of note, the PI3K/Akt/mTOR signaling cascade is one of the most studied for the treatment of PC [47], for which novel therapeutic agents are being developed [48].
Heat Shock Protein 90 (Hsp90) is a chaperone protein with a recognized role in the regulation of folding and degradation of many oncogenic signaling proteins. Several preclinical and clinical studies assessing Hsp90 inhibitors have been reported so far, although none of them have received approval for therapeutic use to date [49,50,51]. Interestingly, Hsp90 has recently come to light again as a promising anti-cancer target against several types of tumors including PC, especially in multi-target therapies [52,53]. In addition, we also identified two members of the Histone deacetylase family (i.e., HDAC1 and HDAC4), which are known to be key regulators of chromatin structure and post-translation modifiers on several proteins in different tissues [54]. In particular, HDAC1 is a class I HDAC member that plays a crucial role in cellular epigenetic landscaping and transcriptional repression [55,56]. HDAC1 pathological expression is recognized in different types of cancers including PC [57], where its overexpression increases proliferation and differentiation [55,58,59,60,61], making it an interesting therapeutic target for several diseases. HDAC4 is a class IIA HDAC that exhibits tissue-specific patterns of expression. Alterations of HDAC4 activity, such as its overexpression and its repression effects on the tumor-suppressor gene p21, are well documented in different types of tumors [62,63,64,65,66], with its role in PC development and survival being reported in the literature [67,68].
Kinesin family member 11 (KIF11) is a molecular motor protein essential in mitosis and its inactivation causes cell cycle arrest and apoptotic cell death [69]. This protein also has a central role in several cancers [70,71,72,73], and its overexpression is a biomarker that could be associated with poor outcomes [74,75]. Of note, several studies suggest that KIF11 expression levels might be related to PC aggressiveness and bone metastasis occurrence, with this protein being potentially involved in the metastatic process [76,77]. Various KIF11 inhibitors are currently under development (i.e., Ispinesib, S-(methoxytrityl)-L-cysteine, Monastrol) for the treatment of tumors including prostate cancer [78,79,80,81]. Although the modulation of KIF11 did not result in the expected therapeutic efficacy in some cases [82], it remains an interesting target for PC with a proven prognostic and a potential therapeutic role [83,84].
T-type Ca2+ channels are transporters for Ca2+, which is a fundamental requirement for tumor progression and proliferation [85,86,87,88]. One of the three isoforms of T-type Ca2+ channels is Cav3.1 (also known as α1G), which is expressed in different cancers, such as breast cancer [89], small-cell lung cancer [90], and retinoblastoma [91]. Moreover, Cav3.1 downregulation by complementary shRNA has been recently observed to lead to a decrease in cell growth and migration, through the targeting of the AKT signaling in PC cells. These results open to the possibility that Cav3.1 may be a novel potential target for PC treatment [92,93].
Cyclin-Dependent Kinase 1 (CDK1, also known as p34, or CDC2) is a promoter of cell cycle progression, which is deeply involved in cell growth progression [94]. CDK1 activity is also linked to apoptosis induction in different pathways [95]. Moreover, it has been reported to play a central role in different types of cancers [96,97,98,99,100,101,102,103]. CDK1 has also a role in the regulation of AR phosphorylation and expression, suggesting a potential enhancement in the response to androgen deprivation therapy through CDK1 inhibition [104,105].
DNA topoisomerase I (i.e., TOP1) is a ubiquitous enzyme involved in the relaxation of DNA supercoiling, which is often targeted, along with topoisomerase II, by a wide variety of antimicrobial and anticancer agents [106,107]. Moreover, the inhibition of TOP1 with consequent DNA damage and apoptosis induction has been reported in PC cells [108,109,110,111]. Besides the ones described above, additional targets showing correlation coefficients slightly below the selected threshold were also identified (see Table S4 and Figure S4) such as PI3Kα/β [48,112], Androgen Receptor [113], Glycogen Synthase Kinase-3 β [114], STAT3 [115], and Serine/Threonine-Protein Kinase Aurora-A [116], which, however, present a recognized role in PC development and progression. These targets showed lower correlations to PC antiproliferative activity most likely because they lack exhaustive target activity annotations in ChEMBL.
2.2. Similarity Estimations and Selection of the Candidates for Drug Repurposing
Extensive 2D MACCS and ECFP4 fingerprints-based, followed by 3D-shape-based, similarity estimations with ROCS (Rapid Overlay of Chemical Structures, OpenEye) [117] were performed between the DrugBank and ChEMBL datasets, as detailed in the Methods section. These analyses were performed to identify DrugBank ligands already assessed at pre-clinical and clinical stages similar to ChEMBL compounds with known activity against prostate cancer cell lines. The performed similarity analyses highlighted an overall low degree of similarity between the compounds in the two datasets, which is a consequence of the high diversity between the chemical scaffolds of the ligands considered (Figure S5). However, these analyses allowed us to identify 138 DrugBank ligands with a high degree of similarity with molecules having antiproliferative activity on PC cells below 1 µM, according to both 2D and 3D similarity indexes (see Table S5). The 138 identified DrugBank ligands (Figure 1, panel A) included molecules investigated for their efficacy and toxicity at preclinical levels, and more importantly drug candidates already tested in clinical settings, with these latter being the most interesting in the view of fast repurposing against PC. In addition, the identified DrugBank compounds have also reported activity data on biological targets and on cell lines that were in a number of cases already tested against prostate cancer cells. On these premises, the identification of the best candidates for drug repurposing on PC required a further step of integration and analysis of their literature data (Figure 1, panel B). In particular, our attention focused on: (i) in vitro activity data records on PC cells; (ii) in vivo activity data records on PC; (iii) clinical trials on PC or solid tumors with PC patients; (iv) bioactivity records on targets with a proven relation to PC. Of note, the performed analyses allowed us to provide a validation of the adopted workflow (see below), and more importantly to identify 48 DrugBank compounds that meet at least one or more of the above-mentioned criteria, which could be, therefore, considered of high interest for their potential repurposing against PC. The list of the identified 48 DrugBank ligands with their similarity statistics is reported in Table S6.
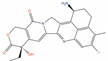
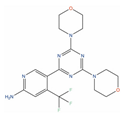
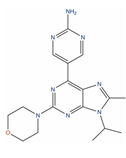
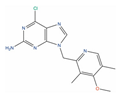
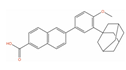
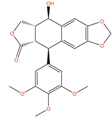
Of the 48 DrugBank ligands, 10 molecules with reported activity on PC cells lower than 1 µM, and whose safety profile had already been assessed at least in a phase I study, were selected as the most suitable candidates for fast repurposing against PC (Table 2). Interestingly, some of the identified ligands have been previously included in trials including PC patients, although this type of cancer was not the main focus of these studies. Moreover, some of these compounds have also been reported to be active on targets relevant to PC (e.g., Vistusertib, Bimiralisib, Onatasertib, VS-5584, and BIIB021 in Table 2) and could exert potential beneficial polypharmacological effects. Of note, 5 of the 48 identified DrugBank molecules have already been assessed in clinical trials for PC as a single agent or as a combination of drugs (underlined in Table 2). These molecules have been assessed in phase I/II clinical trials, together with other therapeutic agents, such as Docetaxel or Androgen Receptor inhibitors (e.g., LAE001) to assess their overall efficacy and potentially unwanted side-effects when used in combination. The remaining 33 ligands were excluded at the final stage of candidates’ selection, due to the poor activity on PC cells, lack of novelty of the molecules, or toxicity in humans. Information related to these latter ligands is reported in Table S7. A detailed discussion of the drug candidates and the molecules already assessed in PC trials is reported below (i.e., “Molecules already under clinical evaluation for PC” and “New potential Candidates for drug repurposing”).

Table 2.
Compounds identified from the similarity screenings and relevant association to PC, proposed as potential repurposing candidates. The first 5 compounds underlined in the table have already completed or are currently enrolled in clinical trials against PC. “Max Phase” was obtained from ChEMBL. “Disease” and “Primary target” were obtained from TTD. “Primary target” also contains the target UniProt name and the molecule’s mechanism of activity on the target in square brackets.
2.3. Molecules Already under Clinical Evaluation for PC
An analysis of literature data reported for the selected candidates highlighted five molecules that have already been included in clinical trials for prostate cancer therapy, either as single agents or in combination with other drugs (Table 2). To some extent, this result provides an internal validation of the applied methodology and raised interesting considerations regarding their enrollment in PC clinical trials. In particular, Azacitidine (DB00928) is a cytosine analog and hypomethylating agent, which is used as an antineoplastic agent in the therapy of myelodysplastic syndromes [132]. Azacitidine efficacy as a single agent was assessed in a phase II clinical trial on chemo-naive CRPC patients, resulting in favorable disease-modifying activities [119]. Azacitidine has also entered a phase I/II clinical trial (clinicaltrials.gov ID: NCT00503984), in combination with docetaxel and prednisone in mCRPC patients previously treated with docetaxel-based therapy, albeit conclusive results have not been delivered yet.
Trimetretexate (TMTX, DB01157) is a lipophilic inhibitor of DHFR that has been approved for the management of Pneumocystis jiroveci pneumonia in patients with AIDS [140]. TMTX efficacy against advanced hormone-refractory prostate cancer was assessed in a phase II trial in the early 1990s, although its clinical usefulness for PC treatment resulted as limited [120].
Afuresertib (GSK2110183, DB11648) is a pan-AKT kinase inhibitor with low nanomolar potency against AKT2 (IC50 = 10 nM) [141], AKT3 (IC50 = 16.9 nM) [141], and AKT1 (IC50 = 1 nM) [141], with the latter target being often hyperactivated in mCRPC [142]. Afuresertib has been demonstrated to inhibit the proliferation of various cancer cells, with a particular effect on hematological diseases [121]. More importantly, Afuresertib has recently been included in phase I/II trials against mCRPC in combination with LAE001, which is an androgen synthesis inhibitor, and Prednisone (clinicaltrials.gov ID: NCT04060394), providing promising results [143].
Vistusertib (AZD2014, DB11925) is an orally bioavailable, low-nanomolar inhibitor of mTOR (IC50 = 2.8 nM) [144], which demonstrated promising antineoplastic activity [145,146,147]. Vistusertib has also shown low nanomolar activity against PI3Kα (Kd = 33 nM) [148], which is a target implicated in the PI3K/AKT/mTOR signaling pathway, recognized to play a role in PC development and progression [46]. Vistusertib has also been enrolled in clinical trials for prostate cancer patients prior to radical prostatectomy (clinicaltrials.gov ID: NCT02064608) and in patients with CRPC in combination with AZD8186 (clinicaltrials.gov ID: NCT01884285). The results of these studies suggest that Vistusertib is tolerated in humans and inhibits mTOR1/2 in tumor tissues [149]. Preliminary results showed efficacy when it is used in combination with AZD8186 on a CRPC patient, with the study of the safety and tolerability of this combination still in place [150].
Exatecan (DB12185) is a novel camptothecin derivative and a TOP1 inhibitor, which has shown antiproliferative effects against various cancer cell lines [151]. In a phase II trial, DX-8951F (Exatecan mesylate) effectiveness was assessed for the treatment of patients with metastatic prostate cancer that has not responded to hormone therapy (clinicaltrials.gov ID: NCT00004045); however, clinical development has been discontinued.
