Structural Insight into the Working Mechanism of the FAD Synthetase from the Human Pathogen Streptococcus pneumoniae: A Molecular Docking Simulation Study
Abstract
1. Introduction
2. Results and Discussion
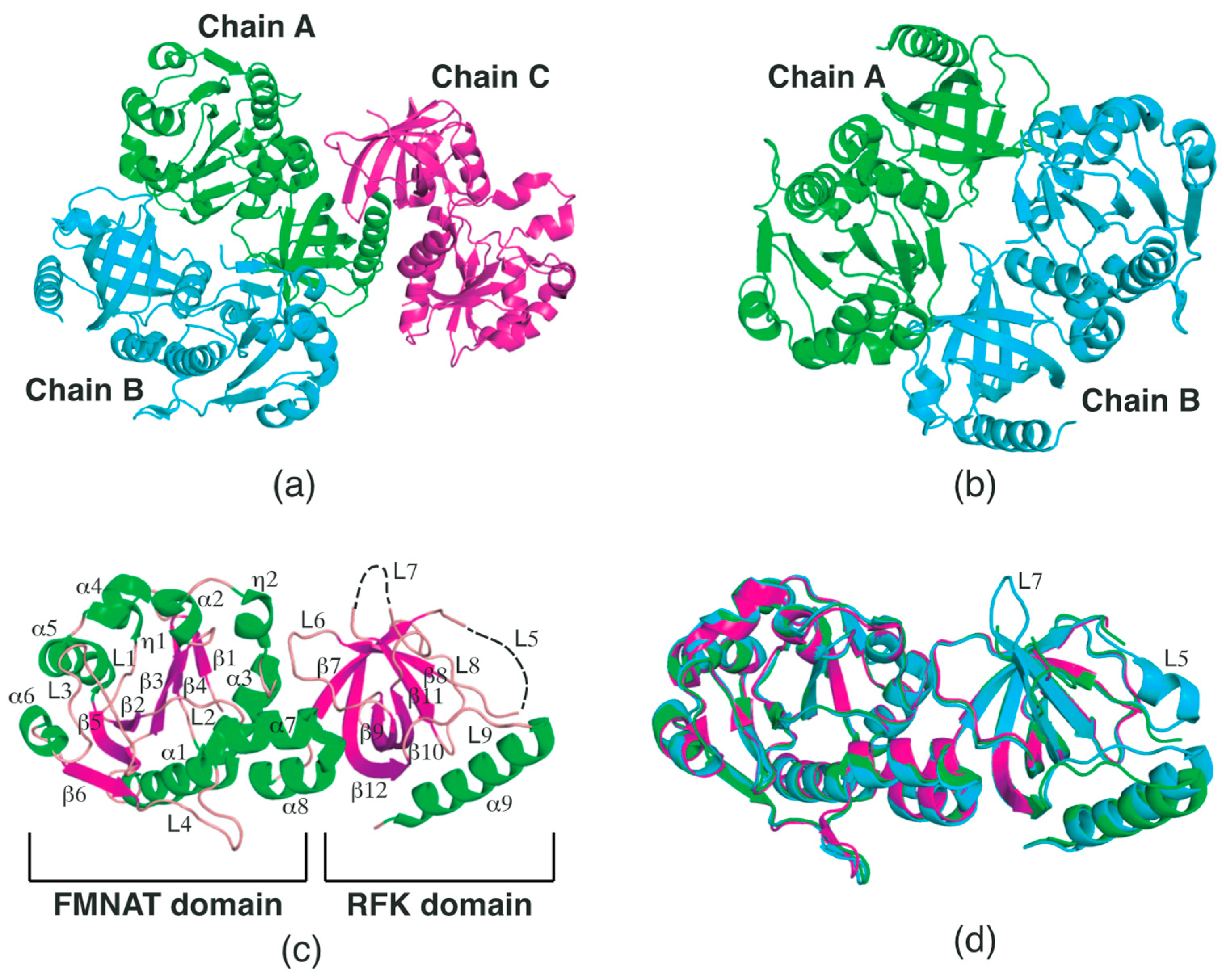
2.1. Structural Features of SpFADS
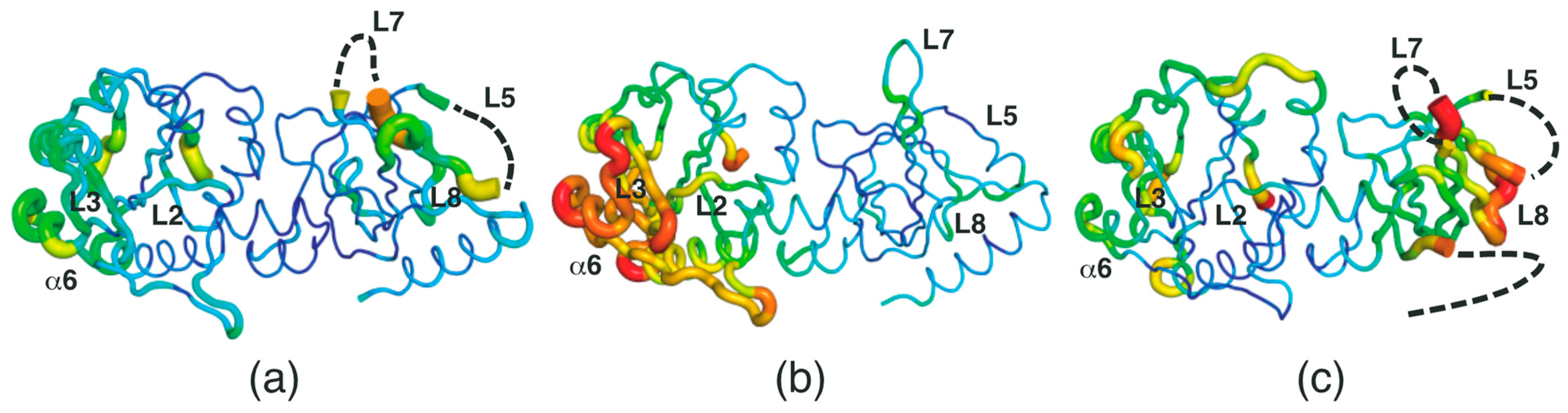
2.2. Conformational Changes in the RFK Domain
2.3. Surface Properties of SpFADS
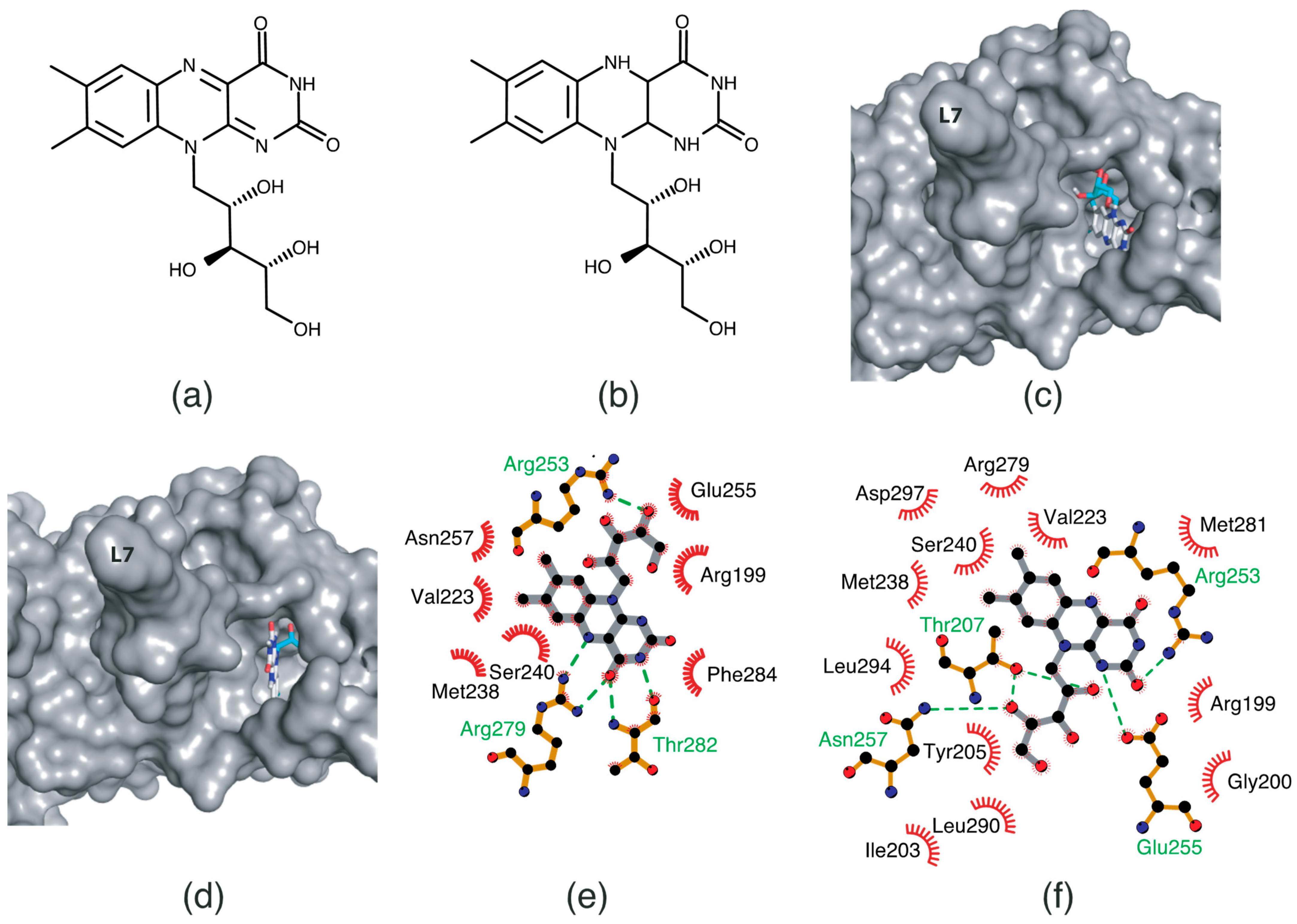
2.4. Oxidized and Reduced RF-Binding Modes in the RFK Domain
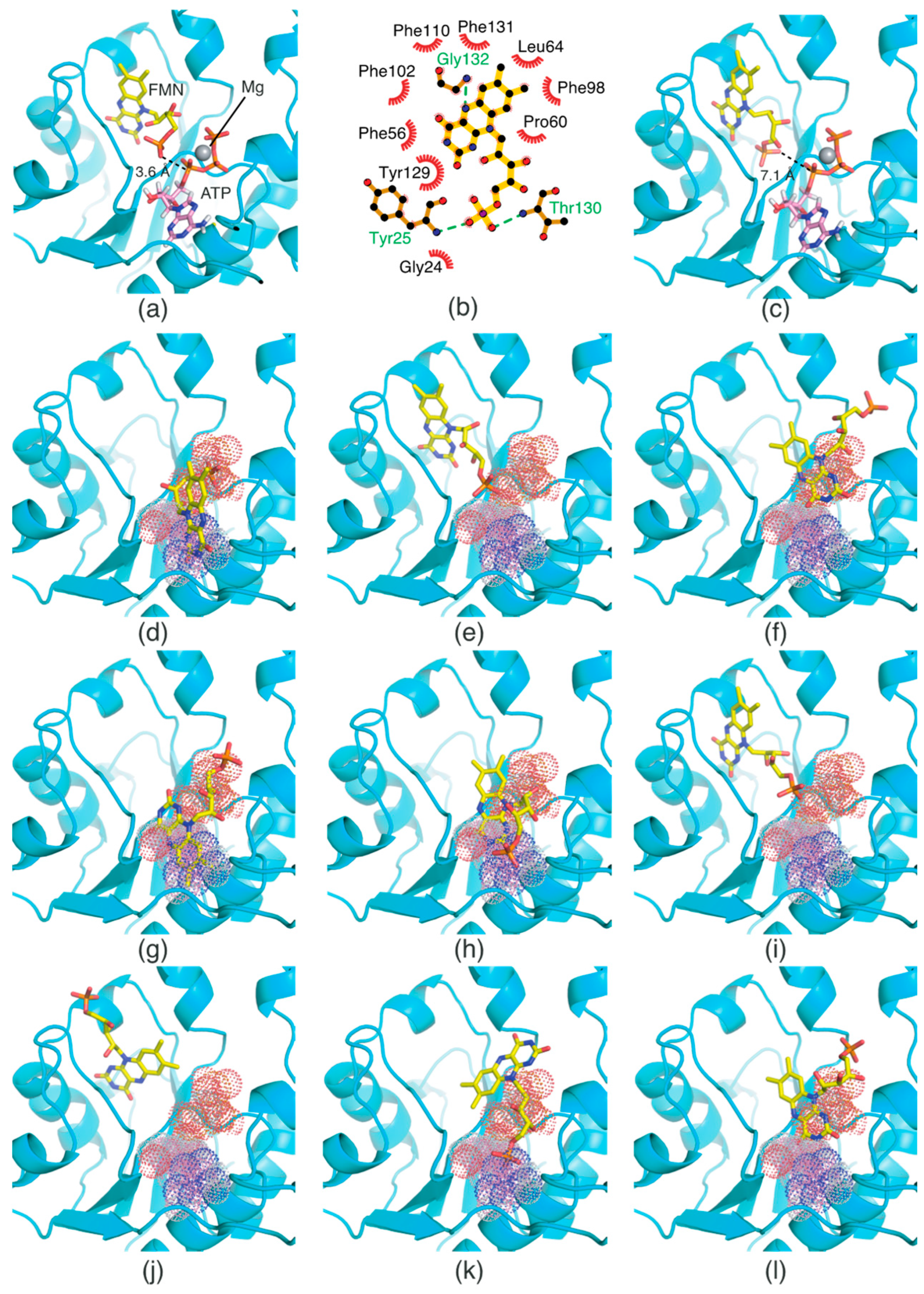
2.5. FMN-Binding Mode in the FMNAT Domain
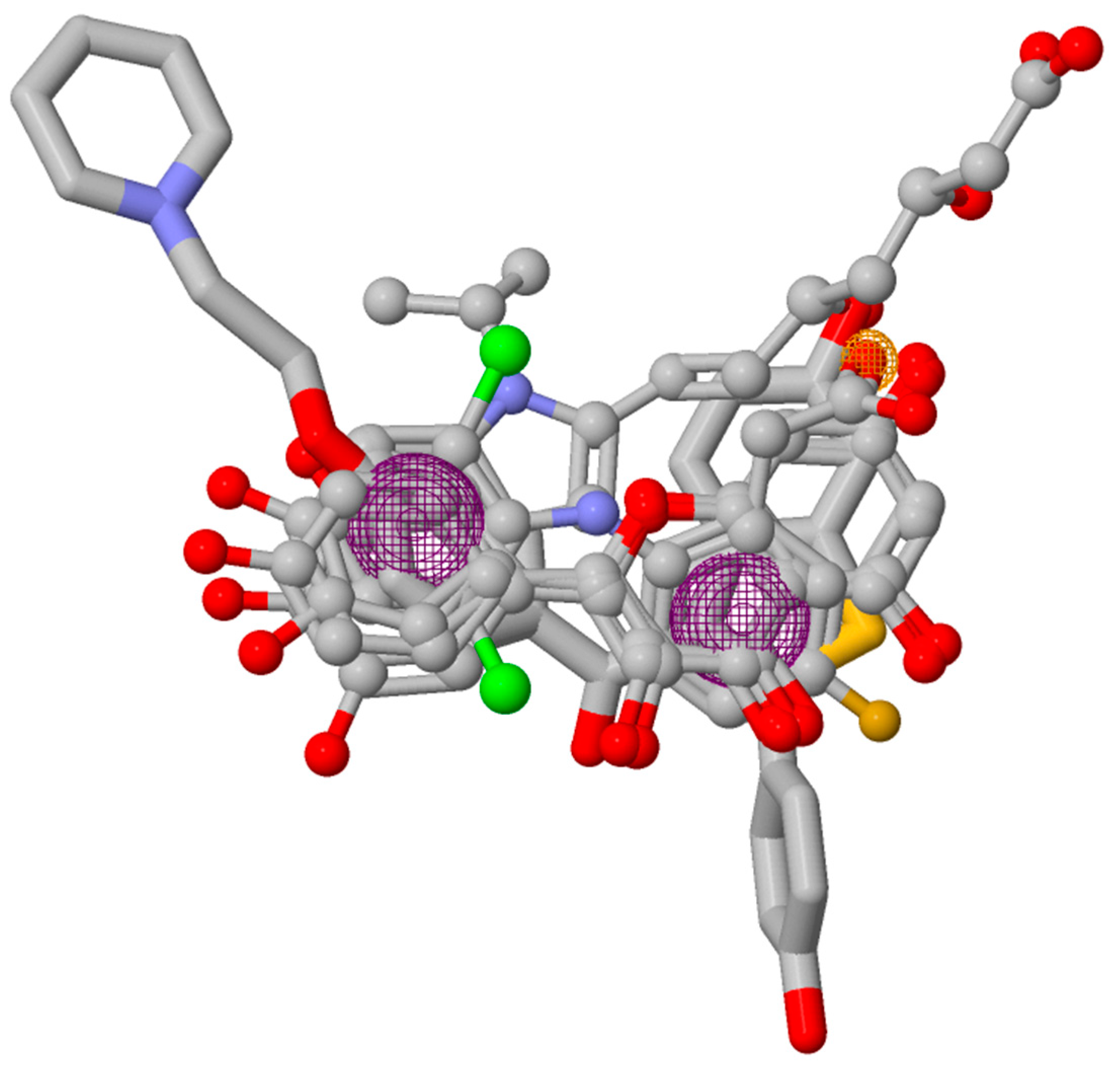
2.6. Pharmacophore Model
3. Materials and Methods
3.1. Protein and Substrate Preparation
3.2. Molecular Docking Simulation
3.3. 3D-Pharmacophore Modeling
3.4. Structure Visualization
4. Conclusions and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansoorabadi, S.O.; Thibodeaux, C.J.; Liu, H.W. The diverse roles of flavin coenzymes—nature’s most versatile thespians. J. Org. Chem. 2007, 72, 6329–6342. