Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation
Abstract
:1. Introduction
2. Results
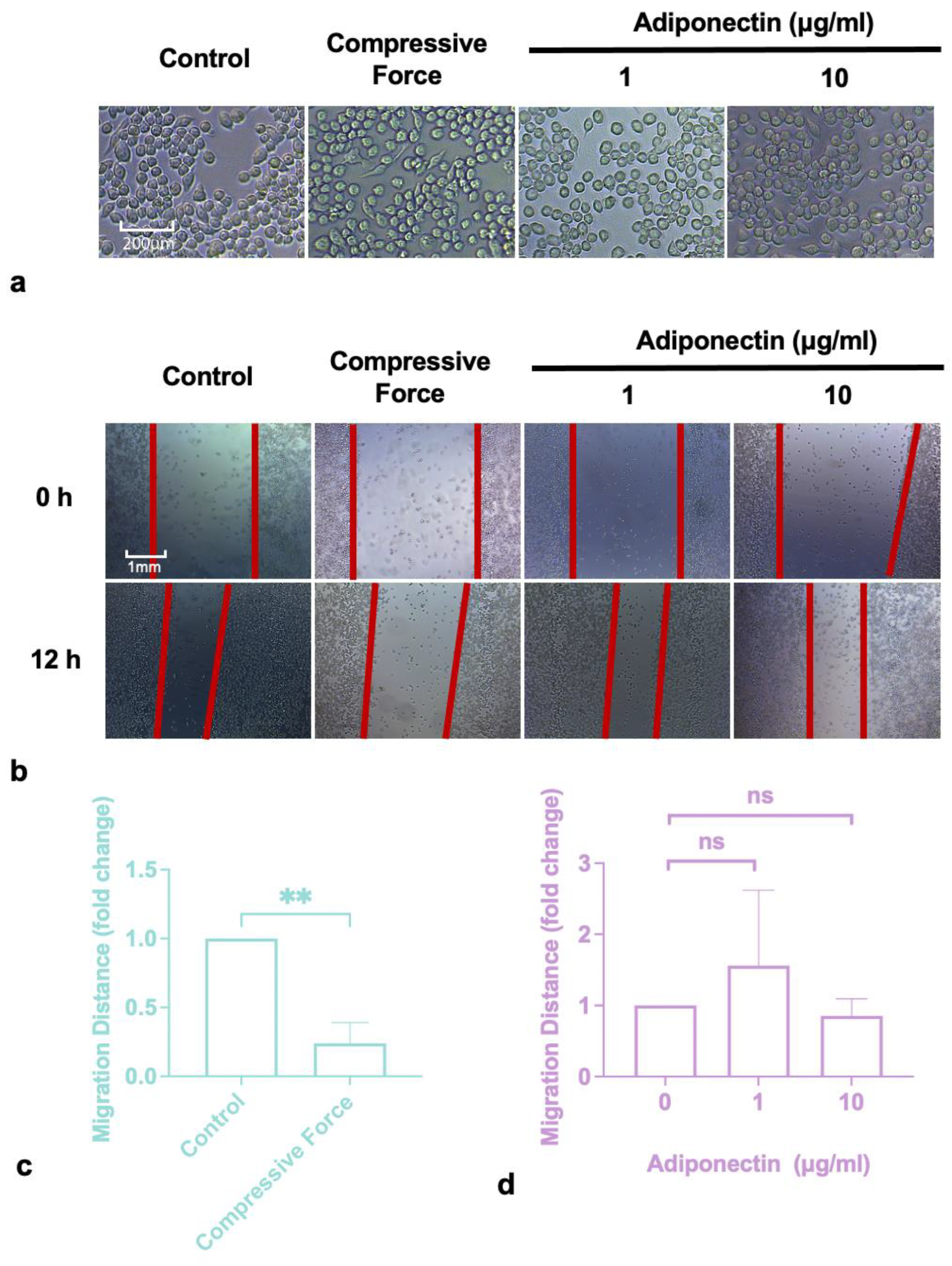
2.1. Compressive Force Inhibited Macrophages Migration
2.2. Compressive Force Promotes Macrophage Expression of Polarization Markers
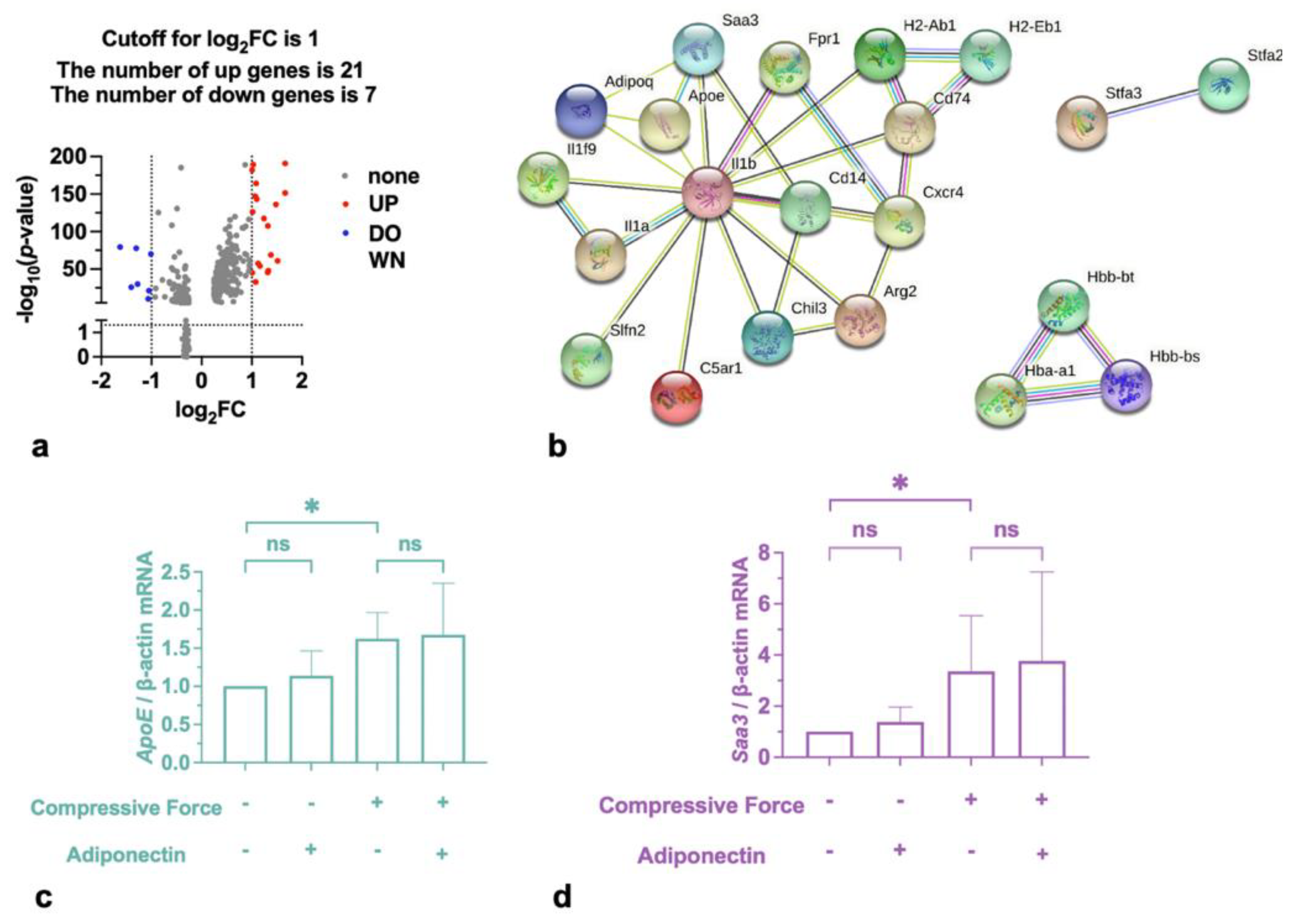
2.3. Compressive Force Promoted Saa3 and ApoE in Macrophages
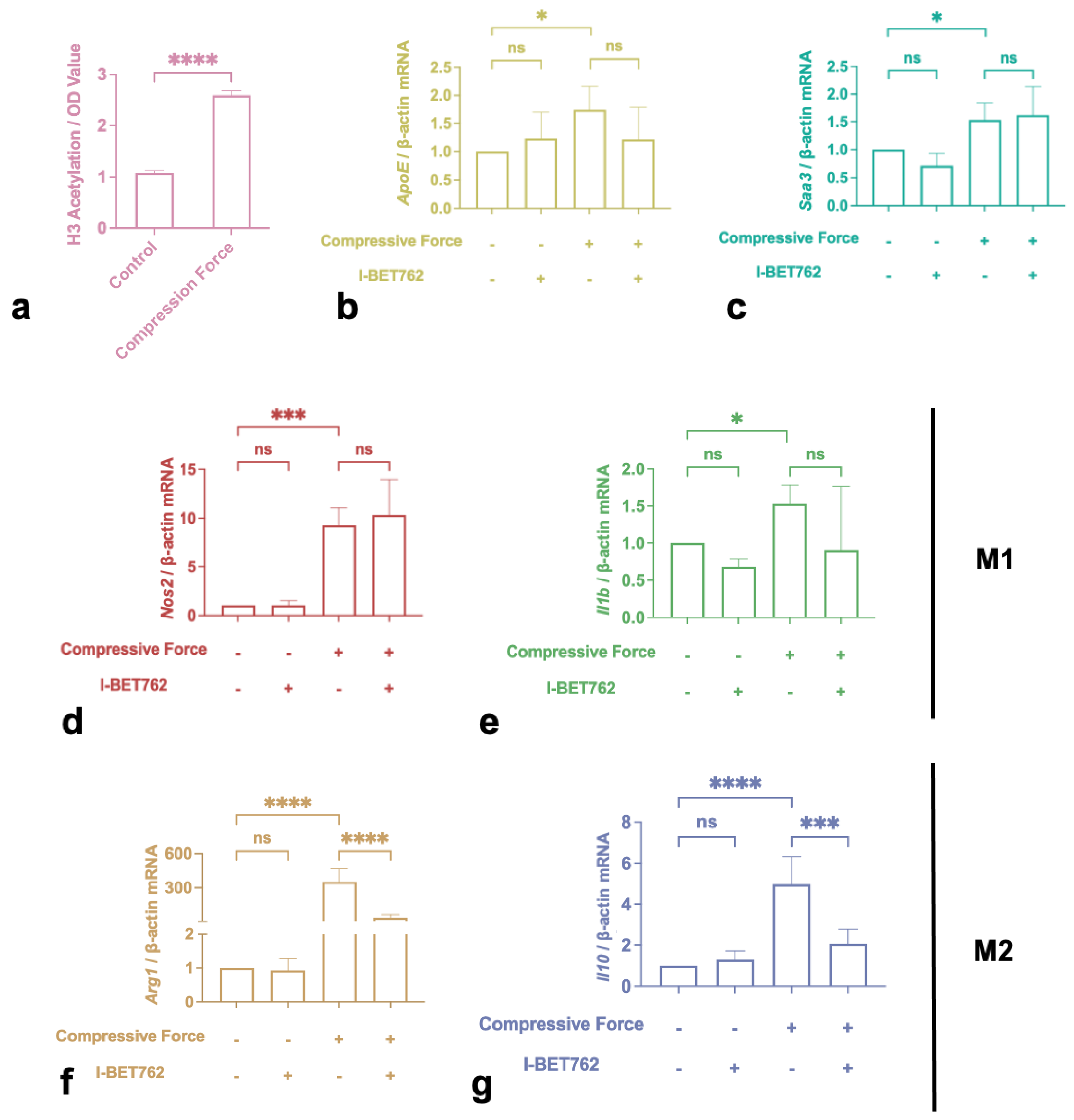
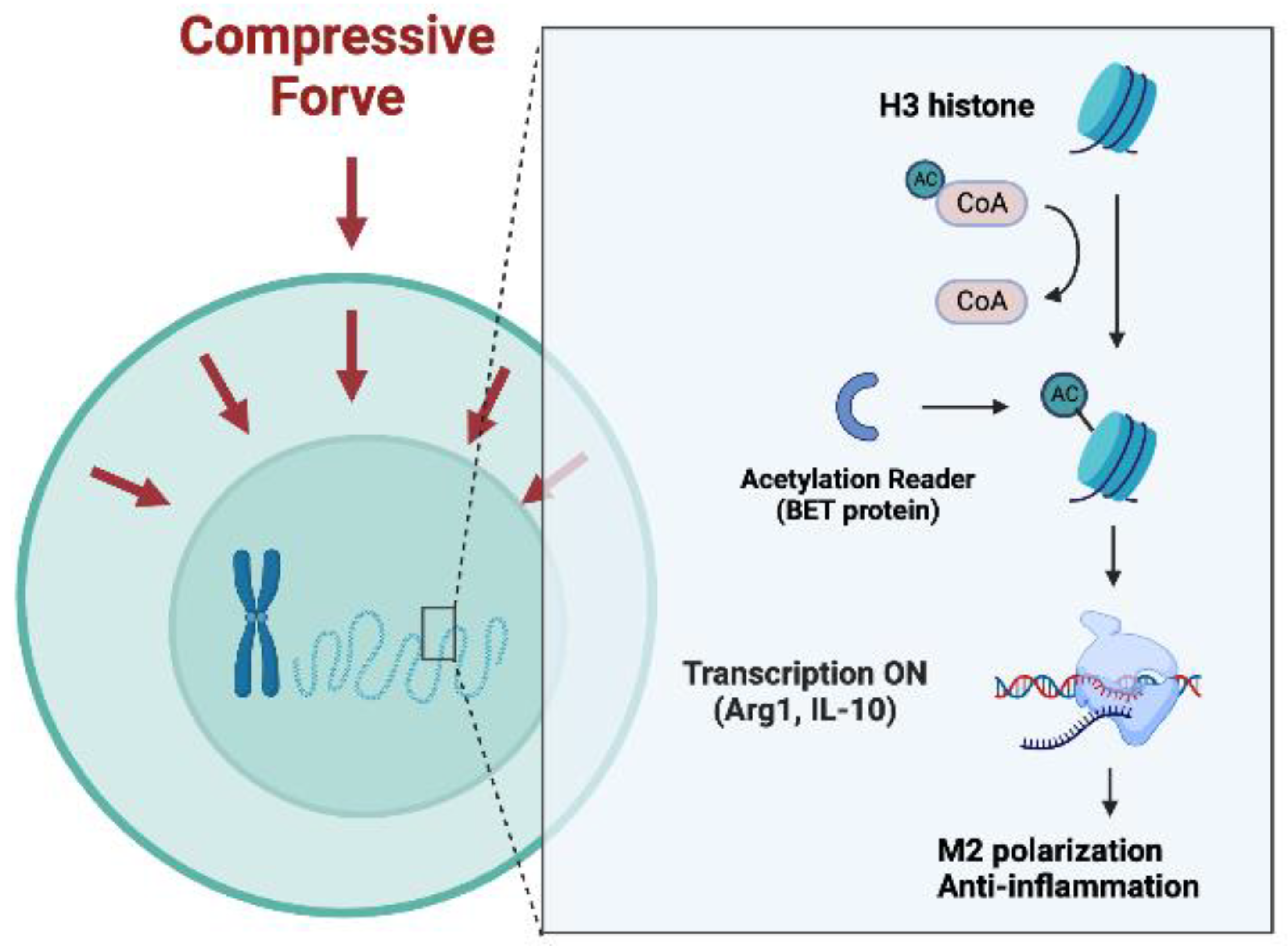
2.4. H3 Histone Acetylation Mediated M2 Polarization Induced by Compression
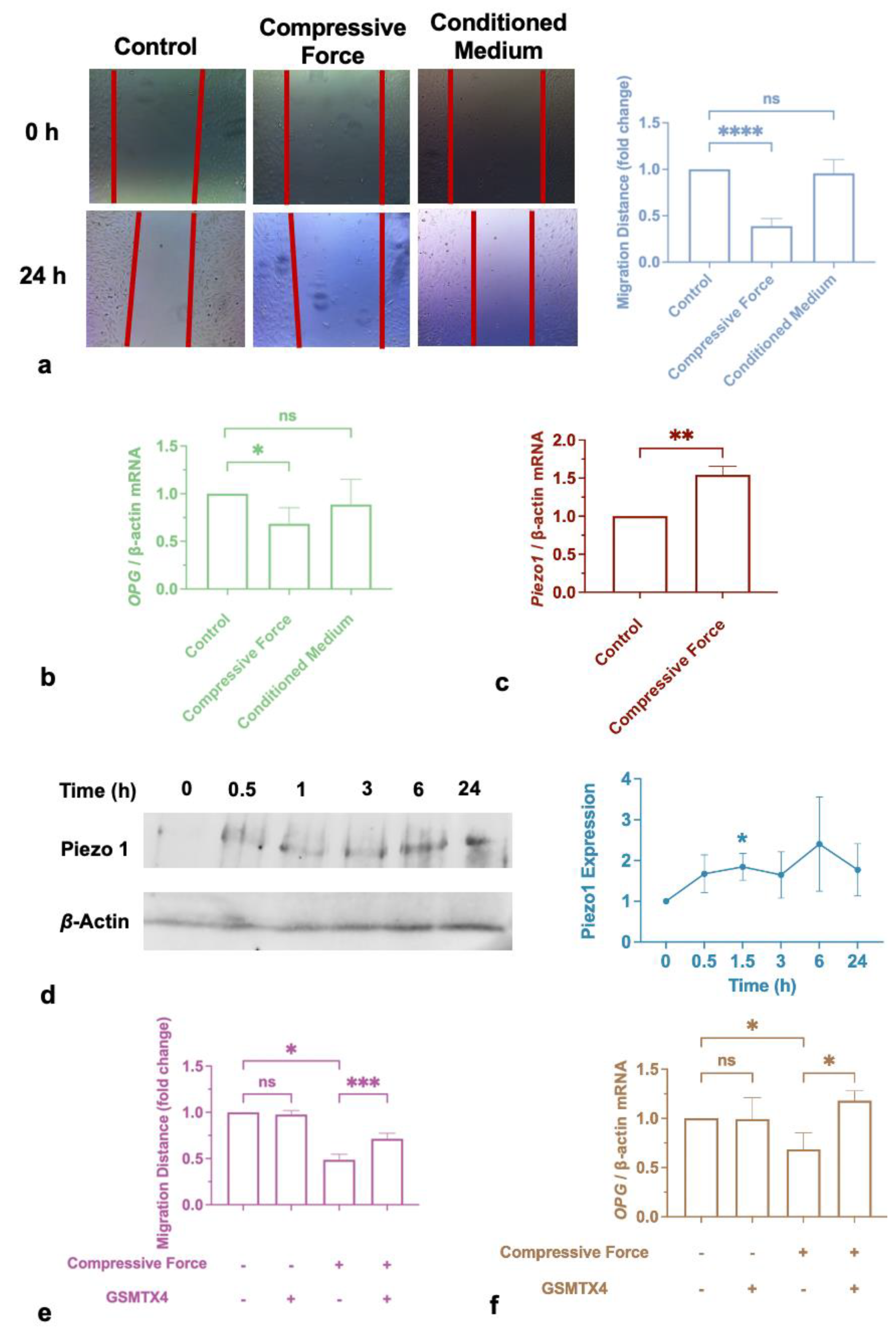
2.5. Cementum Repair was Impaired by Compression, but Not by Incubation with Macrophage-Conditioned Medium
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
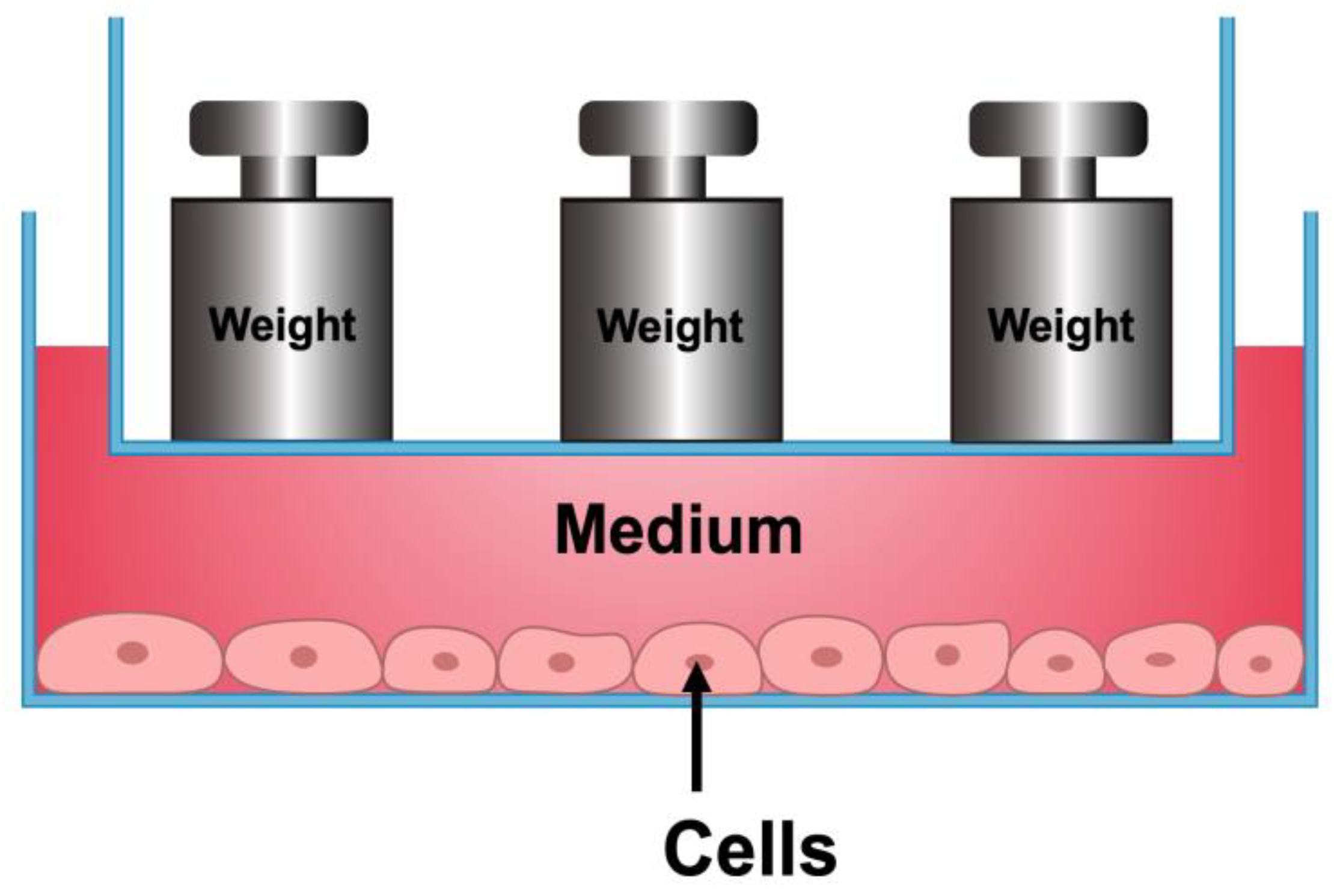
4.2. Compressive Force-Loading
4.3. Scratch Assay
4.4. Identification of DEGs and PPI Analysis
4.5. Quantitative Real-Time Reverse Transcriptase–Polymerase Chain Reaction (qRT-PCR)
4.6. Protein Extraction and Western Blot Analysis
4.7. H3 Histone Global Acetylation Levels
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krishnan, V.; Davidovitch, Z. On a path to unfolding the biological mechanisms of orthodontic tooth movement. J. Dent. Res. 2009, 88, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jacox, L.A.; Little, S.H.; Ko, C.C. Orthodontic tooth movement: The biology and clinical implications. Kaohsiung J. Med. Sci. 2018, 34, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Chaushu, S.; Klein, Y.; Mandelboim, O.; Barenholz, Y.; Fleissig, O. Immune Changes Induced by Orthodontic Forces: A Critical Review. J. Dent. Res. 2022, 101, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Fukasawa, S. Is Inflammation a Friend or Foe for Orthodontic Treatment?: Inflammation in Orthodontically Induced Inflammatory Root Resorption and Accelerating Tooth Movement. Int. J. Mol. Sci. 2021, 22, 2388. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Y.; Liu, H.; Zheng, Y.; Jia, L.; Li, W. Compressive force regulates cementoblast migration via downregulation of autophagy. J. Periodontol. 2021, 92, e128–e138. [Google Scholar] [CrossRef] [PubMed]
- Diercke, K.; Kohl, A.; Lux, C.J.; Erber, R. IL-1β and compressive forces lead to a significant induction of RANKL-expression in primary human cementoblasts. J. Orofac. Orthop. 2012, 73, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Feller, L.; Khammissa, R.A.; Thomadakis, G.; Fourie, J.; Lemmer, J. Apical External Root Resorption and Repair in Orthodontic Tooth Movement: Biological Events. Biomed Res. Int. 2016, 2016, 4864195. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Wan, C.; Liu, Y.; Wang, Y.; Meng, C.; Zhang, Y.; Jiang, C. NLRP3 inflammasome mediates M1 macrophage polarization and IL-1β production in inflammatory root resorption. J. Clin. Periodontol. 2020, 47, 451–460. [Google Scholar] [CrossRef]
- He, D.; Kou, X.; Luo, Q.; Yang, R.; Liu, D.; Wang, X.; Song, Y.; Cao, H.; Zeng, M.; Gan, Y.; et al. Enhanced M1/M2 macrophage ratio promotes orthodontic root resorption. J. Dent. Res. 2015, 94, 129–139. [Google Scholar] [CrossRef]
- Lee, S.Y.; Yoo, H.I.; Kim, S.H. CCR5-CCL Axis in PDL during Orthodontic Biophysical Force Application. J. Dent. Res. 2015, 94, 1715–1723. [Google Scholar] [CrossRef]