2.4. New Potential Candidates for Drug Repurposing
Besides compounds already enrolled in clinical trials against prostate cancer, we also identified 10 compounds with tested safety profiles, significant activity on PC cell lines, and more advanced experimentation. The molecules reported in the lower part of Table 2 represent valuable putative candidates for fast repurposing against PC. Interestingly, some of these molecules previously entered clinical trials for their therapeutic assessment against different types of cancers and they have been demonstrated to exert their therapeutic effects through the modulation of targets and pathways highly relevant also in PC. Moreover, some of the compounds have already been tested in trials focusing on solid tumors, also including few PC patients, and have associated bioactivity data on several cell lines of different tumors (Table S8). In addition, some of the identified compounds have been clinically tested or approved in completely different therapeutic areas and represent candidates in which more novelty comes into play.
One among the most promising ligands of this class is Bimiralisib (PQR-309, DB14846), which has been reported to potently inhibit the α, β, and γ isoforms of kinase PI3K (IC50 = 33 nM, IC50 = 661 nM, and Kd = 25 nM, respectively), and mTOR (IC50 = 89 nM) [123]. Interestingly, both mTOR and PI3Kγ are two of the targets that demonstrated a significant correlation with the activity on PC cells, based on the analyses conducted on the activity records obtained from ChEMBL (see Table 1). The ability of this compound to inhibit different key players of the PI3K/AKT/mTOR signaling cascade is of particular relevance for PC treatment, with this signaling pathway being one of the most commonly disrupted in cancer, and being crucial for cell motility, growth, survival, metabolism, and the establishment of drug resistance [46,152]. Furthermore, inhibitors of the PI3K/mTOR/AKT pathway have been proven to be effective against several different PC cells [153,154], including DU-145. Bimiralisib is in phase II clinical trials for the treatment of head and neck cancer (clinicaltrials.gov ID: NCT03740100), glioblastoma (clinicaltrials.gov ID: NCT02850744), lymphoma (clinicaltrials.gov ID: NCT02249429), and breast cancer (clinicaltrials.gov ID: NCT02723877). Importantly, this compound has demonstrated potent antiproliferative effects on PC-3 mice xenografts, and it also has low nanomolar activity on mTOR and sub-micromolar activity on PI3Kγ (Table 2). Moreover, it can also cross the blood–brain barrier (BBB) [155], making it one of the most promising molecules for drug repurposing on PC, especially for treatment of PC patients with brain metastases. Based on the safety profile of this molecule, specific clinical trials on PC could reveal its potential as a therapeutic agent for this disease.
Onatasertib (CC-223, DB12570) is an orally available, low nanomolar inhibitor of mTOR (IC50 = 10 nM) [156], with putative antineoplastic activity [133,157,158]. The inhibition of mTOR by Onatasertib has been reported to induce tumor cell apoptosis and to decrease tumor cell proliferation [124,159,160,161]. Onatasertib is currently in phase II trials for the treatment of multiple myeloma, non-Hodgkin’s lymphoma (clinicaltrials.gov ID: NCT01177397), and hepatocellular carcinoma (clinicaltrials.gov ID: NCT03591965) and is in phase I trials for non-small-cell lung cancer (clinicaltrials.gov ID: NCT01545947). Of note, CC-223 has also reported sub-micromolar activity on Vascular Endothelial Growth Factor Receptor 3 (VEGR3, IC50 = 651.0 nM) [156], a target implicated in PC development [162]. Moreover, Onatasertib has also been found to inhibit PC-3 proliferation both in vitro and in vivo [124]. Interestingly, Onatasertib has also been found to exhibit antitumor effects in a murine xenograft model of glioblastoma multiforme (U87MG cells) [124], demonstrating the ability to cross the blood–brain barrier. Considering the reported multi-target activity profile, the results observed in the in vitro and in vivo data, and the tolerability observed in patients at the clinical trials [163,164,165], Onatasertib can be considered a valuable candidate for fast repurposing against PC, especially for patients that present metastases spread at different sites.
VS-5584 (DB12986) is a potent and selective dual inhibitor of PI3K/mTOR, with potential antineoplastic activity. In particular, VS-5584 has reported low nanomolar activity against α, β, γ, and δ PI3K isoforms (IC50 = 16 nM, IC50 = 68 nM, IC50 = 25 nM, IC50 = 42 nM, respectively) [126]. Moreover, it was demonstrated to inhibit mTOR low in the low nanomolar range (IC50 = 37 nM), and PC-3 cells in in vitro (IC50 = 180 nM) and in vivo experiments [125,126]. Although phase I clinical trials of VS-5584 against mesothelioma (clinicaltrials.gov ID: NCT02372227) and non-hematological malignancies or lymphoma (clinicaltrials.gov ID: NCT01991938) have been terminated due to study de-prioritization issues, the activity profiles and good tolerability of VS-5584 make it an optimal candidate for PC treatment [125,126].
BIIB021 (DB12359) is a potent, orally active inhibitor of Heat Shock Protein 90 (Hsp90) with anticancer activity [157,158,166]; BIIB021 has been demonstrated to inhibit both Hsp90 isoforms α and β (IC50 = 5.1 nM, IC50 = 17 nM, respectively) [167,168]. The ability of this compound to inhibit Hsp90 is of primary interest for prostate cancer treatment, considering the role of this target in PC development and progression [50]. Moreover, this compound also inhibits the mitochondrial isoform TRAP1 (Hsp75kDa, Ki = 62 nM), which is often upregulated in prostate cancer [169,170]. Interestingly, BIIB021 has been extensively evaluated in clinical trials for breast cancer (phase II, clinicaltrials.gov ID: NCT01004081) and solid tumors (phase I, clinicaltrials.gov ID: NCT00618735). BIIB021 showed also growth inhibition activity on PC-3 cells in the nanomolar range in vitro and tumor growth inhibition on CW22R xenografts in vivo [127,128]. Taking into account its multi-target activity profile and the potent antiproliferative effects on different PC cell models, BIIB021 can be considered another promising candidate for fast repurposing against prostate cancer.
Adapalene (CD-271, DB00210) is a synthetic retinoid that activates the retinoic acid receptor gamma subtype (RARγ), which is currently approved for topical treatment of Acne Vulgaris [171]. Adapalene exhibits Ki = 130 nM against RARy, and Ki = 1100 nM and Ki = 34 nM against RARα and RARβ, respectively [172]. Moreover, retinoic acid receptors (RARs) are known to play a role in cell differentiation and tumor suppression; therefore, they could represent relevant targets for the treatment of prostate cancer [173,174,175,176,177]. According to recent research data, Adapalene exerts strong in vitro and in vivo antiproliferative effects on different PC cell lines (i.e., DU-145, PC-3, RM-1), by inducing DNA damage, S-phase cell cycle arrest, and apoptosis [129,130]. One of the major limitations of Adapalene is the poor water solubility, which hampers in vivo bioavailability, with this effect being common to retinoids. However, recent studies have demonstrated that advanced delivery systems can significantly improve distribution, delivery, and efficacy of insoluble drugs, such as Adapalene [178], making them valuable therapeutics also for systemic treatment. On these premises, Adapalene can be a valuable candidate for fast repurposing against PC.
Picropodophyllin (AXL1717, PPP, DB12802) is a potent inhibitor of the insulin-like growth factor 1 receptor (IGF1R), which is overexpressed in a variety of human cancers, and it plays a critical role in the growth and survival of many types of tumors [179]. PPP is the cis-isomer of podophyllotoxin (PPT), which retains the antineoplastic activity of PPT, but without the toxicity derived from the β-tubulin binding and the modulation of DNA Topoisomerase II [180,181]. According to literature data, PPP potently suppresses tumor cell proliferation and induction of apoptosis in several types of tumor cells [182,183,184,185]. Moreover, a recent study has also demonstrated that PPP can induce antiproliferative effects, promoting cell cycle arrest and apoptosis on LNCaP and DU-145 cells, through the production of ROS species and the inhibition of the PI3K/AKT signaling pathway [131]. PPP was also tested in vitro and in vivo on PC-3 cells and other IGF1R-bearing cancerous cells, demonstrating good antiproliferative and tumor-inhibiting effects [133]. Moreover, Picropodophyllin showed also good BBB permeability in intracerebral xenograft models of glioblastoma [186], suggesting potential efficacy against PC metastases in the brain. PPP has completed phase II trials on squamous cell carcinoma (clinicaltrials.gov ID: NCT01561456) and non-small-cell lung cancer (clinicaltrials.gov ID: NCT01466647), demonstrating good tolerability. The observed in vitro and in vivo efficacy, in combination with the already established safety profile, make PPP an attractive candidate for fast repurposing against prostate cancer.
VS-4718 (DB15273) is an inhibitor of Focal Adhesion Kinase (FAK, IC50 = 1.5 nM) [134], which is a target often overexpressed in PC, and it is involved in the regulation activation of several major tumorigenic pathways behind pathological growth and survival of prostate cancer [187]. While phase I trials of VS-4718 against metastatic non-hematologic malignancies (clinicaltrials.gov ID: NCT01849744) and advanced pancreatic cancer (clinicaltrials.gov ID: NCT02651727) have been discontinued due to study de-prioritization issues, it remains a valuable repurposing candidate for prostate cancer treatment, according to its antiproliferative activity on PC-3 cells and the low nanomolar activity on the FAK kinase.
BMS-214662 (DB12234) is a potent inhibitor of the enzyme farnesyltransferase, which is a target involved in the post-translational regulation of several proteins involved in signal transduction [188]. According to recent data, BMS-214662 presents low micromolar antiproliferative activity on PC-3 and LNCaP cells [135]. Results of phase I trials of this compound against acute promyelocytic leukemia (clinicaltrials.gov ID: NCT00006213) and adult solid tumors in combination with Paclitaxel (clinicaltrials.gov ID: NCT00006018) including 2 PC patients [136] suggest that further clinical assessments on BMS-214662, either as a single agent or as a combination of drugs, could support its repurposing on PC.
Another valuable candidate is Flubendazole (DB08974), which was demonstrated to potently inhibit the proliferation of CRPC cells in different in vitro and in vivo models [137]. This compound acts by inducing cell cycle arrest in the G2/M phase while promoting ferroptosis in CRPC cells [137]. In PC-3 and DU-145, a cell viability decrease was observed after treatment with 0.1 µM of Flubendazole [137]. Moreover, it also strongly upregulates the expression of p53 and its downstream effector p21, which are genes frequently mutated in several types of cancer [189]. Of note, Flubendazole has been found to exert synergistic effects with 5-fluorouracil against CRPC in PC-3 xenograft models [137], and it has been reported to exert anti-tumor effects on different types of cancers [190]. Notably, this compound is closely related to mebendazole, another anthelmintic drug that has recently been repurposed in combination with docetaxel against prostate cancer [191]. On these premises, Flubendazole is worth further assessments for potential repurposing against PC.
Another broad-spectrum anthelminthic agent potentially repositionable against PC is Albendazole (DB00518), which showed potent antiproliferative effects toward PC-3 and DU-145 at 0.1 µM concentration and 0.5 µM against LNCaP [138]. Unfortunately, recent studies showed that high doses of Albendazole for a prolonged time could result in toxicity according to results of the phase I trial of this compound including 2 PC patients, suggesting frequent dose monitoring [139]. However, the fact that this compound presents potent PC antiproliferative activity and passed all phases of clinical trials as an antihelmintic drug makes it very interesting for repositioning against prostate cancer.
