Circulating Microbial Cell-Free DNA in Health and Disease
Abstract
1. Introduction
2. Circulating cfmDNA or Contamination?
3. The Presence of Circulating cfmDNA in Patients and Healthy Individuals
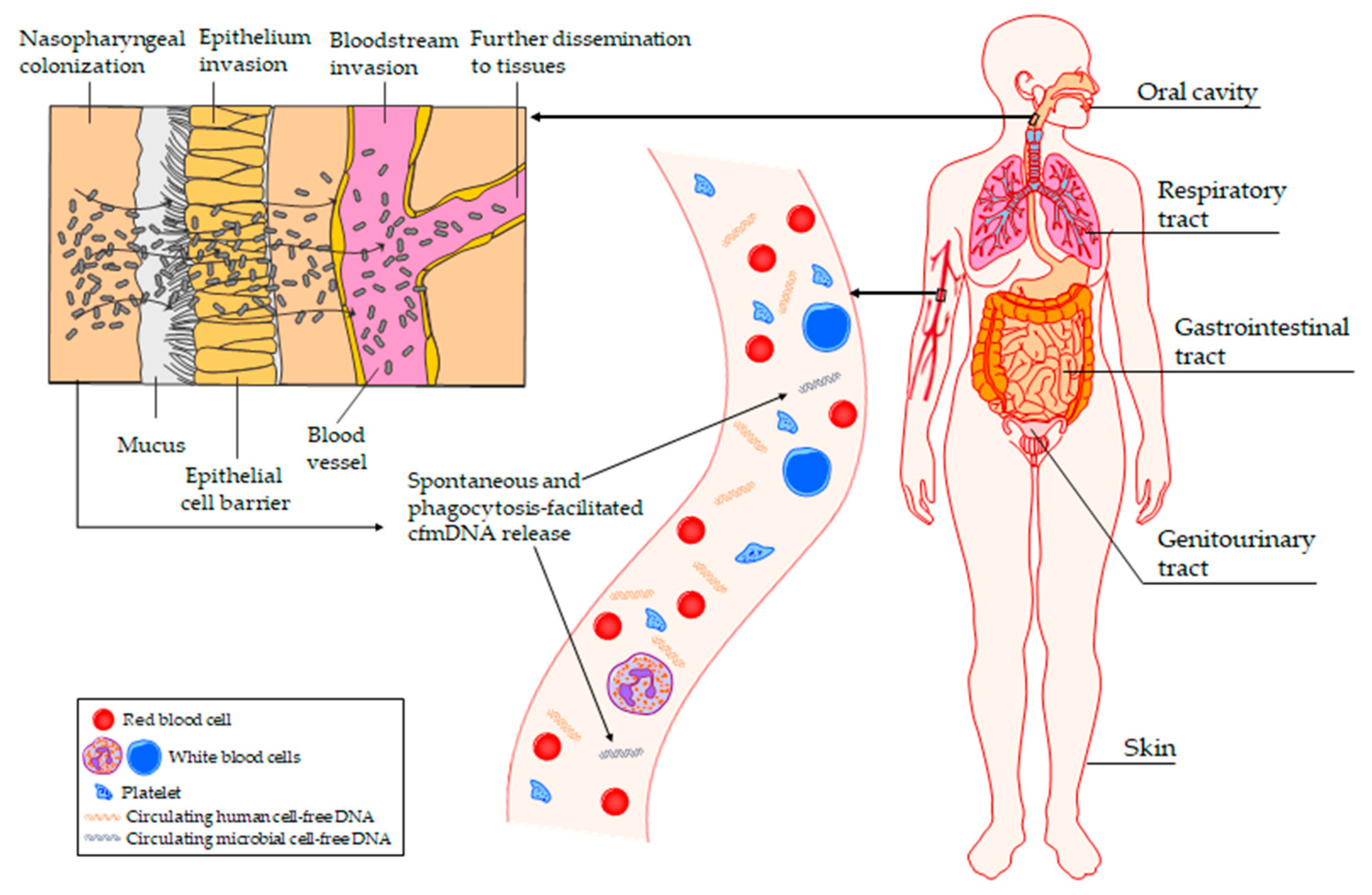
4. The Potential Origin of Circulating cfmDNA
5. Potential of Circulating cfmDNA in Clinical Applications
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ranucci, R. Cell-Free DNA: Applications in Different Diseases. Methods Mol. Biol. 2019, 1909, 3–12. [Google Scholar] [PubMed]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme [Nuclear Acids in Human Blood Plasma]. Comptes Rendus Des Seances Soc. Biol. Ses Fil. 1948, 142, 241–243. [Google Scholar]
- Bronkhorst, A.J.; Ungerer, V.; Oberhofer, A.; Gabriel, S.; Polatoglou, E.; Randeu, H.; Uhlig, C.; Pfister, H.; Mayer, Z.; Holdenrieder, S. New Perspectives on the Importance of Cell-Free DNA Biology. Diagnostics 2022, 12, 2147. [Google Scholar] [CrossRef] [PubMed]
- Wadden, J.; Ravi, K.; John, V.; Babila, C.M.; Koschmann, C. Cell-Free Tumor DNA (Cf-TDNA) Liquid Biopsy: Current Methods and Use in Brain Tumor Immunotherapy. Front. Immunol. 2022, 13, 882452. [Google Scholar] [CrossRef]
- Wang, J.-W.; Lyu, Y.-N.; Qiao, B.; Li, Y.; Zhang, Y.; Dhanyamraju, P.K.; Bamme, Y.; Yu, M.D.; Yang, D.; Tong, Y.-Q. Cell-Free Fetal DNA Testing and Its Correlation with Prenatal Indications. BMC Pregnancy Childbirth 2021, 21, 585. [Google Scholar] [CrossRef]
- Martuszewski, A.; Paluszkiewicz, P.; Król, M.; Banasik, M.; Kepinska, M. Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review. J. Clin. Med. 2021, 10, 193. [Google Scholar] [CrossRef]
- Jing, Q.; Hung, C.; Leung, C.; Wu, A.R. Cell-Free DNA as Biomarker for Sepsis by Integration of Microbial and Host Information. Clin. Chem. 2022, 68, 1184–1195. [Google Scholar] [CrossRef]
- Grabuschnig, S.; Bronkhorst, A.J.; Holdenrieder, S.; Rodriguez, I.R.; Schliep, K.P.; Schwendenwein, D.; Ungerer, V.; Sensen, C.W. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. J. Mol. Sci. 2020, 21, 8062. [Google Scholar] [CrossRef]
- Han, D.S.; Lo, Y.D. The Nexus of CfDNA and Nuclease Biology. Trends Genet. 2021, 37, 758–770. [Google Scholar] [CrossRef]
- Kowarsky, M.; Camunas-Soler, J.; Kertesz, M.; de Vlaminck, I.; Koh, W.; Pan, W.; Martin, L.; Neff, N.F.; Okamoto, J.; Wong, R.J.; et al. Numerous Uncharacterized and Highly Divergent Microbes Which Colonize Humans Are Revealed by Circulating Cell-Free DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 9623–9628. [Google Scholar] [CrossRef]
- Xiao, Q.; Lu, W.; Kong, X.; Shao, Y.W.; Hu, Y.; Wang, A.; Bao, H.; Cao, R.; Liu, K.; Wang, X.; et al. Alterations of Circulating Bacterial DNA in Colorectal Cancer and Adenoma: A Proof-of-Concept Study. Cancer Lett. 2021, 499, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Yu, X.; Du, Y.; Su, F.; Liu, Y.; Li, H.; Liu, Y.; Mu, K.; Liu, Q.; Li, H.; et al. Peripheral Blood Microbiome Analysis via Noninvasive Prenatal Testing Reveals the Complexity of Circulating Microbial Cell-Free DNA. Microbiol. Spectr. 2022, 10, e0041422. [Google Scholar] [CrossRef] [PubMed]