- Janjic Rankovic, M.; Docheva, D.; Wichelhaus, A.; Baumert, U. Effect of static compressive force on in vitro cultured PDL fibroblasts: Monitoring of viability and gene expression over 6 days. Clin. Oral Investig. 2020, 24, 2497–2511. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mo, S.; Hua, Y. Compressive force-induced autophagy in periodontal ligament cells downregulates osteoclastogenesis during tooth movement. J. Periodontol. 2019, 90, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Hind, L.E.; Dembo, M.; Hammer, D.A. Macrophage motility is driven by frontal-towing with a force magnitude dependent on substrate stiffness. Integr. Biol. 2015, 7, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Mennens, S.F.B.; van den Dries, K.; Cambi, A. Role for Mechanotransduction in Macrophage and Dendritic Cell Immunobiology. Results Probl. Cell Differ. 2017, 62, 209–242. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, S.; Yoshinaga, Y.; Kuramoto, A.; Nagano, F.; Ichimura, I.; Oshino, K.; Yoshimura, A.; Yano, Y.; Hara, Y. Occlusal trauma accelerates attachment loss at the onset of experimental periodontitis in rats. J. Periodontal Res. 2014, 49, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.L.; Negrini, D.; Rocco, P.R. Mechanisms of ventilator-induced lung injury in healthy lungs. Best Pract. Res. Clin. Anaesthesiol. 2015, 29, 301–313. [Google Scholar] [CrossRef]
- Zanotelli, M.R.; Zhang, J.; Reinhart-King, C.A. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab. 2021, 33, 1307–1321. [Google Scholar] [CrossRef]
- Funes, S.C.; Rios, M.; Escobar-Vera, J.; Kalergis, A.M. Implications of macrophage polarization in autoimmunity. Immunology 2018, 154, 186–195. [Google Scholar] [CrossRef]