3. Materials and Methods
3.1. Curation of the Ligands and Targets Dataset
Bioactivity records of molecules assayed on RWPE1 (androgen responsive adult b prostatic epithelial cells) and malignant prostate cell lines PC-3, DU-145, LNCaP, LNCaP clone FGC, Vcap, 22Rv-1, LAPC4, and PWR1E were first acquired from the ChEMBL database (https://www.ebi.ac.uk/chembl/, accessed on 1 November 2021) [25,192]. Then, the molecules were filtered to retain only those with bioactivity data expressed by means of: (i) “Standard Type” equal to GI50, EC50, or IC50; (ii) “Standard Relation” equal to “<”, “>”, or “=”; (iii) “Standard Unit” in the nanomolar range (i.e., “nM”). For each ligand, duplicate records deriving from multiple assays conducted under different experimental conditions were removed, retaining the one with the best activity value (i.e., 54871 unique compounds). Finally, only ChEMBL compounds that showed an activity below 1 µM on at least one PC cell line (herein considered as “highly active”) and a molecular weight in the range of 180–850 Da were retained for the subsequent calculations. The performed filtration resulted in 6626 compounds, each of them with reported activity data on at least one PC cell line.
Compounds reported in the DrugBank database (https://go.drugbank.com/, accessed on 1 November 2021) [24] were downloaded (i.e.,11912 ligands) and filtered to remove those already present in the curated dataset of ChEMBL ligands. The two datasets were compared by means of their molecular properties (i.e., LogP, polar surface area (PSA), number of hydrogen bond donors (HBD) and acceptors (HBA), molecular weight (MW), volume, and ovality), calculated by using the “RDKit Descriptor Calculation” and “CDK Molecular properties” KNIME nodes [193]. Then, the curated datasets of ChEMBL and DrugBank ligands were prepared for the similarity assessments, as follows. Canonical smiles were first generated for all the compounds. Then, hydrogen atoms were added to the structures of the compounds and salt counterions were removed. This phase of the ligands preparation was performed with the OpenEye Python toolkits [194]. The preparation of the compounds for the 3D similarity estimations required also a further step of conformational sampling, which was performed with the OMEGA2 software, adopting the same parameters used in our previous study [195,196].
3.2. Ligand-Based Calculations
The similarity degree of the ligands reported in the curated ChEMBL and DrugBank datasets were evaluated as follows. First, an all-against-all 2D-fingerprint-based similarity assessment was performed by using an in-house-developed Python script. The degree of 2D similarity was calculated according to the MACCS and circular (i.e., ECFP4) type of fingerprints available on OpenEye Python toolkits [194], and by using the Tanimoto coefficient (Tc) as an index for ligands similarity measurement. Then, the results of the 2D similarity calculations were filtered to retain only those with MACCS and ECFP4 Tanimoto coefficients equal to or higher than 0.8 and 0.3, respectively, in agreement with previously reported studies [196]. Afterward, extensive 3D shape and atom type similarity calculations of compounds resulting as significantly similar in the 2D estimations were performed by using the ROCS software [117]. In this case, the similarity degree between ligands was assessed according to the Tanimoto Combo (TCc) coefficient [117]. Finally, the results of the 3D-shape-based similarity calculations were filtered to retain only similarity records with a TCc ≥ 1.5, which is a commonly accepted threshold of similarity [196].
3.3. Data Integration
The results obtained in the similarity estimations were associated with information on targets and PC-cell-based bioactivity data and filtered to retain only DrugBank ligands that resulted as significantly similar to ChEMBL compounds with potent antiproliferative activity against prostate cancer cells. Our investigations focused on compounds that already possessed annotations on prostate cancer according to literature data, which are more likely to be assessed in clinical trials and potentially repurposed against PC. In particular, data related to trials on PC or solid tumors including patients with prostate cancer were retrieved from the clinicaltrials.gov website (https://clinicaltrials.gov/, accessed on 1 November 2021) [29]. In addition, bioactivity records on molecular targets reported on ChEMBL were also associated with the DrugBank ligands that emerged from the similarity estimations. In this phase, DrugBank ligands were associated with activity data deriving from target-based experiments on human proteins and expressed by means of the IC50, Ki, Kd, or EC50 types. Duplicate records reported for the same targets were removed, retaining the best value, and the resulting activity data were classified as “highly active” (IC50, Ki, Kd, or EC50 ≤ 1 µM), “scarcely active” (1 µM ≥ IC50, Ki, Kd, or EC50 ≤ 10 µM), and “inactive” (IC50, Ki, Kd, or EC50 ≥ 10 µM). All steps of data curation were performed with the Pandas Python library [197].
3.4. Analysis of Biological Target Activity Annotations
An analysis of the activity annotations on biological targets associated with ChEMBL compounds with known antiproliferative records on PC-3, DU-145, and LNCaP PC cancer cell lines (54773 unique molecules) was also carried out. In particular, bioactivity data related to these compounds were first retrieved from ChEMBL. Then, these records were filtered to retain only those with bioactivity data expressed by means of: (i) “Standard Type” equal to IC50, Ki, Kd, or EC50; (ii) “Standard Relation” equal to “=”; (iii) “Standard Unit” in the nanomolar range (i.e., “nM”); (iv) “Target type” equal to “Single protein”; (v) “Target Organism” equal to “Homo Sapiens”; vi) “Assay Type” equal to “B” (i.e., binding assay type). Duplicate records reported for the same target were removed, retaining the best value. Afterward, only targets with at least 10 bioactivity annotations were retained. The correlation between the activity on the target and antiproliferative activity on PC cells was assessed with Spearman’s Rho (ρs) correlation coefficient, which was calculated with the Scipy Python library [198]. A p-value equal to or lower than 0.05 was considered statistically significant for the analyses. Biological targets with ρs ≥ 0.40 were finally retained [199].
4. Conclusions
In this study, an integrated in silico workflow based on extensive similarity estimations, and data integration and analysis were devised in the search for compounds with favorable safety and tolerability profiles for repurposing against PC therapy. In particular, the candidates were first selected according to their similarity degree with respect to potent molecules with reported potent antiproliferative activity against PC cells (below 1 µM).
Moreover, an analysis of the activity annotations reported for compounds also allowed us to identify biological targets whose modulation presents a significant correlation with known antiproliferative activities reported on different PC cell lines (see Table 1 and Figure 2). Of note, some of the identified biological target-cell activity associations have previously been reported in the literature, while others have never been described, potentially providing clues for future drug discovery efforts against PC.
Then, extensive integration and analyses on compounds annotations reported in different databases and literature searches allowed us to select 10 particularly interesting candidates for fast repurposing against prostate cancer (see Table 2). The identified candidates have already been assessed in clinical trials for other pathologies, and most of them showed good safety and tolerability profiles when tested on patients. Remarkably, they have potent in vitro and in vivo activity on PC cells, as well as activity on targets relevant to PC. Some of the identified compounds have demonstrated to be able to cross the BBB, thus being potentially useful for the treatment of forms of prostate cancer that metastasize to neuronal tissues. These compounds represent ideal candidates for fast enrollment in clinical trials against PC, either alone or in combination with other drugs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24043135/s1.
Author Contributions
Conceptualization, G.R.; data curation, analysis, visualization, L.B.; methodology, L.B. and L.P. The manuscript was drafted, reviewed, and edited through the contributions of all authors. All authors have read and agreed to the published version of the manuscript.
Funding
The research leading to these results has received funding from AIRC (Fondazione Italiana per la Ricerca sul Cancro) under IG 2019- I.D. 23635 project—P.I. Giulio Rastelli, and by a PhD fellowship from the Regione Emilia Romagna on “Drug Repurposing against Advanced-Stage Prostate Cancer through Big Data Analysis and Artificial Intelligence” to L.B.; L.P. would like to thank the Italian funding programme Fondo Sociale Europeo REACT-EU—PON “Ricerca e Innovazione” 2014–2020—Azione IV.4 “Dottorati e contratti di ricerca su tematiche dell’innovazione” for supporting his research. The APC was funded by MDPI.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data presented in this study is available within the article or supplementary material. Further inquiries can be provided by the authors upon request.
Acknowledgments
We thank OpenEye Scientific Software, Inc., for a free academic license.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wong, M.C.S.; Goggins, W.B.; Wang, H.H.X.; Fung, F.D.H.; Leung, C.; Wong, S.Y.S.; Ng, C.F.; Sung, J.J.Y. Global Incidence and Mortality for Prostate Cancer: Analysis of Temporal Patterns and Trends in 36 Countries. Eur. Urol. 2016, 70, 862–874. [Google Scholar] [CrossRef]
- Howrey, B.T.; Kuo, Y.-F.; Lin, Y.-L.; Goodwin, J.S. The Impact of PSA Screening on Prostate Cancer Mortality and Overdiagnosis of Prostate Cancer in the United States. J. Gerontol. Ser. A 2013, 68, 56–61. [Google Scholar] [CrossRef]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate Cancer. Nat. Rev. Dis. Prim. 2021, 7, 9. [Google Scholar] [CrossRef]
- Sammon, J.D.; Abdollah, F.; D’Amico, A.; Gettman, M.; Haese, A.; Suardi, N.; Vickers, A.; Trinh, Q.-D. Predicting Life Expectancy in Men Diagnosed with Prostate Cancer. Eur. Urol. 2015, 68, 756–765. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Kyriakopoulos, C.E.; Chen, Y.-H.; Carducci, M.A.; Liu, G.; Jarrard, D.F.; Hahn, N.M.; Shevrin, D.H.; Dreicer, R.; Hussain, M.; Eisenberger, M.; et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer: Long-Term Survival Analysis of the Randomized Phase III E3805 CHAARTED Trial. J. Clin. Oncol. 2018, 36, 1080–1087. [Google Scholar] [CrossRef]
- Fang, D.; Zhou, L. Androgen Deprivation Therapy in Nonmetastatic Prostate Cancer Patients: Indications, Treatment Effects, and New Predictive Biomarkers. Asia-Pac. J. Clin. Oncol. 2019, 15, 108–120. [Google Scholar] [CrossRef]
- Banapour, P.; Schumacher, A.; Lin, J.C.; Finley, D.S. Radical Prostatectomy and Pelvic Lymph Node Dissection in Kaiser Permanente Southern California: 15-Year Experience. Perm. J. 2019, 23, 17–233. [Google Scholar] [CrossRef]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Théodore, C.; James, N.D.; Turesson, I.; et al. Docetaxel plus Prednisone or Mitoxantrone plus Prednisone for Advanced Prostate Cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef]
- Gillessen, S.; Attard, G.; Beer, T.M.; Beltran, H.; Bjartell, A.; Bossi, A.; Briganti, A.; Bristow, R.G.; Chi, K.N.; Clarke, N.; et al. Management of Patients with Advanced Prostate Cancer: Report of the Advanced Prostate Cancer Consensus Conference 2019. Eur. Urol. 2020, 77, 508–547. [Google Scholar] [CrossRef]
- Sayegh, N.; Swami, U.; Agarwal, N. Recent Advances in the Management of Metastatic Prostate Cancer. JCO Oncol. Pract. 2022, 18, 45–55. [Google Scholar] [CrossRef]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate Cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef]
- Beatson, E.L.; Chau, C.H.; Price, D.K.; Figg, W.D. PARP Inhibitors on the Move in Prostate Cancer: Spotlight on Niraparib & Update on PARP Inhibitor Combination Trials. Am. J. Clin. Exp. Urol. 2022, 10, 252–257. [Google Scholar] [CrossRef]
- Yamada, Y.; Beltran, H. The Treatment Landscape of Metastatic Prostate Cancer. Cancer Lett. 2021, 519, 20–29. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Kumar, R.; Harilal, S.; Gupta, S.V.; Jose, J.; Thomas Parambi, D.G.; Uddin, M.S.; Shah, M.A.; Mathew, B. Exploring the New Horizons of Drug Repurposing: A Vital Tool for Turning Hard Work into Smart Work. Eur. J. Med. Chem. 2019, 182, 111602. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Dickson, M.; Gagnon, J.P. Key Factors in the Rising Cost of New Drug Discovery and Development. Nat. Rev. Drug Discov. 2004, 3, 417–429. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Demus, T.; Moubarak, M.M.; Daher, D.; Alvarez Moreno, J.C.; Polit, F.; Lopez, O.; Merhe, A.; Abou-Kheir, W.; Nieder, A.M.; et al. Overcoming Drug Resistance in Advanced Prostate Cancer by Drug Repurposing. Med. Sci. 2022, 10, 15. [Google Scholar] [CrossRef]
- Anighoro, A.; Bajorath, J.; Rastelli, G. Polypharmacology: Challenges and Opportunities in Drug Discovery. J. Med. Chem. 2014, 57, 7874–7887. [Google Scholar] [CrossRef]
- Novac, N. Challenges and Opportunities of Drug Repositioning. Trends Pharmacol. Sci. 2013, 34, 267–272. [Google Scholar] [CrossRef]
- Rastelli, G.; Pellati, F.; Pinzi, L.; Gamberini, M.C. Repositioning Natural Products in Drug Discovery. Molecules 2020, 25, 1154. [Google Scholar] [CrossRef]
- March-Vila, E.; Pinzi, L.; Sturm, N.; Tinivella, A.; Engkvist, O.; Chen, H.; Rastelli, G. On the Integration of In Silico Drug Design Methods for Drug Repurposing. Front. Pharmacol. 2017, 8, 298. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Qiu, Y.; Chen, Y. Therapeutic Target Database Update 2022: Facilitating Drug Discovery with Enriched Comparative Data of Targeted Agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef]
- Home-ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/home (accessed on 4 December 2022).
