Genome-Wide Identification and Characterization of R2R3-MYB Provide Insight into Anthocyanin Biosynthesis Regulation Mechanism of Ananas comosus var. bracteatus
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of AbR2R3-MYB Genes
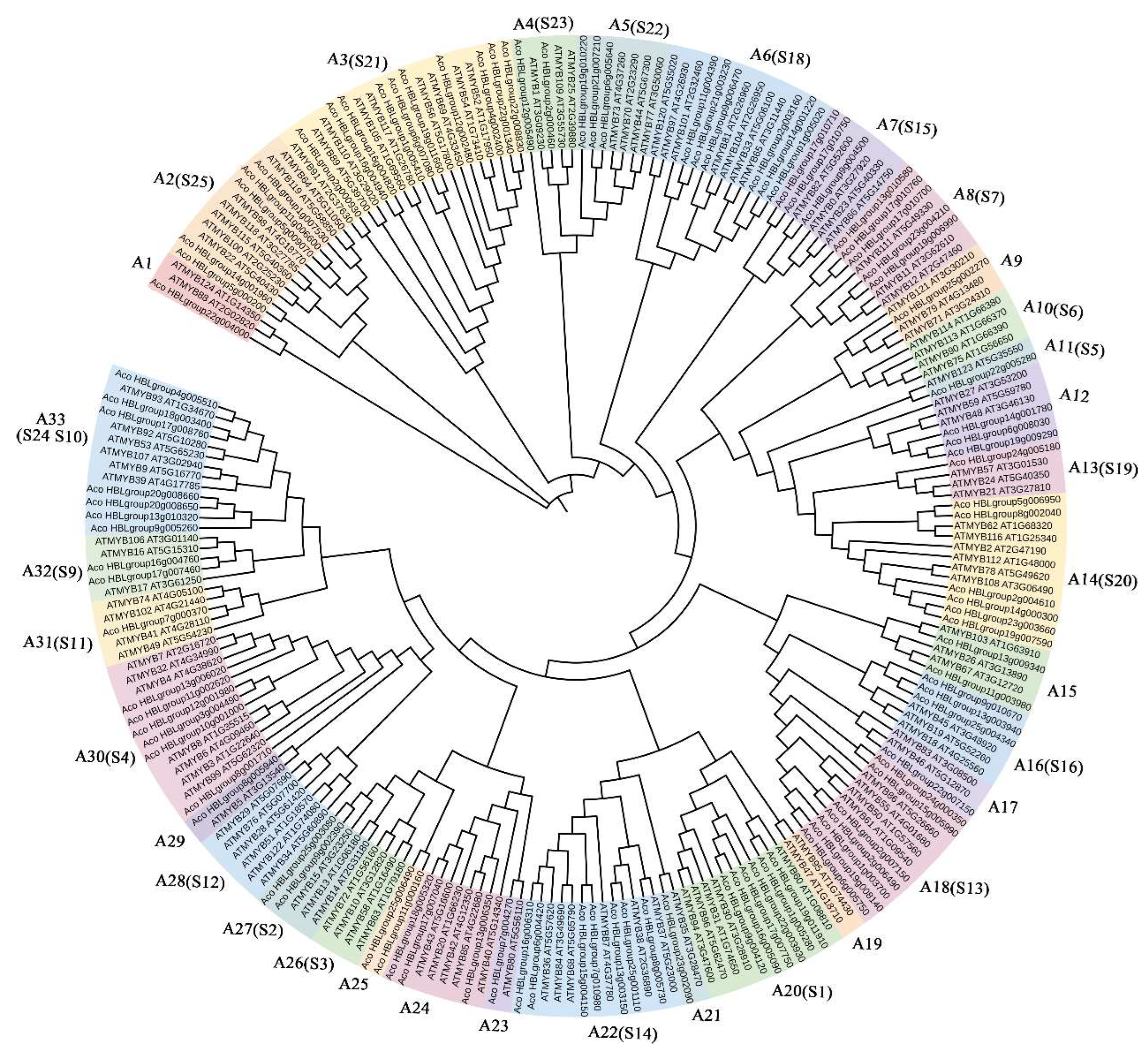
2.2. Phylogenetic Analysis and Sequences Feature of AbR2R3-MYB Genes
2.3. The Motif Composition and Gene Structure of AbR2R3-MYB Gene Family
2.4. Chromosomal Distribution and Gene Duplication of AbR2R3-MYB Genes
2.5. Analysis of the Cis-Acting Elements of the AbR2R3-MYB Genes
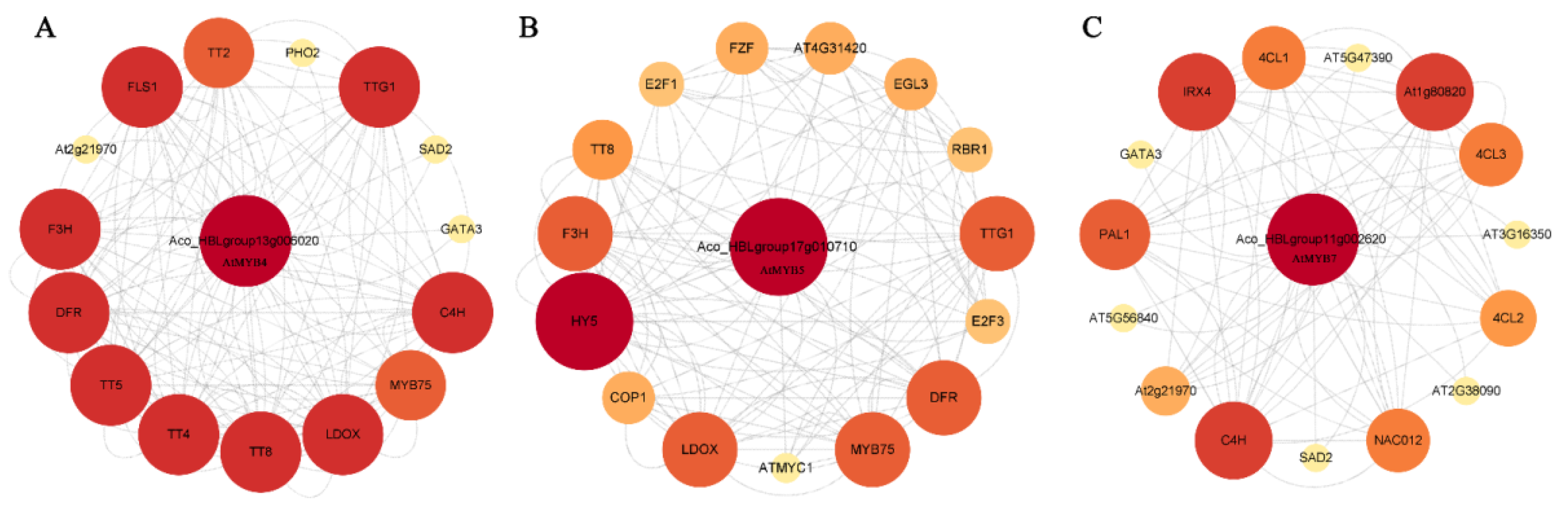
2.6. Protein Interaction Network
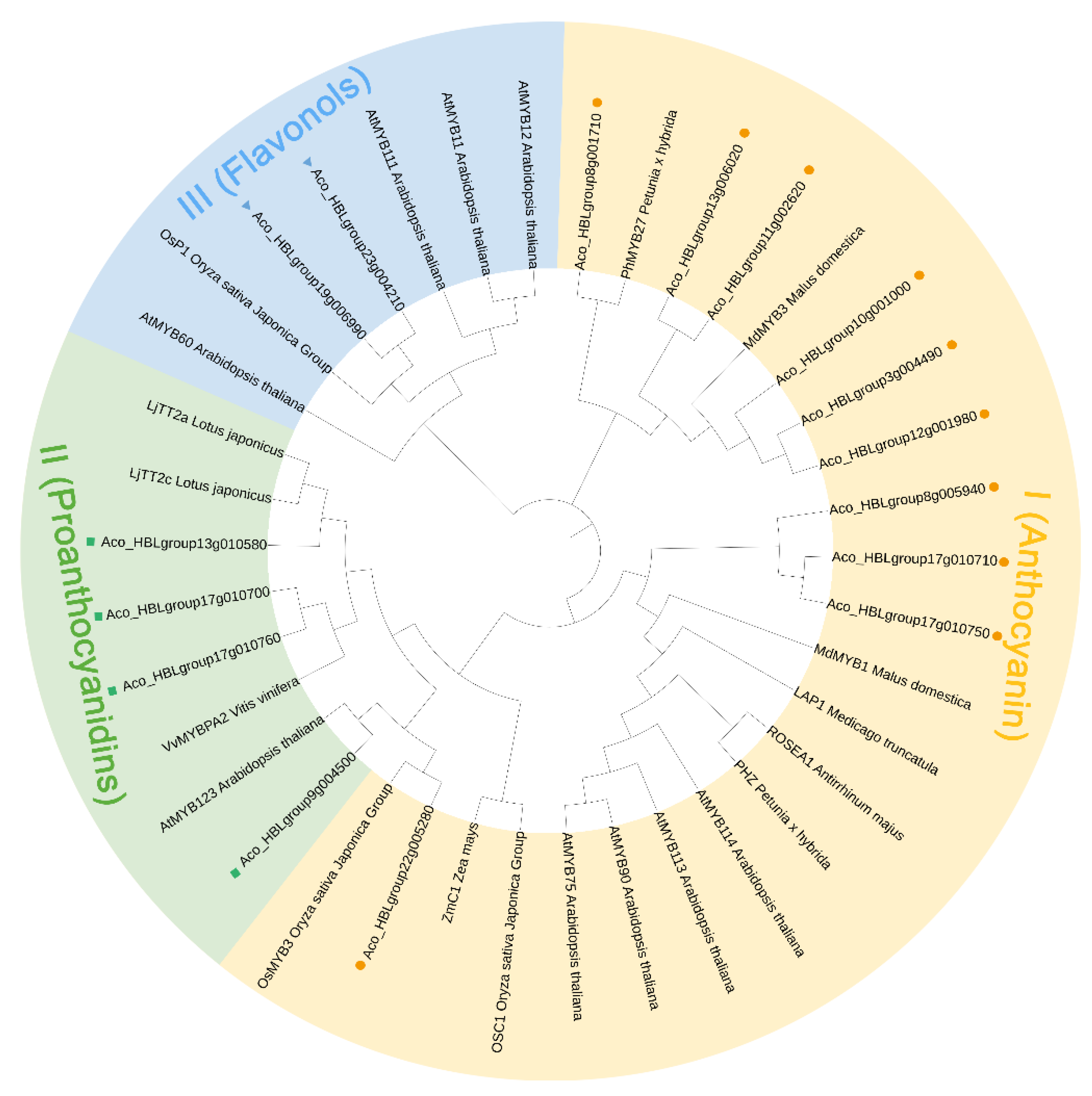
2.7. Screening of the Anthocyanin-Related AbR2R3-MYB Genes by Phylogenetic Analysis
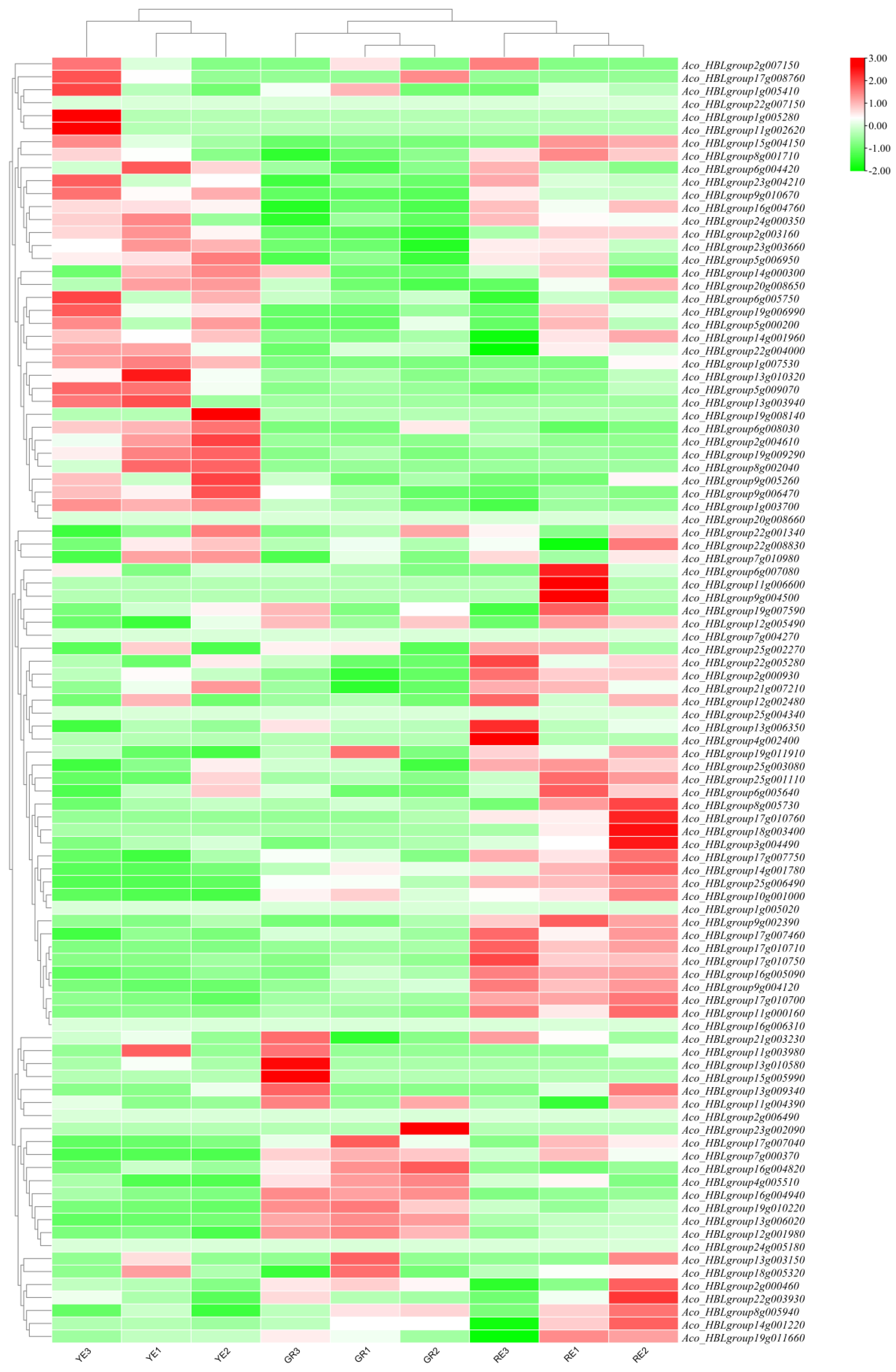
2.8. Expression Patterns of AbR2R3-MYB Genes in Different Phenotypic Leaves
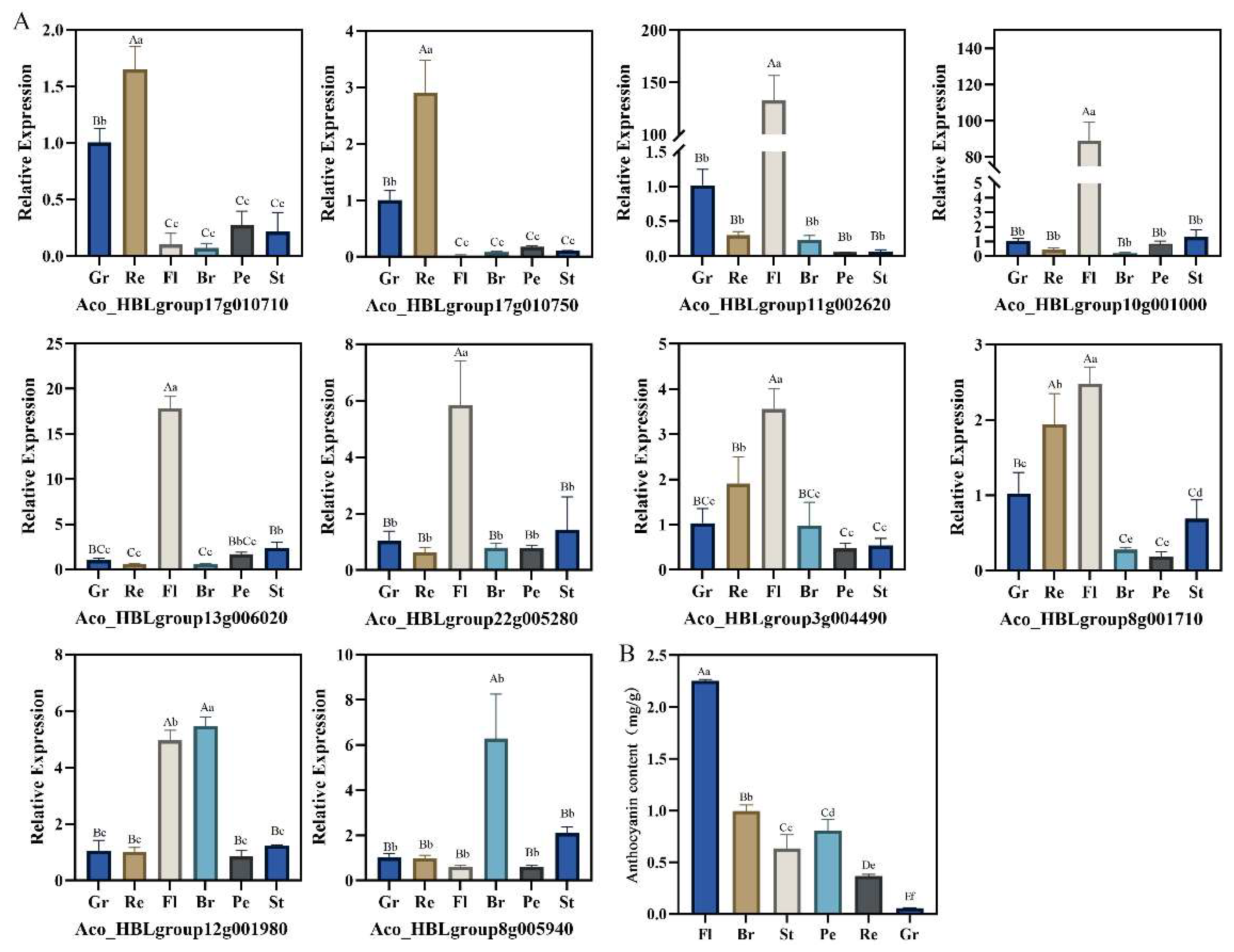
2.9. Expressions of Anthocyanin Biosynthesis-Related Candidate AbR2R3-MYB Genes in Different Tissues
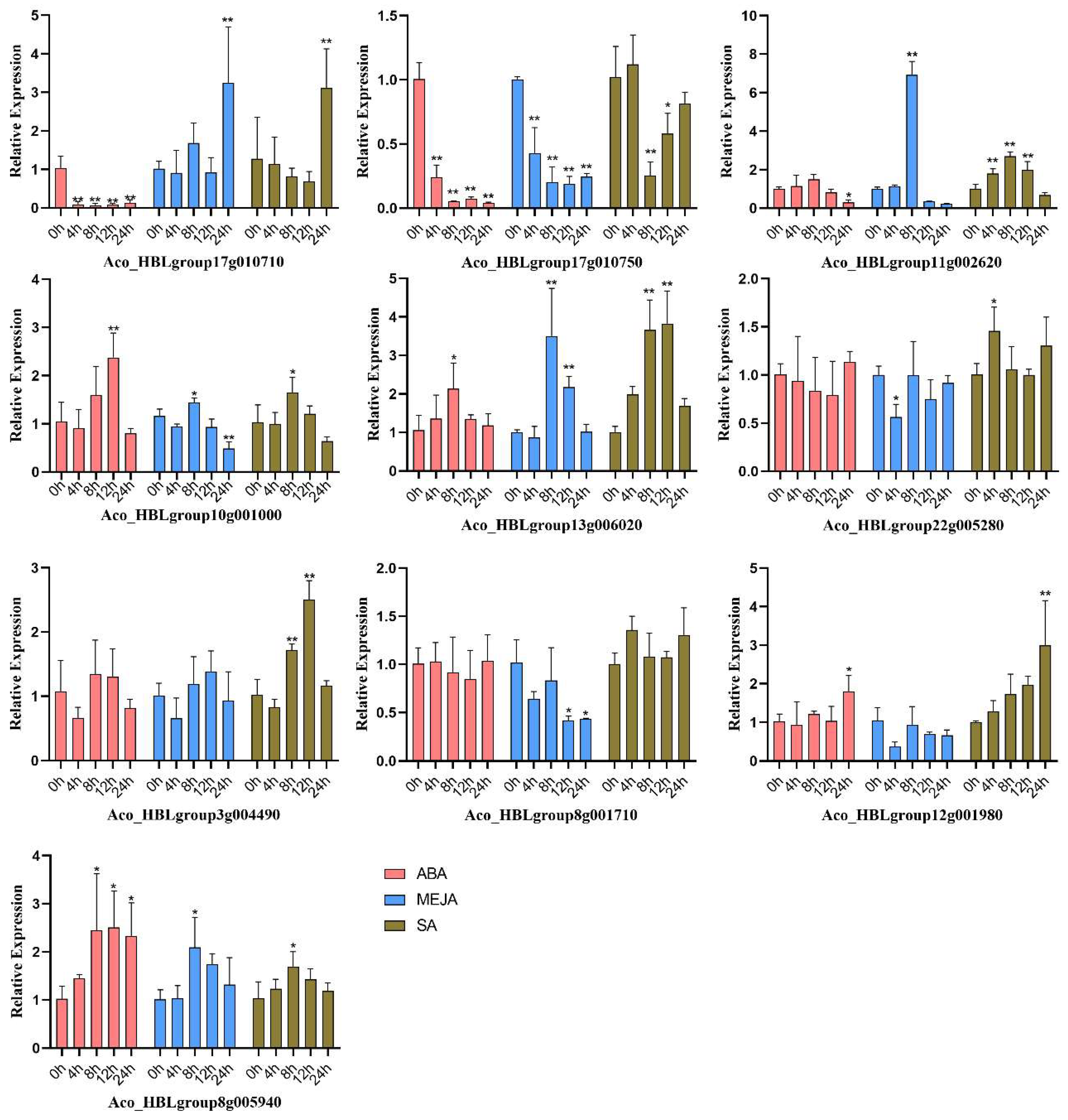