- Yong, J.; von Bremen, J.; Ruiz-Heiland, G.; Ruf, S. Adiponectin Interacts In-Vitro With Cementoblasts Influencing Cell Migration, Proliferation and Cementogenesis Partly Through the MAPK Signaling Pathway. Front. Pharmacol. 2020, 11, 585346. [Google Scholar] [CrossRef]
- Xu, N.; Li, X.; Weng, J.; Wei, C.; He, Z.; Doycheva, D.M.; Lenahan, C.; Tang, W.; Zhou, J.; Liu, Y.; et al. Adiponectin Ameliorates GMH-Induced Brain Injury by Regulating Microglia M1/M2 Polarization Via AdipoR1/APPL1/AMPK/PPARγ Signaling Pathway in Neonatal Rats. Front. Immunol. 2022, 13, 873382. [Google Scholar] [CrossRef]
- Rizzo, M.R.; Fasano, R.; Paolisso, G. Adiponectin and Cognitive Decline. Int. J. Mol. Sci. 2020, 21, 2010. [Google Scholar] [CrossRef]
- Orsini, E.M.; Perelas, A.; Southern, B.D.; Grove, L.M.; Olman, M.A.; Scheraga, R.G. Stretching the Function of Innate Immune Cells. Front. Immunol. 2021, 12, 767319. [Google Scholar] [CrossRef] [PubMed]
- Veerasubramanian, P.K.; Shao, H.; Meli, V.S.; Phan, T.A.Q.; Luu, T.U.; Liu, W.F.; Downing, T.L. A Src-H3 acetylation signaling axis integrates macrophage mechanosensation with inflammatory response. Biomaterials 2021, 279, 121236. [Google Scholar] [CrossRef]
- Maruyama, K.; Nemoto, E.; Yamada, S. Mechanical regulation of macrophage function—Cyclic tensile force inhibits NLRP3 inflammasome-dependent IL-1β secretion in murine macrophages. Inflamm. Regen. 2019, 39, 3. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, F.Y.; Wang, T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef] [PubMed]
- Wosik, J.; Chen, W.; Qin, K.; Ghobrial, R.M.; Kubiak, J.Z.; Kloc, M. Magnetic Field Changes Macrophage Phenotype. Biophys. J. 2018, 114, 2001–2013. [Google Scholar] [CrossRef]