- Costa, F.F. Big data in biomedicine. Drug Discovery Today. 2014, 19, 433–440. [Google Scholar] [CrossRef]
- Pinzi, L.; Lherbet, C.; Baltas, M.; Pellati, F.; Rastelli, G. In Silico Repositioning of Cannabigerol as a Novel Inhibitor of the Enoyl Acyl Carrier Protein (ACP) Reductase (InhA). Molecules 2019, 24, E2567. [Google Scholar] [CrossRef]
- Carrella, D.; Manni, I.; Tumaini, B.; Dattilo, R.; Papaccio, F.; Mutarelli, M.; Sirci, F.; Amoreo, C.A.; Mottolese, M.; Iezzi, M.; et al. Computational Drugs Repositioning Identifies Inhibitors of Oncogenic PI3K/AKT/P70S6K-Dependent Pathways among FDA-Approved Compounds. Oncotarget 2016, 7, 58743–58758. [Google Scholar] [CrossRef]
- Maruca, A.; Rocca, R.; Catalano, R.; Mesiti, F.; Costa, G.; Lanzillotta, D.; Salatino, A.; Ortuso, F.; Trapasso, F.; Alcaro, S.; et al. Natural Products Extracted from Fungal Species as New Potential Anti-Cancer Drugs: A Structure-Based Drug Repurposing Approach Targeting HDAC7. Molecules 2020, 25, 5524. [Google Scholar] [CrossRef]
- Pinzi, L.; Tinivella, A.; Gagliardelli, L.; Beneventano, D.; Rastelli, G. LigAdvisor: A Versatile and User-Friendly Web-Platform for Drug Design. Nucleic Acids Res. 2021, 49, W326–W335. [Google Scholar] [CrossRef]
- Drugpro. Available online: https://drugrepo.org/ (accessed on 4 December 2022).
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-Generation Drug Library and Information Resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef]
- Pinzi, L.; Tinivella, A.; Caporuscio, F.; Rastelli, G. Drug Repurposing and Polypharmacology to Fight SARS-CoV-2 Through Inhibition of the Main Protease. Front. Pharmacol. 2021, 12, 636989. [Google Scholar] [CrossRef]
- Masoudi-Sobhanzadeh, Y.; Omidi, Y.; Amanlou, M.; Masoudi-Nejad, A. Drug Databases and Their Contributions to Drug Repurposing. Genomics 2020, 112, 1087–1095. [Google Scholar] [CrossRef]
- Matsson, P.; Kihlberg, J. How Big Is Too Big for Cell Permeability? J. Med. Chem. 2017, 60, 1662–1664. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Saranyutanon, S.; Deshmukh, S.K.; Dasgupta, S.; Pai, S.; Singh, S.; Singh, A.P. Cellular and Molecular Progression of Prostate Cancer: Models for Basic and Preclinical Research. Cancers 2020, 12, 2651. [Google Scholar] [CrossRef]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and Characterization of a Human Prostatic Carcinoma Cell Line (PC-3). Investig. Urol. 1979, 17, 16–23. [Google Scholar]
- Stone, K.R.; Mickey, D.D.; Wunderli, H.; Mickey, G.H.; Paulson, D.F. Isolation of a Human Prostate Carcinoma Cell Line (DU 145). Int. J. Cancer 1978, 21, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Horoszewicz, J.S.; Leong, S.S.; Kawinski, E.; Karr, J.P.; Rosenthal, H.; Chu, T.M.; Mirand, E.A.; Murphy, G.P. LNCaP Model of Human Prostatic Carcinoma. Cancer Res. 1983, 43, 1809–1818. [Google Scholar] [PubMed]
- Bitting, R.L.; Armstrong, A.J. Targeting the PI3K/Akt/MTOR Pathway in Castration-Resistant Prostate Cancer. Endocr. Relat. Cancer 2013, 20, R83–R99. [Google Scholar] [CrossRef] [PubMed]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-MTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, Y.S.; Kim, D.Y.; So, I.; Jeon, J.-H. PI3K Pathway in Prostate Cancer: All Resistant Roads Lead to PI3K. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2018, 1870, 198–206. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/MTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef]
- Jego, G.; Hazoumé, A.; Seigneuric, R.; Garrido, C. Targeting Heat Shock Proteins in Cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef]
- Taldone, T.; Gozman, A.; Maharaj, R.; Chiosis, G. Targeting Hsp90: Small-Molecule Inhibitors and Their Clinical Development. Curr. Opin. Pharmacol. 2008, 8, 370–374. [Google Scholar] [CrossRef]
- Gomez-Monterrey, I.; Sala, M.; Musella, S.; Campiglia, P. Heat Shock Protein 90 Inhibitors as Therapeutic Agents. Recent Pat. Anticancer Drug Discov. 2012, 7, 313–336. [Google Scholar] [CrossRef]
- Pinzi, L.; Benedetti, R.; Altucci, L.; Rastelli, G. Design of Dual Inhibitors of Histone Deacetylase 6 and Heat Shock Protein 90. ACS Omega 2020, 5, 11473–11480. [Google Scholar] [CrossRef]
- Anighoro, A.; Pinzi, L.; Marverti, G.; Bajorath, J.; Rastelli, G. Heat Shock Protein 90 and Serine/Threonine Kinase B-Raf Inhibitors Have Overlapping Chemical Space. RSC Adv. 2017, 7, 31069–31074. [Google Scholar] [CrossRef]
- Cairns, B.R. Emerging Roles for Chromatin Remodeling in Cancer Biology. Trends Cell Biol. 2001, 11, S15–S21. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Gupta, S. The Role of Histone Deacetylases in Prostate Cancer. Epigenetics 2008, 3, 300–309. [Google Scholar] [CrossRef]
- Li, G.; Tian, Y.; Zhu, W.-G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef] [PubMed]
- Willis-Martinez, D.; Richards, H.W.; Timchenko, N.A.; Medrano, E.E. Role of HDAC1 in senescence, aging, and cancer. Exp. Gerontol. 2010, 45, 279–285. [Google Scholar] [CrossRef]
- Weichert, W.; Röske, A.; Gekeler, V.; Beckers, T.; Stephan, C.; Jung, K.; Fritzsche, F.R.; Niesporek, S.; Denkert, C.; Dietel, M.; et al. Histone Deacetylases 1, 2 and 3 Are Highly Expressed in Prostate Cancer and HDAC2 Expression Is Associated with Shorter PSA Relapse Time after Radical Prostatectomy. Br. J. Cancer 2008, 98, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Halkidou, K.; Gaughan, L.; Cook, S.; Leung, H.Y.; Neal, D.E.; Robson, C.N. Upregulation and Nuclear Recruitment of HDAC1 in Hormone Refractory Prostate Cancer. Prostate 2004, 59, 177–189. [Google Scholar] [CrossRef]
- Kim, N.H.; Kim, S.-N.; Kim, Y.K. Involvement of HDAC1 in E-Cadherin Expression in Prostate Cancer Cells; Its Implication for Cell Motility and Invasion. Biochem. Biophys. Res. Commun. 2011, 404, 915–921. [Google Scholar] [CrossRef]
- Kuo, P.-L.; Shen, K.-H.; Hung, S.-H.; Hsu, Y.-L. CXCL1/GROα Increases Cell Migration and Invasion of Prostate Cancer by Decreasing Fibulin-1 Expression through NF-ΚB/HDAC1 Epigenetic Regulation. Carcinogenesis 2012, 33, 2477–2487. [Google Scholar] [CrossRef]
- Wang, Z.; Qin, G.; Zhao, T.C. HDAC4: Mechanism of Regulation and Biological Functions. Epigenomics 2014, 6, 139–150. [Google Scholar] [CrossRef]
- Jin, K.; Zhao, W.; Xie, X.; Pan, Y.; Wang, K.; Zhang, H. MiR-520b Restrains Cell Growth by Targeting HDAC4 in Lung Cancer. Thorac. Cancer 2018, 9, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.-H.; Wang, C.-Y.; Zhang, W.-L.; Zhang, J.-T.; Yuan, C.-H.; Zhao, P.-W.; Lin, Y.-Y.; Hong, S.; Li, C.-Y.; Wang, L. Histone Deacetylase HDAC4 Promotes Gastric Cancer SGC-7901 Cells Progression via P21 Repression. PLoS ONE 2014, 9, e98894. [Google Scholar] [CrossRef] [PubMed]
- Mottet, D.; Pirotte, S.; Lamour, V.; Hagedorn, M.; Javerzat, S.; Bikfalvi, A.; Bellahcène, A.; Verdin, E.; Castronovo, V. HDAC4 Represses P21WAF1/Cip1 Expression in Human Cancer Cells through a Sp1-Dependent, P53-Independent Mechanism. Oncogene 2009, 28, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Byun, D.-S.; Nasser, S.; Murray, L.B.; Ayyanar, K.; Arango, D.; Figueroa, M.; Melnick, A.; Kao, G.D.; Augenlicht, L.H.; et al. HDAC4 Promotes Growth of Colon Cancer Cells via Repression of P21. Mol. Biol. Cell 2008, 19, 4062–4075. [Google Scholar] [CrossRef] [PubMed]
- Halkidou, K.; Cook, S.; Leung, H.Y.; Neal, D.E.; Robson, C.N. Nuclear Accumulation of Histone Deacetylase 4 (HDAC4) Coincides with the Loss of Androgen Sensitivity in Hormone Refractory Cancer of the Prostate. Eur. Urol. 2004, 45, 382–389. [Google Scholar] [CrossRef]
- Ren, G.; Zhang, G.; Dong, Z.; Liu, Z.; Li, L.; Feng, Y.; Su, D.; Zhang, Y.; Huang, B.; Lu, J. Recruitment of HDAC4 by Transcription Factor YY1 Represses HOXB13 to Affect Cell Growth in AR-Negative Prostate Cancers. Int. J. Biochem. Cell Biol. 2009, 41, 1094–1101. [Google Scholar] [CrossRef]
- Wojcik, E.J.; Buckley, R.S.; Richard, J.; Liu, L.; Huckaba, T.M.; Kim, S. Kinesin-5: Cross-Bridging Mechanism to Targeted Clinical Therapy. Gene 2013, 531, 133–149. [Google Scholar] [CrossRef]
- Gu, X.; Zhu, Q.; Tian, G.; Song, W.; Wang, T.; Wang, A.; Chen, X.; Qin, S. KIF11 Manipulates SREBP2-Dependent Mevalonate Cross Talk to Promote Tumor Progression in Pancreatic Ductal Adenocarcinoma. Cancer Med. 2022, 11, 3282–3295. [Google Scholar] [CrossRef]