2.10. Expression of Anthocyanin Biosynthesis-Related Candidate AbR2R3-MYB to Hormone Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of R2R3-MYB Genes in A. comosus var. bracteatus
4.2. Phylogenetic Analysis and Sequence Analysis
4.3. Gene Sructure and Conserved Motif Analysis of AbR2R3-MYB Genes
4.4. Chromosomal Distribution, Synteny Analysis, and the Selection Pressure of AbR2R3-MYB Genes
4.5. Analysis of the Cis Elements of AbR2R3-MYB
4.6. Function Prediction of AbR2R3-MYBs
4.7. Expression Patterns of AbR2R3-MYB in the Different Pigmented Leaves
4.8. Plant Materials and Treatments
4.9. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Du, H.; Feng, B.R.; Yang, S.S.; Huang, Y.B.; Tang, Y.X. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 2012, 7, e37463. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Li, G.L.; Lin, Z.; Zhang, H.; Liu, Z.; Xu, Y.; Xu, G.; Li, H.; Ji, R.; Luo, W.; Qiu, Y.; et al. Anthocyanin Accumulation in the Leaves of the Purple Sweet Potato (Ipomoea batatas L.) Cultivars. Molecules 2019, 24, 3743. [Google Scholar] [CrossRef]
- Nuraini, L.; Ando, Y.; Kawai, K.; Tatsuzawa, F.; Tanaka, K.; Ochiai, M.; Suzuki, K.; Aragonés, V.; Daròs, J.A.; Nakatsuka, T. Anthocyanin regulatory and structural genes associated with violet flower color of Matthiola incana. Planta 2020, 251, 61. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Han, L.; Yuan, M.; Zhang, M.; Zhou, L.; Wang, Y. Sequence Analysis and Functional Verification of the Effects of Three Key Structural Genes, PdTHC2’GT, PdCHS and PdCHI, on the Isosalipurposide Synthesis Pathway in Paeonia delavayi var. lutea. Int. J. Mol. Sci. 2022, 23, 5696. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Quattrocchio, F.; Wing, J.F.; Leppen, H.T.C.; Mol, J.N.M.; Koes, R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 1993, 5, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.M.; Koes, R. Anthocyanin1 of Petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Saitoh, M.; Hoshino, A.; Nitasaka, E.; Iida, S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006, 47, 457–470. [Google Scholar] [CrossRef]
- Yamagishi, M.; Shimoyamada, Y.; Nakatsuka, T.; Masuda, K. Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid Lily. Plant Cell Physiol. 2010, 51, 463–474. [Google Scholar] [CrossRef]