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Winge, D.R. Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim. Biophys. Acta 2013, 1827, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Garma, L.D.; Medina, M.; Juffer, A.H. Structure-based classification of FAD binding sites: A comparative study of structural alignment tools. Proteins 2016, 84, 1728–1747. [Google Scholar] [CrossRef] [PubMed]
- Manstein, D.J.; Pai, E.F. Purification and characterization of FAD synthetase from Brevibacterium ammoniagenes. J. Biol. Chem. 1986, 261, 16169–16173. [Google Scholar] [CrossRef]
- Efimov, I.; Kuusk, V.; Zhang, X.; McIntire, W.S. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. J. Biol. Chem. 1998, 37, 9716–9723. [Google Scholar] [CrossRef] [PubMed]
- Barile, M.; Brizio, C.; Valenti, D.; De Virgilio, C.; Passarella, S. The riboflavin/FAD cycle in rat liver mitochondria. Eur. J. Biochem. 2000, 267, 4888–4900. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, F.J.; Zhang, Y.; Roje, S. Flavin nucleotide metabolism in plants: Monofunctional enzymes synthesize fad in plastids. J. Biol. Chem. 2008, 283, 30890–30900. [Google Scholar] [CrossRef] [PubMed]
- Giancaspero, T.A.; Locato, V.; de Pinto, M.C.; De Gara, L.; Barile, M. The occurrence of riboflavin kinase and FAD synthetase ensures FAD synthesis in tobacco mitochondria and maintenance of cellular redox status. FEBS J. 2009, 276, 219–231. [Google Scholar] [CrossRef]
- Mashhadi, Z.; Xu, H.; Grochowski, L.L.; White, R.H. Archaeal RibL: A new FAD synthetase that is air sensitive. Biochemistry 2010, 49, 8748–8755. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.; Ferreira, P.; Martínez-Júlvez, M.; Medina, M. The prokaryotic FAD synthetase family: A potential drug target. Curr. Pharm. Des. 2013, 19, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Kim, R.; Jancarik, J.; Yokota, H.; Kim, S.H. Crystal structure of a flavin-binding protein from Thermotoga maritima. Proteins 2003, 52, 633–635. [Google Scholar] [CrossRef]
- Wang, W.; Kim, R.; Yokota, H.; Kim, S.H. Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima. Proteins 2005, 58, 246–248. [Google Scholar] [CrossRef]