- Han, Y.; Chen, J.; Wei, D.; Wang, B. KIF11 Is a Promising Therapeutic Target for Thyroid Cancer Treatment. Comput. Math. Methods Med. 2022, 2022, e6426800. [Google Scholar] [CrossRef]
- Ling, J.; Wang, Y.; Ma, L.; Zheng, Y.; Tang, H.; Meng, L.; Zhang, L. KIF11, a plus End-Directed Kinesin, as a Key Gene in Benzo(a)Pyrene-Induced Non-Small Cell Lung Cancer. Environ. Toxicol. Pharmacol. 2022, 89, 103775. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, G.; Cui, S.; Du, G. Upregulation of KIF11 in TP53 Mutant Glioma Promotes Tumor Stemness and Drug Resistance. Cell. Mol. Neurobiol. 2022, 42, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, Y.; Yi, L.; Gao, Z.; Lou, M.; Yuan, K. High KIF11 Expression Is Associated with Poor Outcome of NSCLC. Tumori J. 2022, 108, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-F.; Zeng, H.-J.; Shan, Z.; Ye, R.-Y.; Cheang, T.-Y.; Zhang, Y.-J.; Lu, S.-H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of Kinesin Superfamily Members as Prognostic Biomarkers of Breast Cancer. Cancer Cell Int. 2020, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, S.; Liu, B.; Wei, S.; Wang, T.; Li, T.; Lin, J.; Ni, X. KIF11: A Potential Prognostic Biomarker for Predicting Bone Metastasis-free Survival of Prostate Cancer. Oncol. Lett. 2022, 24, 312. [Google Scholar] [CrossRef]
- Piao, X.-M.; Byun, Y.J.; Jeong, P.; Ha, Y.-S.; Yoo, E.S.; Yun, S.J.; Kim, W.-J. Kinesin Family Member 11 MRNA Expression Predicts Prostate Cancer Aggressiveness. Clin. Genitourin. Cancer 2017, 15, 450–454. [Google Scholar] [CrossRef]
- Xing, N.-D.; Ding, S.-T.; Saito, R.; Nishizawa, K.; Kobayashi, T.; Inoue, T.; Oishi, S.; Fujii, N.; Lv, J.-J.; Ogawa, O.; et al. A Potent Chemotherapeutic Strategy in Prostate Cancer: S-(Methoxytrityl)--Cysteine, a Novel Eg5 Inhibitor. Asian J. Androl. 2011, 13, 236–241. [Google Scholar] [CrossRef]
- Cochran, J.C.; Gatial, J.E.; Kapoor, T.M.; Gilbert, S.P. Monastrol Inhibition of the Mitotic Kinesin Eg5 *. J. Biol. Chem. 2005, 280, 12658–12667. [Google Scholar] [CrossRef]
- Lad, L.; Luo, L.; Carson, J.D.; Wood, K.W.; Hartman, J.J.; Copeland, R.A.; Sakowicz, R. Mechanism of Inhibition of Human KSP by Ispinesib. Biochemistry 2008, 47, 3576–3585. [Google Scholar] [CrossRef]
- Blagden, S.; Molife, L.R.; Seebaran, A.; Payne, M.; Reid, A.H.; Protheroe, A.S.; Vasist, L.S.; Williams, D.D.; Bowen, C.; Kathman, S.J.; et al. A phase I trial of ispinesib, a kinesin spindle protein inhibitor, with docetaxel in patients with advanced solid tumours. Br. J. Cancer 2008, 98, 894–899. [Google Scholar] [CrossRef]
- Beer, T.M.; Goldman, B.; Synold, T.W.; Ryan, C.W.; Vasist, L.S.; Van Veldhuizen, P.J.; Dakhil, S.R.; Lara, P.N.; Drelichman, A.; Hussain, M.H.A.; et al. Southwest Oncology Group Phase II Study of Ispinesib in Androgen-Independent Prostate Cancer Previously Treated with Taxanes. Clin. Genitourin. Cancer 2008, 6, 103–109. [Google Scholar] [CrossRef]
- Davis, D.A.; Sarkar, S.H.; Hussain, M.; Li, Y.; Sarkar, F.H. Increased Therapeutic Potential of an Experimental Anti-Mitotic Inhibitor SB715992 by Genistein in PC-3 Human Prostate Cancer Cell Line. BMC Cancer 2006, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Koller, E.; Fazli, L.; Gleave, M.E. Effects of Eg5 Knockdown on Human Prostate Cancer Xenograft Growth and Chemosensitivity. Prostate 2008, 68, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Panner, A.; Wurster, R.D. T-Type Calcium Channels and Tumor Proliferation. Cell Calcium 2006, 40, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Ho, C.; Ohe-Toyota, M.; Baylin, S.B.; Issa, J.-P.J. Inactivation of CACNA1G, a T-Type Calcium Channel Gene, by Aberrant Methylation of Its 5′ CpG Island in Human Tumors1. Cancer Res. 1999, 59, 4535–4541. [Google Scholar]
- Panner, A.; Cribbs, L.L.; Zainelli, G.M.; Origitano, T.C.; Singh, S.; Wurster, R.D. Variation of T-Type Calcium Channel Protein Expression Affects Cell Division of Cultured Tumor Cells. Cell Calcium 2005, 37, 105–119. [Google Scholar] [CrossRef]
- Barnes, S.; Haynes, L.W. Low-Voltage-Activated Calcium Channels in Human Retinoblastoma Cells. Brain Res. 1992, 598, 19–22. [Google Scholar] [CrossRef]
- Ohkubo, T.; Yamazaki, J. T-Type Voltage-Activated Calcium Channel Cav3.1, but Not Cav3.2, Is Involved in the Inhibition of Proliferation and Apoptosis in MCF-7 Human Breast Cancer Cells. Int. J. Oncol. 2012, 41, 267–275. [Google Scholar] [CrossRef]
- Banderali, U.; Jain, M.; Thakur, S.; Jayanthan, A.; Belke, D.D.; Giles, W.R.; Narendran, A. The T-Type Calcium Channel Cav3.1 in Y79 Retinoblastoma Cells Is Regulated by the Epidermal Growth Factor Receptor via the MAPK Signaling Pathway. Curr. Eye Res. 2022, 47, 426–435. [Google Scholar] [CrossRef]
- Suo, A.; Childers, A.; D’Silva, A.; Petersen, L.F.; Otsuka, S.; Dean, M.; Li, H.; Enwere, E.K.; Pohorelic, B.; Klimowicz, A.; et al. Cav3.1 Overexpression Is Associated with Negative Characteristics and Prognosis in Non-Small Cell Lung Cancer. Oncotarget 2018, 9, 8573–8583. [Google Scholar] [CrossRef]
- Haverstick, D.M.; Heady, T.N.; Macdonald, T.L.; Gray, L.S. Inhibition of Human Prostate Cancer Proliferation in Vitro and in a Mouse Model by a Compound Synthesized to Block Ca2+ Entry1. Cancer Res. 2000, 60, 1002–1008. [Google Scholar]
- Hu, S.; Li, L.; Huang, W.; Liu, J.; Lan, G.; Yu, S.; Peng, L.; Xie, X.; Yang, L.; Nian, Y.; et al. CAV3.1 Knockdown Suppresses Cell Proliferation, Migration and Invasion of Prostate Cancer Cells by Inhibiting AKT. Cancer Manag. Res. 2018, 10, 4603–4614. [Google Scholar] [CrossRef] [PubMed]
- Enserink, J.M.; Kolodner, R.D. An Overview of Cdk1-Controlled Targets and Processes. Cell Div. 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Castedo, M.; Perfettini, J.-L.; Roumier, T.; Kroemer, G. Cyclin-Dependent Kinase-1: Linking Apoptosis to Cell Cycle and Mitotic Catastrophe. Cell Death Differ. 2002, 9, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Izadi, S.; Nikkhoo, A.; Hojjat-Farsangi, M.; Namdar, A.; Azizi, G.; Mohammadi, H.; Yousefi, M.; Jadidi-Niaragh, F. CDK1 in Breast Cancer: Implications for Theranostic Potential. Anti-Cancer Agents Med. Chem. 2020, 20, 758–767. [Google Scholar] [CrossRef]
- Wijnen, R.; Pecoraro, C.; Carbone, D.; Fiuji, H.; Avan, A.; Peters, G.J.; Giovannetti, E.; Diana, P. Cyclin Dependent Kinase-1 (CDK-1) Inhibition as a Novel Therapeutic Strategy against Pancreatic Ductal Adenocarcinoma (PDAC). Cancers 2021, 13, 4389. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. Cell Cycle, CDKs and Cancer: A Changing Paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef]
- Xiao, D.; Johnson, C.S.; Trump, D.L.; Singh, S.V. Proteasome-Mediated Degradation of Cell Division Cycle 25C and Cyclin-Dependent Kinase 1 in Phenethyl Isothiocyanate-Induced G2-M-Phase Cell Cycle Arrest in PC-3 Human Prostate Cancer Cells. Mol. Cancer Ther. 2004, 3, 567–576. [Google Scholar] [CrossRef]
- Agarwal, C.; Tyagi, A.; Agarwal, R. Gallic Acid Causes Inactivating Phosphorylation of Cdc25A/Cdc25C-Cdc2 via ATM-Chk2 Activation, Leading to Cell Cycle Arrest, and Induces Apoptosis in Human Prostate Carcinoma DU145 Cells. Mol. Cancer Ther. 2006, 5, 3294–3302. [Google Scholar] [CrossRef]
- Kan, S.-F.; Yu, C.-H.; Pu, H.-F.; Hsu, J.-M.; Chen, M.-J.; Wang, P.S. Anti-Proliferative Effects of Evodiamine on Human Prostate Cancer Cell Lines DU145 and PC3. J. Cell. Biochem. 2007, 101, 44–56. [Google Scholar] [CrossRef]
- Tsaur, I.; Makarević, J.; Hudak, L.; Juengel, E.; Kurosch, M.; Wiesner, C.; Bartsch, G.; Harder, S.; Haferkamp, A.; Blaheta, R.A. The Cdk1-Cyclin B Complex Is Involved in Everolimus Triggered Resistance in the PC3 Prostate Cancer Cell Line. Cancer Lett. 2011, 313, 84–90. [Google Scholar] [CrossRef]
- Tang, Z.; Pilié, P.G.; Geng, C.; Manyam, G.C.; Yang, G.; Park, S.; Wang, D.; Peng, S.; Wu, C.; Peng, G.; et al. ATR Inhibition Induces CDK1–SPOP Signaling and Enhances Anti–PD-L1 Cytotoxicity in Prostate Cancer. Clin. Cancer Res. 2021, 27, 4898–4909. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, Y.; Yuan, X.; Bubley, G.J.; Balk, S.P. Androgen Receptor Phosphorylation and Stabilization in Prostate Cancer by Cyclin-Dependent Kinase 1. Proc. Natl. Acad. Sci. 2006, 103, 15969–15974. [Google Scholar] [CrossRef] [PubMed]