- Fang, X.Y.; Zhan, Y.X.; Zhou, X.M.; Wu, L.N.; Lin, J.; Yi, Y.T.; Jiang, C.M.; Wang, J.; Liu, J. CXCL12/CXCR4 Mediates Orthodontic Root Resorption via Regulating the M1/M2 Ratio. J. Dent. Res. 2022, 101, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef]
- Modolell, M.; Corraliza, I.M.; Link, F.; Soler, G.; Eichmann, K. Reciprocal regulation of the nitric oxide synthase/arginase balance in mouse bone marrow-derived macrophages by TH1 and TH2 cytokines. Eur. J. Immunol. 1995, 25, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.; Akhavan, N.S.; Mullins, A.P.; Arjmandi, B.H. Macrophage Polarization and Osteoporosis: A Review. Nutrients 2020, 12, 2999. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, S.; Sathe, A.A.; Jin, Z.; Guan, J.; Sun, W.; Xing, C.; Zhang, H.; Yan, B. CCR2(+) Macrophages Promote Orthodontic Tooth Movement and Alveolar Bone Remodeling. Front. Immunol. 2022, 13, 835986. [Google Scholar] [CrossRef]
- Oishi, Y.; Manabe, I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Choi, S.M.; Kim, B.C. Adiponectin Regulates the Polarization and Function of Microglia via PPAR-γ Signaling Under Amyloid β Toxicity. Front. Cell. Neurosci. 2017, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Lovren, F.; Pan, Y.; Quan, A.; Szmitko, P.E.; Singh, K.K.; Shukla, P.C.; Gupta, M.; Chan, L.; Al-Omran, M.; Teoh, H.; et al. Adiponectin primes human monocytes into alternative anti-inflammatory M2 macrophages. Am. J. Physiol. Heart Circ. Physiol. 2010, 299, H656–H663. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Pratt, B.T.; Barnes, M.; McMullen, M.R.; Nagy, L.E. Molecular mechanism for adiponectin-dependent M2 macrophage polarization: Link between the metabolic and innate immune activity of full-length adiponectin. J. Biol. Chem. 2011, 286, 13460–13469. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Luo, Y.; Luo, L.; Wu, D.; Ding, X.; Zheng, H.; Wu, H.; Liu, B.; Yang, X.; Silva, F.; et al. Adiponectin restrains ILC2 activation by AMPK-mediated feedback inhibition of IL-33 signaling. J. Exp. Med. 2021, 218, e20191054. [Google Scholar] [CrossRef]
- Dikmen, K.; Bostanci, H.; Gobut, H.; Yavuz, A.; Alper, M.; Kerem, M. Recombinant adiponectin inhibits inflammation processes via NF-kB pathway in acute pancreatitis. Bratisl. Lek. Listy 2018, 119, 619–624. [Google Scholar] [CrossRef]
- Tannock, L.R.; De Beer, M.C.; Ji, A.; Shridas, P.; Noffsinger, V.P.; den Hartigh, L.; Chait, A.; De Beer, F.C.; Webb, N.R. Serum amyloid A3 is a high density lipoprotein-associated acute-phase protein. J. Lipid Res. 2018, 59, 339–347. [Google Scholar] [CrossRef]
- Kim, W.S.; Kim, H.J.; Lee, Z.H.; Lee, Y.; Kim, H.H. Apolipoprotein E inhibits osteoclast differentiation via regulation of c-Fos, NFATc1 and NF-κB. Exp. Cell Res. 2013, 319, 436–446. [Google Scholar] [CrossRef]
- Lanfranco, M.F.; Sepulveda, J.; Kopetsky, G.; Rebeck, G.W. Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia 2021, 69, 1478–1493. [Google Scholar] [CrossRef] [PubMed]
- Lakshmaiah, K.C.; Jacob, L.A.; Aparna, S.; Lokanatha, D.; Saldanha, S.C. Epigenetic therapy of cancer with histone deacetylase inhibitors. J. Cancer Res. Ther. 2014, 10, 469–478. [Google Scholar] [CrossRef]