- Chiou, C.Y.; Yeh, K.W. Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 2008, 66, 379–388. [Google Scholar] [CrossRef]
- Sun, S.S.; Gugger, P.F.; Wang, Q.F.; Chen, J.M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 2016, e2369. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Niu, S.S.; Xu, C.J.; Zhang, W.S.; Zhang, B.; Li, X.; Lin-Wang, K.; Ferguson, I.B.; Allan, A.C.; Chen, K.S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta 2010, 231, 887–899. [Google Scholar] [CrossRef]
- Xi, W.; Feng, J.; Liu, Y.; Zhang, S.; Zhao, G. The R2R3-MYB transcription factor PaMYB10 is involved in anthocyanin biosynthesis in apricots and determines red blushed skin. BMC Plant Biol. 2019, 19, 289. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.; et al. Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 2015, 10, e136365. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; Kim, T.S.; De Jong, W.S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009, 120, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, Y.; He, Y.; Li, L.; Li, D.; Chen, H. R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci. Hortic. (Amsterdam) 2021, 282, 110020. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef]
- Deng, J.; Li, M.; Huang, L.; Yang, M.; Yang, P. Genome-Wide Analysis of the R2R3 MYB Subfamily Genes in Lotus (Nelumbo nucifera). Plant Mol. Biol. Rep. 2016, 34, 1016–1026. [Google Scholar] [CrossRef]
- Xie, R.; Li, Y.; He, S.; Zheng, Y.; Yi, S.; Lv, Q.; Deng, L. Genome-wide analysis of citrus R2R3MYB genes and their spatiotemporal expression under stresses and hormone treatments. PLoS ONE 2014, 9, e113971. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Man, Y.; Wang, Y. Light-and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int. J. Mol. Sci. 2019, 20, 5228. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Bao, Y. Genome-Wide Mining of MYB Transcription Factors in the Anthocyanin Biosynthesis Pathway of Gossypium Hirsutum. Biochem. Genet. 2021, 59, 678–696. [Google Scholar] [CrossRef]
- Zhang, D.W.; Liu, L.L.; Zhou, D.; Liu, X.; Liu, Z.; Yan, M. Genome-wide identification and expression analysis of anthocyanin biosynthetic genes in Brassica juncea. J. Integr. Agric. 2020, 19, 1250–1260. [Google Scholar] [CrossRef]