- Willder, J.M.; Heng, S.J.; McCall, P.; Adams, C.E.; Tannahill, C.; Fyffe, G.; Seywright, M.; Horgan, P.G.; Leung, H.Y.; Underwood, M.A.; et al. Androgen Receptor Phosphorylation at Serine 515 by Cdk1 Predicts Biochemical Relapse in Prostate Cancer Patients. Br. J. Cancer 2013, 108, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Topoisomerase I Inhibitors: Camptothecins and Beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Hevener, K.; Verstak, T.A.; Lutat, K.E.; Riggsbee, D.L.; Mooney, J.W. Recent Developments in Topoisomerase-Targeted Cancer Chemotherapy. Acta Pharm. Sin. B 2018, 8, 844–861. [Google Scholar] [CrossRef]
- Malathi, K.; Paranjape, J.M.; Ganapathi, R.; Silverman, R.H. HPC1/RNASEL Mediates Apoptosis of Prostate Cancer Cells Treated with 2′,5′-Oligoadenylates, Topoisomerase I Inhibitors, and Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand. Cancer Res. 2004, 64, 9144–9151. [Google Scholar] [CrossRef]
- Yu, C.-C.; Pan, S.-L.; Chao, S.-W.; Liu, S.-P.; Hsu, J.-L.; Yang, Y.-C.; Li, T.-K.; Huang, W.-J.; Guh, J.-H. A Novel Small Molecule Hybrid of Vorinostat and DACA Displays Anticancer Activity against Human Hormone-Refractory Metastatic Prostate Cancer through Dual Inhibition of Histone Deacetylase and Topoisomerase I. Biochem. Pharmacol. 2014, 90, 320–330. [Google Scholar] [CrossRef]
- Roy, J.; Nguyen, T.X.; Kanduluru, A.K.; Venkatesh, C.; Lv, W.; Reddy, P.V.N.; Low, P.S.; Cushman, M. DUPA Conjugation of a Cytotoxic Indenoisoquinoline Topoisomerase I Inhibitor for Selective Prostate Cancer Cell Targeting. J. Med. Chem. 2015, 58, 3094–3103. [Google Scholar] [CrossRef]
- Tan, K.W.; Seng, H.L.; Lim, F.S.; Cheah, S.-C.; Ng, C.H.; Koo, K.S.; Mustafa, M.R.; Ng, S.W.; Maah, M.J. Towards a Selective Cytotoxic Agent for Prostate Cancer: Interaction of Zinc Complexes of Polyhydroxybenzaldehyde Thiosemicarbazones with Topoisomerase I. Polyhedron 2012, 38, 275–284. [Google Scholar] [CrossRef]
- Willems, L.; Tamburini, J.; Chapuis, N.; Lacombe, C.; Mayeux, P.; Bouscary, D. PI3K and MTOR Signaling Pathways in Cancer: New Data on Targeted Therapies. Curr. Oncol. Rep. 2012, 14, 129–138. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen Receptor in Prostate Cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Thrasher, J.B.; Terranova, P. Glycogen Synthase Kinase-3: A Potential Preventive Target for Prostate Cancer Management. Urol. Oncol. Semin. Orig. Investig. 2015, 33, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Abdulghani, J.; Gu, L.; Dagvadorj, A.; Lutz, J.; Leiby, B.; Bonuccelli, G.; Lisanti, M.P.; Zellweger, T.; Alanen, K.; Mirtti, T.; et al. Stat3 Promotes Metastatic Progression of Prostate Cancer. Am. J. Pathol. 2008, 172, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.C.Y.; Frolov, A.; Li, R.; Ayala, G.; Greenberg, N.M. Targeting Aurora Kinases for the Treatment of Prostate Cancer. Cancer Res. 2006, 66, 4996–5002. [Google Scholar] [CrossRef]
- ROCS; 3.5.0.1. OpenEye Scientific Software: Santa Fe, NM, USA. Available online: https://www.eyesopen.com/news/2015/09/rocs-v3.2.1 (accessed on 19 November 2022).
- Gravina, G.L.; Marampon, F.; Sanità, P.; Mancini, A.; Colapietro, A.; Scarsella, L.; Jitariuc, A.; Biordi, L.; Ficorella, C.; Festuccia, C. Increased Expression and Activity of P75NTR Are Crucial Events in Azacitidine-Induced Cell Death in Prostate Cancer. Oncol. Rep. 2016, 36, 125–130. [Google Scholar] [CrossRef]
- Sonpavde, G.; Aparicio, A.M.; Zhan, F.; North, B.; DeLaune, R.; Garbo, L.E.; Rousey, S.R.; Weinstein, R.E.; Xiao, L.; Boehm, K.A.; et al. Azacitidine Favorably Modulates PSA Kinetics Correlating with Plasma DNA LINE-1 Hypomethylation in Men with Chemonaïve Castration-Resistant Prostate Cancer. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 682–689. [Google Scholar] [CrossRef]
- Witte, R.S.; Yeap, B.Y.; Trump, D.L. Trimetrexate in Advanced Hormone-Refractory Prostate Cancer. Investig. New Drugs 1994, 12, 255–258. [Google Scholar] [CrossRef]
- Dumble, M.; Crouthamel, M.-C.; Zhang, S.-Y.; Schaber, M.; Levy, D.; Robell, K.; Liu, Q.; Figueroa, D.J.; Minthorn, E.A.; Seefeld, M.A.; et al. Discovery of Novel AKT Inhibitors with Enhanced Anti-Tumor Effects in Combination with the MEK Inhibitor. PLoS ONE 2014, 9, e100880. [Google Scholar] [CrossRef]
- Lynch, J.T.; Polanska, U.M.; Hancox, U.; Delpuech, O.; Maynard, J.; Trigwell, C.; Eberlein, C.; Lenaghan, C.; Polanski, R.; Avivar-Valderas, A.; et al. Combined Inhibition of PI3Kβ and MTOR Inhibits Growth of PTEN-Null Tumors. Mol. Cancer Ther. 2018, 17, 2309–2319. [Google Scholar] [CrossRef]
- Beaufils, F.; Cmiljanovic, N.; Cmiljanovic, V.; Bohnacker, T.; Melone, A.; Marone, R.; Jackson, E.; Zhang, X.; Sele, A.; Borsari, C.; et al. 5-(4,6-Dimorpholino-1,3,5-Triazin-2-Yl)-4-(Trifluoromethyl)Pyridin-2-Amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/MTOR Inhibitor as Clinical Candidate in Oncology. J. Med. Chem. 2017, 60, 7524–7538. [Google Scholar] [CrossRef]
- Mortensen, D.S.; Fultz, K.E.; Xu, S.; Xu, W.; Packard, G.; Khambatta, G.; Gamez, J.C.; Leisten, J.; Zhao, J.; Apuy, J.; et al. CC-223, a Potent and Selective Inhibitor of MTOR Kinase: In Vitro and In Vivo Characterization. Mol. Cancer Ther. 2015, 14, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Soh, C.K.; Goh, W.H.; Wang, H. Design, Synthesis, and Preclinical Evaluation of Fused Pyrimidine-Based Hydroxamates for the Treatment of Hepatocellular Carcinoma. J. Med. Chem. 2018, 61, 1552–1575. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.; Novotny-Diermayr, V.; Goh, K.C.; Williams, M.; Tan, Y.C.; Ong, L.C.; Cheong, A.; Ng, B.K.; Amalini, C.; Madan, B.; et al. VS-5584, a Novel and Highly Selective PI3K/MTOR Kinase Inhibitor for the Treatment of Cancer. Mol. Cancer Ther. 2013, 12, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Nepali, K.; Lin, M.-H.; Chao, M.-W.; Peng, S.-J.; Hsu, K.-C.; Eight Lin, T.; Chen, M.-C.; Lai, M.-J.; Pan, S.-L.; Liou, J.-P. Amide-Tethered Quinoline-Resorcinol Conjugates as a New Class of HSP90 Inhibitors Suppressing the Growth of Prostate Cancer Cells. Bioorganic Chem. 2019, 91, 103119. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, K.; Zhang, H.; Brekken, J.; Huser, N.; Powell, R.E.; Timple, N.; Busch, D.J.; Neely, L.; Sensintaffar, J.L.; Yang, Y.; et al. BIIB021, an Orally Available, Fully Synthetic Small-Molecule Inhibitor of the Heat Shock Protein Hsp90. Mol. Cancer Ther. 2009, 8, 921–929. [Google Scholar] [CrossRef]
- Nong, H.; Zhang, Y.; Bai, Y.; Zhang, Q.; Liu, M.; Zhou, Q.; Shi, Z.; Zeng, G.; Zong, S.-H. Adapalene Inhibits Prostate Cancer Cell Proliferation In Vitro and In Vivo by Inducing DNA Damage, S-Phase Cell Cycle Arrest, and Apoptosis. Front. Pharmacol. 2022, 13, 801624. [Google Scholar] [CrossRef]
- Lu, X.P.; Fanjul, A.; Picard, N.; Shroot, B.; Pfahl, M. A Selective Retinoid with High Activity against an Androgen-resistant Prostate Cancer Cell Type. Int. J. Cancer. 1999, 80, 272–278. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, X.; Wang, G.; Lei, D.; Chen, X.; Lin, K.; Li, M.; Lin, H.; Li, D.; Zheng, Q. Picropodophyllin Inhibits the Proliferation of Human Prostate Cancer DU145 and LNCaP Cells via ROS Production and PI3K/AKT Pathway Inhibition. Biol. Pharm. Bull. 2022, 45, 1027–1035. [Google Scholar] [CrossRef]
- Scott, L.J. Azacitidine: A Review in Myelodysplastic Syndromes and Acute Myeloid Leukaemia. Drugs 2016, 76, 889–900. [Google Scholar] [CrossRef]
- Girnita, A.; Girnita, L.; del Prete, F.; Bartolazzi, A.; Larsson, O.; Axelson, M. Cyclolignans as Inhibitors of the Insulin-like Growth Factor-1 Receptor and Malignant Cell Growth. Cancer Res 2004, 64, 236–242. [Google Scholar] [CrossRef]
- Xie, H.; Lin, X.; Zhang, Y.; Tan, F.; Chi, B.; Peng, Z.; Dong, W.; An, D. Design, Synthesis and Biological Evaluation of Ring-Fused Pyrazoloamino Pyridine/Pyrimidine Derivatives as Potential FAK Inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127459. [Google Scholar] [CrossRef] [PubMed]
- Rose, W.C.; Lee, F.Y.F.; Fairchild, C.R.; Lynch, M.; Monticello, T.; Kramer, R.A.; Manne, V. Preclinical Antitumor Activity of BMS-214662, a Highly Apoptotic and Novel Farnesyltransferase Inhibitor. Cancer Res. 2001, 61, 7507–7517. [Google Scholar] [PubMed]