- Schuldt, L.; von Brandenstein, K.; Jacobs, C.; Symmank, J. Oleic acid-related anti-inflammatory effects in force-stressed PdL fibroblasts are mediated by H3 lysine acetylation associated with altered IL10 expression. Epigenetics 2022, 17, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Vogel, V. Spatial confinement downsizes the inflammatory response of macrophages. Nat. Mater. 2018, 17, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wu, R.; Tang, D.; Kang, R. The BET family in immunity and disease. Signal Transduct. Target. Ther. 2021, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Chen, Y.; Wang, H.Y.; Wang, R.F. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum. Vac. Immunother. 2014, 10, 3270–3285. [Google Scholar] [CrossRef] [PubMed]
- Burdette, B.E.; Esparza, A.N.; Zhu, H.; Wang, S. Gasdermin D in pyroptosis. Acta Pharm. Sin. B 2021, 11, 2768–2782. [Google Scholar] [CrossRef]
- Martinac, B.; Nikolaev, Y.A.; Silvani, G.; Bavi, N.; Romanov, V.; Nakayama, Y.; Martinac, A.D.; Rohde, P.; Bavi, O.; Cox, C.D. Cell membrane mechanics and mechanosensory transduction. Curr. Top. Membr. 2020, 86, 83–141. [Google Scholar] [CrossRef]
- Shibasaki, K. TRPV4 activation by thermal and mechanical stimuli in disease progression. Lab. Investig. 2020, 100, 218–223. [Google Scholar] [CrossRef]
- Yu, J.L.; Liao, H.Y. Piezo-type mechanosensitive ion channel component 1 (Piezo1) in human cancer. Biomed. Pharmacother. 2021, 140, 111692. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, D.; Zhang, C.; Yang, W.; Li, C.; Gao, Z.; Pei, K.; Li, Y. Piezo1 mediates endothelial atherogenic inflammatory responses via regulation of YAP/TAZ activation. Hum. Cell 2022, 35, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Solis, A.G.; Bielecki, P.; Steach, H.R.; Sharma, L.; Harman, C.C.D.; Yun, S.; de Zoete, M.R.; Warnock, J.N.; To, S.D.F.; York, A.G.; et al. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature 2019, 573, 69–74. [Google Scholar] [CrossRef] [PubMed]
- D’Errico, J.A.; MacNeil, R.L.; Takata, T.; Berry, J.; Strayhorn, C.; Somerman, M.J. Expression of bone associated markers by tooth root lining cells, in situ and in vitro. Bone 1997, 20, 117–126. [Google Scholar] [CrossRef] [PubMed]

| DEGs | Gene Name |
|---|---|
| Upregulated | Saa3, BC100530, Lrg1, Chil3, Stfa2, Il1b, Ifitm1, Asprv1, Fpr1, Gm5483, Il1a, Apoe, Il1f9, Marcksl1, Arg2, Stfa2l1, C5ar1, Cd14, Ms4a8a, Stfa3, Slfn2 |
| Downregulated | Cxcr4, Cd74, H2-Eb1, H2-Ab1, Hbb-bs, Hbb-bt, Hba-a1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Groeger, S.; Yong, J.; Ruf, S. Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation. Int. J. Mol. Sci. 2023, 24, 3117. https://doi.org/10.3390/ijms24043117
Wang Y, Groeger S, Yong J, Ruf S. Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation. International Journal of Molecular Sciences. 2023; 24(4):3117. https://doi.org/10.3390/ijms24043117
Chicago/Turabian StyleWang, Yao, Sabine Groeger, Jiawen Yong, and Sabine Ruf. 2023. "Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation" International Journal of Molecular Sciences 24, no. 4: 3117. https://doi.org/10.3390/ijms24043117
APA StyleWang, Y., Groeger, S., Yong, J., & Ruf, S. (2023). Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation. International Journal of Molecular Sciences, 24(4), 3117. https://doi.org/10.3390/ijms24043117