- Chen, L.Y.; VanBuren, R.; Paris, M.; Zhou, H.; Zhang, X.; Wai, C.M.; Yan, H.; Chen, S.; Alonge, M.; Ramakrishnan, S.; et al. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 2019, 51, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lin, Y.; Xue, Y.; Mao, M.; Zhou, X.; Hu, H.; Liu, J.; Feng, L.; Zhang, H.; Luo, J.; et al. Light Intensity Affects the Coloration and Structure of Chimeric Leaves of Ananas comosus var. bracteatus. Phyton (B. Aires) 2022, 91, 333–348. [Google Scholar] [CrossRef]
- Feng, L.; Wang, J.; Mao, M.; Yang, W.; Adje, M.O.; Xue, Y.; Zhou, X.; Zhang, H.; Luo, J.; Tang, R.; et al. The highly continuous reference genome of a leaf-chimeric red pineapple (Ananas comosus var. bracteatus f. tricolor) provides insights into elaboration of leaf color. G3 Genes Genomes Genet. 2022, 12, jkab452. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhou, X.; Xue, Y.; Mao, M.; He, Y.; Adjei, M.O.; Yang, W.; Hu, H.; Liu, J.; Feng, L.; Zhang, H.; et al. Metabolome and transcriptome profiling reveals anthocyanin contents and anthocyanin-related genes of chimeric leaves in Ananas comosus var. bracteatus. BMC Genom. 2021, 22, 331. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, H.; Cui, J.; You, C.; Li, Y. Plant Science Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021, 312, 111024. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Rosleff Sörensen, T.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Schlueter, J.A.; Lin, J.Y.; Schlueter, S.D.; Vasylenko-Sanders, I.F.; Deshpande, S.; Yi, J.; O’Bleness, M.; Roe, B.A.; Nelson, R.T.; Scheffler, B.E.; et al. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genom. 2007, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.M.; Cui, L.; Wall, P.K.; Zhang, Q.; Zhang, X.; Leebens-Mack, J.; Ma, H.; Altman, N.; DePamphilis, C.W. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006, 23, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Carolina, N. The Evolutionary Fate and Consequences of Duplicate Genes. Science (80-.) 2000, 290, 1151–1155. [Google Scholar]
- Otto, S.P.; Yong, P. The Evolution of Gene Duplicates. Adv. Genet. 2002, 46, 451–483. Available online: http://www.sciencedirect.com/science/article/pii/S0065266002460178 (accessed on 14 December 2022).