- Bailey, H.H.; Alberti, D.B.; Thomas, J.P.; Mulkerin, D.L.; Binger, K.A.; Gottardis, M.M.; Martell, R.E.; Wilding, G. Phase I Trial of Weekly Paclitaxel and BMS-214662 in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2007, 13, 3623–3629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zou, L.; Chen, W.; Yang, T.; Luo, J.; Wu, K.; Shu, F.; Tan, X.; Yang, Y.; Cen, S.; et al. Flubendazole, FDA-Approved Anthelmintic, Elicits Valid Antitumor Effects by Targeting P53 and Promoting Ferroptosis in Castration-Resistant Prostate Cancer. Pharmacol. Res. 2021, 164, 105305. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Shin, C.; Kim, C.-Y.; Ryu, B.; Kim, J.; Bang, J.; Park, J.-H. Albendazole Exerts Antiproliferative Effects on Prostate Cancer Cells by Inducing Reactive Oxygen Species Generation. Oncol. Lett. 2021, 21, 395. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.Y.; Jung, B.K.; Hong, S.J. Albendazole and Mebendazole as Anti-Parasitic and Anti-Cancer Agents: An Update. Korean J Parasitol. 2021, 59(3), 189–225. [Google Scholar] [CrossRef]
- Weinberger, S.E.; Cockrill, B.A.; Mandel, J. 26-Pulmonary Complications in the Immunocompromised Host. In Principles of Pulmonary Medicine, 6th ed.; Weinberger, S.E., Cockrill, B.A., Mandel, J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2014; pp. 331–343. ISBN 978-1-4557-2532-8. [Google Scholar]
- Seefeld, M.A.; Rouse, M.B.; Heerding, D.A. Inhibitors of AKT Activity. Patent number US20110071182A1, 2011. [Google Scholar]
- Herberts, C.; Murtha, A.J.; Fu, S.; Wang, G.; Schönlau, E.; Xue, H.; Lin, D.; Gleave, A.; Yip, S.; Angeles, A.; et al. Activating AKT1 and PIK3CA Mutations in Metastatic Castration-Resistant Prostate Cancer. Eur. Urol. 2020, 78, 834–844. [Google Scholar] [CrossRef]
- Therapeutics, L. Laekna Therapeutics Reported Positive Results in Two Clinical Studies for the Treatment of Various Stages of Prostate Cancer at the ESMO Congress. Available online: https://www.prnewswire.com/news-releases/laekna-therapeutics-reported-positive-results-in-two-clinical-studies-for-the-treatment-of-various-stages-of-prostate-cancer-at-the-esmo-congress-301378428.html (accessed on 13 December 2022).
- Al-Ashmawy, A.A.K.; Ragab, F.A.; Elokely, K.M.; Anwar, M.M.; Perez-Leal, O.; Rico, M.C.; Gordon, J.; Bichenkov, E.; Mateo, G.; Kassem, E.M.M.; et al. Design, Synthesis and SAR of New-Di-Substituted Pyridopyrimidines as ATP-Competitive Dual PI3Kα/MTOR Inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 3117–3122. [Google Scholar] [CrossRef]
- Basu, B.; Krebs, M.G.; Sundar, R.; Wilson, R.H.; Spicer, J.; Jones, R.; Brada, M.; Talbot, D.C.; Steele, N.; Ingles Garces, A.H.; et al. Vistusertib (Dual m-TORC1/2 Inhibitor) in Combination with Paclitaxel in Patients with High-Grade Serous Ovarian and Squamous Non-Small-Cell Lung Cancer. Ann. Oncol. 2018, 29, 1918–1925. [Google Scholar] [CrossRef]
- Westin, S.N.; Litton, J.K.; Williams, R.A.; Shepherd, C.J.; Brugger, W.; Pease, E.J.; Soliman, P.T.; Frumovitz, M.M.; Levenback, C.F.; Sood, A.; et al. Phase I Trial of Olaparib (PARP Inhibitor) and Vistusertib (MTORC1/2 Inhibitor) in Recurrent Endometrial, Ovarian and Triple Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 5504. [Google Scholar] [CrossRef]
- Pancholi, S.; Leal, M.F.; Ribas, R.; Simigdala, N.; Schuster, E.; Chateau-Joubert, S.; Zabaglo, L.; Hills, M.; Dodson, A.; Gao, Q.; et al. Combination of MTORC1/2 Inhibitor Vistusertib plus Fulvestrant in Vitro and in Vivo Targets Oestrogen Receptor-Positive Endocrine-Resistant Breast Cancer. Breast Cancer Res. 2019, 21, 135. [Google Scholar] [CrossRef]
- Rageot, D.; Bohnacker, T.; Melone, A.; Langlois, J.-B.; Borsari, C.; Hillmann, P.; Sele, A.M.; Beaufils, F.; Zvelebil, M.; Hebeisen, P.; et al. Discovery and Preclinical Characterization of 5-[4,6-Bis({3-Oxa-8-Azabicyclo [3.2.1]Octan-8-Yl})-1,3,5-Triazin-2-Yl]-4-(Difluoromethyl)Pyridin-2-Amine (PQR620), a Highly Potent and Selective MTORC1/2 Inhibitor for Cancer and Neurological Disorders. J. Med. Chem. 2018, 61, 10084–10105. [Google Scholar] [CrossRef]
- Pacey, S.; Shah, N.; Davies, B.; Bratt, O.; Warren, A.; Baird, R.D.; Gnanapragasam, V.; Ingle, S.; Stearn, S.; Machin, A.; et al. A Pharmacodynamic Biomarker Study of Vistusertib (AZD2014), an MTORC1/2 Inhibitor, given Prior to Radical Prostatectomy (CANCAP02). J. Clin. Oncol. 2018, 36, 5081. [Google Scholar] [CrossRef]
- Hansen, A.R.; Shapiro, G.; Do, K.T.; Kumar, R.; Martin-Liberal, J.; Higano, C.S.; Wisinski, K.B.; Dean, E.J.; Heath, E.I.; Rathkopf, D.E.; et al. A First in Human Phase I Study of AZD8186, a Potent and Selective Inhibitor of PI3K in Patients with Advanced Solid Tumours as Monotherapy and in Combination with the Dual MTORC1/2 Inhibitor Vistusertib (AZD2014) or Abiraterone Acetate. J. Clin. Oncol. 2017, 35, 2570. [Google Scholar] [CrossRef]
- van Hattum, A.H.; Pinedo, H.M.; Schlüper, H.M.M.; Erkelens, C.A.M.; Tohgo, A.; Boven, E. The Activity Profile of the Hexacyclic Camptothecin Derivative DX-8951f in Experimental Human Colon Cancer and Ovarian Cancer. Biochem. Pharmacol. 2002, 64, 1267–1277. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT Network at the Interface of Oncogenic Signalling and Cancer Metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Li, X.; Tang, Y.; Yu, F.; Sun, Y.; Huang, F.; Chen, Y.; Yang, Z.; Ding, G. Inhibition of Prostate Cancer DU-145 Cells Proliferation by Anthopleura Anjunae Oligopeptide (YVPGP) via PI3K/AKT/MTOR Signaling Pathway. Mar. Drugs 2018, 16, 325. [Google Scholar] [CrossRef]
- Facompre, N.D.; Sinha, I.; El-Bayoumy, K.; Pinto, J.T.; Sinha, R. Remarkable Inhibition of MTOR Signaling by the Combination of Rapamycin and 1,4-Phenylenebis(Methylene)Selenocyanate in Human Prostate Cancer Cells. Int. J. Cancer 2012, 131, 2134–2142. [Google Scholar] [CrossRef]
- Yang, K.; Tang, X.J.; Xu, F.F.; Liu, J.H.; Tan, Y.Q.; Gao, L.; Sun, Q.; Ding, X.; Liu, B.H.; Chen, Q.X. PI3K/mTORC1/2 inhibitor PQR309 inhibits proliferation and induces apoptosis in human glioblastoma cells. Oncol. Rep. 2020, 43, 773–782. [Google Scholar] [CrossRef]
- Mortensen, D.S.; Perrin-Ninkovic, S.M.; Shevlin, G.; Zhao, J.; Packard, G.; Bahmanyar, S.; Correa, M.; Elsner, J.; Harris, R.; Lee, B.G.S.; et al. Discovery of Mammalian Target of Rapamycin (MTOR) Kinase Inhibitor CC-223. J. Med. Chem. 2015, 58, 5323–5333. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, H.; Lundgren, K.; Wilson, L.; Burrows, F.; Shores, C.G. BIIB021, a Novel Hsp90 Inhibitor, Sensitizes Head and Neck Squamous Cell Carcinoma to Radiotherapy. Int. J. Cancer 2010, 126, 1216–1225. [Google Scholar] [CrossRef]
- Zhang, H.; Neely, L.; Lundgren, K.; Yang, Y.-C.; Lough, R.; Timple, N.; Burrows, F. BIIB021, a Synthetic Hsp90 Inhibitor, Has Broad Application against Tumors with Acquired Multidrug Resistance. Int. J. Cancer 2010, 126, 1226–1234. [Google Scholar] [CrossRef]
- Guo, Y.; Zhu, H.; Weng, M.; Zhang, H.; Wang, C.; Sun, L. CC-223, NSC781406, and BGT226 Exerts a Cytotoxic Effect Against Pancreatic Cancer Cells via MTOR Signaling. Front. Pharmacol. 2020, 11, 580407. [Google Scholar] [CrossRef]
- Wang, J.; Jin, X.; Zhang, X.; Li, X. CC-223 Inhibits Human Head and Neck Squamous Cell Carcinoma Cell Growth. Biochem. Biophys. Res. Commun. 2018, 496, 1191–1196. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, J.; Liu, M.; Chen, D.; Qiu, C.; Sun, K. CC-223 Blocks MTORC1/C2 Activation and Inhibits Human Hepatocellular Carcinoma Cells in Vitro and in Vivo. PLoS ONE 2017, 12, e0173252. [Google Scholar] [CrossRef]
- Goodyear, S.m.; Kheyfets, S.b.; Garcia, F.u.; Stearns, M.e. Role of the VEGFR3/VEGFD Receptor Axis in TGFβ1 Activation of Primary Prostate Cell Lines. Prostate 2009, 69, 982–990. [Google Scholar] [CrossRef]
- Bendell, J.C.; Kelley, R.K.; Shih, K.C.; Grabowsky, J.A.; Bergsland, E.; Jones, S.; Martin, T.; Infante, J.R.; Mischel, P.S.; Matsutani, T.; et al. A Phase I Dose-Escalation Study to Assess Safety, Tolerability, Pharmacokinetics, and Preliminary Efficacy of the Dual MTORC1/MTORC2 Kinase Inhibitor CC-223 in Patients with Advanced Solid Tumors or Multiple Myeloma. Cancer 2015, 121, 3481–3490. [Google Scholar] [CrossRef]