- Herguedas, B.; Martínez-Júlvez, M.; Frago, S.; Medina, M.; Hermoso, J.A. Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J. Mol. Biol. 2010, 400, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Herguedas, B.; Lans, I.; Sebastián, M.; Hermoso, J.A.; Martínez-Júlvez, M.; Medina, M. Structural insights into the synthesis of FMN in prokaryotic organisms. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2526–2542. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C.; Frempong, G.A.; Balsera, M.; Medina, M.; Lostao, A. Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. Antioxidants 2021, 10, 1437. [Google Scholar] [CrossRef]
- Villanueva, R.; Ferreira, P.; Marcuello, C.; Usón, A.; Miramar, M.D.; Peleato, M.L.; Lostao, A.; Susin, S.A.; Medina, M. Key Residues Regulating the Reductase Activity of the Human Mitochondrial Apoptosis Inducing Factor. Biochemistry. 2015, 54, 5175–5184. [Google Scholar] [CrossRef] [PubMed]
- Sebastián, M.; Lira-Navarrete, E.; Serrano, A.; Marcuello, C.; Velázquez-Campoy, A.; Lostao, A.; Hurtado-Guerrero, R.; Medina, M.; Martínez-Júlvez, M. The FAD synthetase from the human pathogen Streptococcus pneumoniae: A bifunctional enzyme exhibiting activity-dependent redox requirements. Sci. Rep. 2017, 7, 7609. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Fogolari, F.; Brigo, A.; Molinari, H. The Poisson-Boltzmann equation for biomolecular electrostatics: A tool for structural biology. J. Mol. Recognit. 2002, 15, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Miyanaga, A.; Michaudel, Q.; Stull, F.; Louie, G.; Noel, J.P.; Baran, P.S.; Palfey, B.; Moore, B.S. Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement. Nature 2013, 503, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Barile, M.; Giancaspero, T.A.; Brizio, C.; Panebianco, C.; Indiveri, C.; Galluccio, M.; Vergani, L.; Eberini, I.; Gianazza, E. Biosynthesis of flavin cofactors in man: Implications in health and disease. Curr. Pharm. Des. 2013, 19, 2649–2675. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Stull, F.; Meehan, M.J.; Michaudel, Q.; Dorrestein, P.C.; Palfey, B.; Moore, B.S. Biochemical Establishment and Characterization of EncM’s Flavin-N5-oxide Cofactor. J. Am. Chem. Soc. 2015, 137, 8078–8085. [Google Scholar] [CrossRef]
- Teufel, R.; Agarwal, V.; Moore, B.S. Unusual flavoenzyme catalysis in marine bacteria. Curr. Opin. Chem. Biol. 2016, 31, 31–39. [Google Scholar] [CrossRef]
- Sebastián, M.; Anoz-Carbonell, E.; Gracia, B.; Cossio, P.; Aínsa, J.A.; Lans, I.; Medina, M. Discovery of antimicrobial compounds targeting bacterial type FAD synthetases. J. Enzyme Inhib. Med. Chem. 2018, 33, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. New model for calculating atomic charges in molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Dror, O.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PharmaGist: A webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008, 36, W223–W228. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2020; Available online: http://www.pymol.org (accessed on 26 November 2022).