- Sun, W.; Ma, Z.; Chen, H.; Liu, M. MYB gene family in potato (Solanum tuberosum L.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Skolnick, J.; Fetrow, J.S. From genes to protein structure and function: Novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000, 18, 34–39. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Bhambhani, S.; Bhatia, C.; Trivedi, P.K. Expression of arabidopsis myb transcription factor, atmyb111, in tobacco requires light to modulate flavonol content. Sci. Rep. 2014, 4, 5018. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef] [PubMed]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Peel, G.J.; Pang, Y.; Modolo, L.V.; Dixon, R.A. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 2009, 59, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, H.; Zhao, M.; Yang, Z.; Zhou, Z.; Guo, Y.; Lin, Y.; Chen, H. OsMYB3 is a R2R3-MYB gene responsible for anthocyanin biosynthesis in black rice. Mol. Breed. 2021, 41, 51. [Google Scholar] [CrossRef]
- Xie, T.; Zeng, L.; Chen, X.; Rong, H.; Wu, J.; Batley, J.; Jiang, J.; Wang, Y. Genome-wide analysis of the lateral organ boundaries domain gene family in brassica napus. Genes 2020, 11, 280. [Google Scholar] [CrossRef]
- Chen, X.; Wang, P.; Gu, M.; Lin, X.; Hou, B.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 2021, 113, 1565–1578. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, X.; Ma, Y.; Wang, Y.; Fang, W. Genome-wide analysis reveals stress and hormone responsive patterns of JAZ family genes in camellia sinensis. Int. J. Mol. Sci. 2020, 21, 2433. [Google Scholar] [CrossRef]
- Gagné, S.; Cluzet, S.; Mérillon, J.M.; Gény, L. ABA Initiates Anthocyanin Production in Grape Cell Cultures. J. Plant Growth Regul. 2011, 30, 1–10. [Google Scholar] [CrossRef]
- Ram, M.; Prasad, K.V.; Singh, S.K.; Hada, B.S.; Kumar, S. Influence of salicylic acid and methyl jasmonate elicitation on anthocyanin production in callus cultures of Rosa hybrida L. Plant Cell. Tissue Organ Cult. 2013, 113, 459–467. [Google Scholar] [CrossRef]
- Sudha, G.; Ravishankar, G.A. Elicitation of anthocyanin production in callus cultures of Daucus carota and the involvement of methyl jasmonate and salicylic acid. Acta Physiol. Plant. 2003, 25, 249–256. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Liu, Z.; Chen, W.; Xu, H.; Feng, H. Anthocyanin degrading and chlorophyll accumulation lead to the formation of bicolor leaf in ornamental kale. Int. J. Mol. Sci. 2019, 20, 603. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mao, M.; Xue, Y.; He, Y.; Zhou, X.; Hu, H.; Liu, J.; Feng, L.; Yang, W.; Luo, J.; Zhang, H.; et al. Validation of Reference Genes for Quantitative Real-Time PCR Normalization in Ananas comosus var. bracteatus During Chimeric Leaf Development and Response to Hormone Stimuli. Front. Genet. 2021, 12, 716137. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Feng, L.; Luo, J.; Zhang, H.; Jiang, F.; He, Y.; Li, X.; Du, J.; Owusu Adjei, M.; Luan, A.; et al. Genome-Wide Identification and Characterization of R2R3-MYB Provide Insight into Anthocyanin Biosynthesis Regulation Mechanism of Ananas comosus var. bracteatus. Int. J. Mol. Sci. 2023, 24, 3133. https://doi.org/10.3390/ijms24043133
Yang W, Feng L, Luo J, Zhang H, Jiang F, He Y, Li X, Du J, Owusu Adjei M, Luan A, et al. Genome-Wide Identification and Characterization of R2R3-MYB Provide Insight into Anthocyanin Biosynthesis Regulation Mechanism of Ananas comosus var. bracteatus. International Journal of Molecular Sciences. 2023; 24(4):3133. https://doi.org/10.3390/ijms24043133
Chicago/Turabian StyleYang, Wei, Lijun Feng, Jiaheng Luo, Huiling Zhang, Fuxing Jiang, Yehua He, Xi Li, Juan Du, Mark Owusu Adjei, Aiping Luan, and et al. 2023. "Genome-Wide Identification and Characterization of R2R3-MYB Provide Insight into Anthocyanin Biosynthesis Regulation Mechanism of Ananas comosus var. bracteatus" International Journal of Molecular Sciences 24, no. 4: 3133. https://doi.org/10.3390/ijms24043133
APA StyleYang, W., Feng, L., Luo, J., Zhang, H., Jiang, F., He, Y., Li, X., Du, J., Owusu Adjei, M., Luan, A., & Ma, J. (2023). Genome-Wide Identification and Characterization of R2R3-MYB Provide Insight into Anthocyanin Biosynthesis Regulation Mechanism of Ananas comosus var. bracteatus. International Journal of Molecular Sciences, 24(4), 3133. https://doi.org/10.3390/ijms24043133