- Shih, K.C.; Bendell, J.C.; Reinert, A.; Jones, S.; Kelley, R.K.; Infante, J.R.; Korn, M.; Hege, K.; Chopra, R.; Xu, S.; et al. Phase I Trial of an Oral TORC1/TORC2 Inhibitor (CC-223) in Advanced Solid and Hematologic Cancers. J. Clin. Oncol. 2012, 30, 3006. [Google Scholar] [CrossRef]
- Wolin, E.; Mita, A.; Mahipal, A.; Meyer, T.; Bendell, J.; Nemunaitis, J.; Munster, P.N.; Paz-Ares, L.; Filvaroff, E.H.; Li, S.; et al. A Phase 2 Study of an Oral MTORC1/MTORC2 Kinase Inhibitor (CC-223) for Non-Pancreatic Neuroendocrine Tumors with or without Carcinoid Symptoms. PLoS ONE 2019, 14, e0221994. [Google Scholar] [CrossRef]
- Böll, B.; Eltaib, F.; Reiners, K.S.; Von Tresckow, B.; Tawadros, S.; Simhadri, V.R.; Burrows, F.J.; Lundgren, K.; Hansen, H.P.; Engert, A.; et al. Heat Shock Protein 90 Inhibitor BIIB021 (CNF2024) Depletes NF-ΚB and Sensitizes Hodgkin’s Lymphoma Cells for Natural Killer Cell-Mediated Cytotoxicity. Clin. Cancer Res. 2009, 15, 5108–5116. [Google Scholar] [CrossRef]
- Shi, J.; Van de Water, R.; Hong, K.; Lamer, R.B.; Weichert, K.W.; Sandoval, C.M.; Kasibhatla, S.R.; Boehm, M.F.; Chao, J.; Lundgren, K.; et al. EC144 Is a Potent Inhibitor of the Heat Shock Protein 90. J. Med. Chem. 2012, 55, 7786–7795. [Google Scholar] [CrossRef] [PubMed]
- Taldone, T.; Patel, P.D.; Patel, M.; Patel, H.J.; Evans, C.E.; Rodina, A.; Ochiana, S.; Shah, S.K.; Uddin, M.; Gewirth, D.; et al. Experimental and Structural Testing Module to Analyze Paralogue-Specificity and Affinity in the Hsp90 Inhibitors Series. J. Med. Chem. 2013, 56, 6803–6818. [Google Scholar] [CrossRef] [PubMed]
- Leav, I.; Plescia, J.; Goel, H.L.; Li, J.; Jiang, Z.; Cohen, R.J.; Languino, L.R.; Altieri, D.C. Cytoprotective Mitochondrial Chaperone TRAP-1 as a Novel Molecular Target in Localized and Metastatic Prostate Cancer. Am. J. Pathol. 2010, 176, 393–401. [Google Scholar] [CrossRef]
- Ernst, J.T.; Liu, M.; Zuccola, H.; Neubert, T.; Beaumont, K.; Turnbull, A.; Kallel, A.; Vought, B.; Stamos, D. Correlation between Chemotype-Dependent Binding Conformations of HSP90α/β and Isoform Selectivity—Implications for the Structure-Based Design of HSP90α/β Selective Inhibitors for Treating Neurodegenerative Diseases. Bioorg. Med. Chem. Lett. 2014, 24, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Piskin, S.; Uzunali, E. A Review of the Use of Adapalene for the Treatment of Acne Vulgaris. Ther. Clin. Risk Manag. 2007, 3, 621–624. [Google Scholar] [CrossRef]
- Charpentier, B.; Bernardon, J.-M.; Eustache, J.; Millois, C.; Martin, B.; Michel, S.; Shroot, B. Synthesis, Structure-Affinity Relationships, and Biological Activities of Ligands Binding to Retinoic Acid Receptor Subtypes. J. Med. Chem. 1995, 38, 4993–5006. [Google Scholar] [CrossRef]
- Pasquali, D.; Thaller, C.; Eichele, G. Abnormal Level of Retinoic Acid in Prostate Cancer Tissues. J. Clin. Endocrinol. Metab. 1996, 81, 2186–2191. [Google Scholar] [CrossRef]
- Hammond, L.A.; Brown, G.; Keedwell, R.G.; Durham, J.; Chandraratna, R.A. The Prospects of Retinoids in the Treatment of Prostate Cancer. Anti-Cancer Drugs 2002, 13, 781–790. [Google Scholar] [CrossRef]
- Yu, G.; Corn, P.G.; Shen, P.; Song, J.H.; Lee, Y.-C.; Lin, S.-C.; Pan, J.; Agarwal, S.K.; Panaretakis, T.; Pacifici, M.; et al. Retinoic Acid Receptor Activation Reduces Metastatic Prostate Cancer Bone Lesions by Blocking the Endothelial-to-Osteoblast Transition. Cancer Res. 2022, 82, 3158–3171. [Google Scholar] [CrossRef]
- Sugiura, M.; Sato, H.; Kanesaka, M.; Imamura, Y.; Sakamoto, S.; Ichikawa, T.; Kaneda, A. Epigenetic Modifications in Prostate Cancer. Int. J. Urol. 2021, 28, 140–149. [Google Scholar] [CrossRef]
- Pasquali, D.; Rossi, V.; Bellastella, G.; Bellastella, A.; Sinisi, A.A. Natural and Synthetic Retinoids in Prostate Cancer. Curr. Pharm. Des. 2006, 12, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.X.; Chung, E.P.; Bowser, R.; Sirianni, R.W. Lipid and Polymer Blended Polyester Nanoparticles Loaded with Adapalene for Activation of Retinoid Signaling in the CNS Following Intravenous Administration. J. Drug Deliv. Sci. Technol. 2019, 52, 927–933. [Google Scholar] [CrossRef]
- Yuan, J.; Yin, Z.; Tao, K.; Wang, G.; Gao, J. Function of Insulin-like Growth Factor 1 Receptor in Cancer Resistance to Chemotherapy. Oncol. Lett. 2018, 15, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Cortese, F.; Bhattacharyya, B.; Wolff, J. Podophyllotoxin as a Probe for the Colchicine Binding Site of Tubulin. J. Biol. Chem. 1977, 252, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.K.; Patnaik, S.; Rup, A.R.; Gaurav, A. A Rare Case of Podophyllin Poisoning: Early Intervention Is Lifesaving. Indian J. Crit. Care Med. 2020, 24, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Du, M.; Lv, Q. Picropodophyllin Inhibits Type I Endometrial Cancer Cell Proliferation via Disruption of the PI3K/Akt Pathway. Acta Biochim. Et Biophys. Sin. 2019, 51, 753–760. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Mei, J.; Wang, X.; Zhu, X.; Zhang, Q.; Lv, J. Picropodophyllin Inhibits Epithelial Ovarian Cancer Cells in Vitro and in Vivo. Biochem. Biophys. Res. Commun. 2013, 435, 385–390. [Google Scholar] [CrossRef]
- Ohshima-Hosoyama, S.; Hosoyama, T.; Nelon, L.D.; Keller, C. IGF-1 Receptor Inhibition by Picropodophyllin in Medulloblastoma. Biochem. Biophys. Res. Commun 2010, 399, 727–732. [Google Scholar] [CrossRef]
- Tomizawa, M.; Shinozaki, F.; Sugiyama, T.; Yamamoto, S.; Sueishi, M.; Yoshida, T. Insulin-like Growth Factor-I Receptor in Proliferation and Motility of Pancreatic Cancer. World J. Gastroenterol. 2010, 16, 1854–1858. [Google Scholar] [CrossRef]
- Yin, S.; Girnita, A.; Strömberg, T.; Khan, Z.; Andersson, S.; Zheng, H.; Ericsson, C.; Axelson, M.; Nistér, M.; Larsson, O.; et al. Targeting the Insulin-like Growth Factor-1 Receptor by Picropodophyllin as a Treatment Option for Glioblastoma. Neuro Oncol. 2010, 12, 19–27. [Google Scholar] [CrossRef]
- Figel, S.; Gelman, I.H. Focal Adhesion Kinase Controls Prostate Cancer Progression Via Intrinsic Kinase and Scaffolding Functions. Anti-Cancer Agents Med. Chem. 2011, 11, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Pradines, A.; Favre, G. Perspectives on Farnesyl Transferase Inhibitors in Cancer Therapy. Cancer Lett. 2004, 206, 159–167. [Google Scholar] [CrossRef]
- Sivoňová, M.K.; Vilčková, M.; Kliment, J.; Mahmood, S.; Jurečeková, J.; Dušenková, S.; Waczulíková, I.; Slezák, P.; Dobrota, D. Association of P53 and P21 Polymorphisms with Prostate Cancer. Biomed. Rep. 2015, 3, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ding, Y.; Liu, H.; Sun, M.; Wang, H.; Wu, D. Flubendazole Plays an Important Anti-Tumor Role in Different Types of Cancers. Int. J. Mol. Sci. 2022, 23, 519. [Google Scholar] [CrossRef]
- Rushworth, L.K.; Hewit, K.; Munnings-Tomes, S.; Somani, S.; James, D.; Shanks, E.; Dufès, C.; Straube, A.; Patel, R.; Leung, H.Y. Repurposing Screen Identifies Mebendazole as a Clinical Candidate to Synergise with Docetaxel for Prostate Cancer Treatment. Br. J. Cancer 2020, 122, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Application of Prostate Cancer Models for Preclinical Study: Advantages and Limitations of Cell Lines, Patient-Derived Xenografts, and Three-Dimensional Culture of Patient-Derived Cells. Cells 2019, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. KNIME: The Konstanz Information Miner. In Proceedings of the Data Analysis, Machine Learning and Applications; Preisach; Preisach, C., Burkhardt, H., Schmidt-Thieme, L., Decker, R., Eds.; Springer: Berlin, Heidelberg, 2008; pp. 319–326. [Google Scholar] [CrossRef]
- OpenEye Toolkits; 2020.2.2. OpenEye Scientific Software: Santa Fe, NM, USA. Available online: https://www.eyesopen.com/ (accessed on 19 November 2022).
- OMEGA; 4.2.0.1. OpenEye Scientific Software: Santa Fe, NM, USA. Available online: https://www.eyesopen.com/omega (accessed on 19 November 2022).
- Pinzi, L.; Rastelli, G. Identification of Target Associations for Polypharmacology from Analysis of Crystallographic Ligands of the Protein Data Bank. J. Chem. Inf. Model. 2020, 60, 372–390. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).