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Iamurri, S.M.; Daugherty, A.B.; Edmondson, D.E.; Lutz, S. Truncated FAD synthetase for direct biocatalytic conversion of riboflavin and analogs to their corresponding flavin mononucleotides. Protein Eng. Des. Sel. 2013, 26, 791–795. [Google Scholar] [CrossRef] [PubMed]


| Rank | Binding Energy (kcal/mol) (Oxidized Form) | Binding Energy (kcal/mol) (Reduced Form) |
|---|---|---|
| 1 | −9.0 | −9.4 |
| 2 | −9.0 | −9.2 |
| 3 | −9.0 | −9.1 |
| 4 | −8.9 | −8.9 |
| 5 | −8.8 | −8.8 |
| 6 | −8.6 | −8.7 |
| 7 | −8.6 | −8.2 |
| 8 | −7.8 | −7.3 |
| 9 | −7.4 | −7.3 |
| 10 | −7.1 | −7.2 |
| Rank | Conformer | Binding Energy (kcal/mol) (with ATP) | Binding Energy (kcal/mol) (without ATP) |
|---|---|---|---|
| 1 | C1 | −9.1 | −8.9 |
| 2 | C2 | −9.1 | −8.8 |
| 3 | C3 | −9.0 | −8.8 |
| 4 | C4 | −8.8 | −8.7 |
| 5 | C5 | −8.8 | −8.6 |
| 6 | C6 | −8.7 | −8.5 |
| 7 | C7 | −8.6 | −8.1 |
| 8 | C8 | −8.4 | −8.1 |
| 9 | C9 | −8.3 | −8.1 |
| 10 | C10 | −8.1 | −8.1 |
| Compound | Chemical Structure |
|---|---|
| 1 | 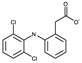 |
| 2 | 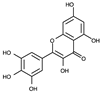 |
| 3 | 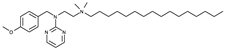 |
| 4 | 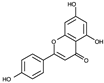 |
| 5 | 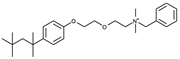 |
| 6 | 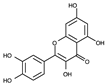 |
| 7 | 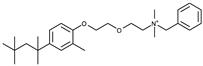 |
| 8 |  |
| 9 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, S. Structural Insight into the Working Mechanism of the FAD Synthetase from the Human Pathogen Streptococcus pneumoniae: A Molecular Docking Simulation Study. Int. J. Mol. Sci. 2023, 24, 3121. https://doi.org/10.3390/ijms24043121
Kwon S. Structural Insight into the Working Mechanism of the FAD Synthetase from the Human Pathogen Streptococcus pneumoniae: A Molecular Docking Simulation Study. International Journal of Molecular Sciences. 2023; 24(4):3121. https://doi.org/10.3390/ijms24043121
Chicago/Turabian StyleKwon, Sunghark. 2023. "Structural Insight into the Working Mechanism of the FAD Synthetase from the Human Pathogen Streptococcus pneumoniae: A Molecular Docking Simulation Study" International Journal of Molecular Sciences 24, no. 4: 3121. https://doi.org/10.3390/ijms24043121
APA StyleKwon, S. (2023). Structural Insight into the Working Mechanism of the FAD Synthetase from the Human Pathogen Streptococcus pneumoniae: A Molecular Docking Simulation Study. International Journal of Molecular Sciences, 24(4), 3121. https://doi.org/10.3390/ijms24043121