- Cebriá-Mendoza, M.; Bracho, M.A.; Arbona, C.; Larrea, L.; Díaz, W.; Sanjuán, R.; Cuevas, J.M. Exploring the Diversity of the Human Blood Virome. Viruses 2021, 13, 2322. [Google Scholar] [CrossRef] [PubMed]
- Whittle, E.; Leonard, M.O.; Harrison, R.; Gant, T.W. Multi-Method Characterization of the Human Circulating Microbiome. Front. Microbiol. 2019, 9, 3266. [Google Scholar] [CrossRef] [PubMed]
- Païssé, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive Description of Blood Microbiome from Healthy Donors Assessed by 16S Targeted Metagenomic Sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zou, S.; Chu, M.; Chen, J.; Zhong, J.; Chen, Y.; Fan, J.; Qi, J.; Wang, Q. Cell Free Bacterial DNAs in Human Plasma Provide Fingerprints for Immune-Related Diseases. Med. Microecol. 2020, 5, 100022. [Google Scholar] [CrossRef]
- Goggin, K.P.; Gonzalez-Pena, V.; Inaba, Y.; Allison, K.J.; Hong, D.K.; Ahmed, A.A.; Hollemon, D.; Natarajan, S.; Mahmud, O.; Kuenzinger, W.; et al. Evaluation of Plasma Microbial Cell-Free DNA Sequencing to Predict Bloodstream Infection in Pediatric Patients with Relapsed or Refractory Cancer. JAMA Oncol. 2020, 6, 552–556. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The Dormant Blood Microbiome in Chronic, Inflammatory Diseases. FEMS Microbiol. Rev. 2015, 013, 567–591. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Methe, B.; Thompson, K.M.; Potgieter, M.; Castillo, D.J.; Rifkin, R.F.; Cowan, D.A. The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell. Infect. Microbiol. 2019, 1, 148. [Google Scholar] [CrossRef]
- Zozaya-Valdés, E.; Wong, S.Q.; Raleigh, J.; Hatzimihalis, A.; Ftouni, S.; Papenfuss, A.T.; Sandhu, S.; Dawson, M.A.; Dawson, S.J. Detection of Cell-Free Microbial DNA Using a Contaminant-Controlled Analysis Framework. Genome Biol. 2021, 22, 187. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhou, H.; Jing, Y.; Dong, C. Association between Blood Microbiome and Type 2 Diabetes Mellitus: A Nested Case-control Study. J. Clin. Lab. Anal. 2019, 33, e22842. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.K.; Blauwkamp, T.A.; Kertesz, M.; Bercovici, S.; Truong, C.; Banaei, N. Liquid Biopsy for Infectious Diseases: Sequencing of Cell-Free Plasma to Detect Pathogen DNA in Patients with Invasive Fungal Disease. Diagn. Microbiol. Infect. Dis. 2018, 92, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Stein, U.; Barault, L.; Forslund, S.; Zhang, L.; Zhang, Y.; Dong, Z.; Chen, B.; Pan, H.; Wang, D.; Liu, M.; et al. Detection of Microbial 16S RRNA Gene in the Serum of Patients with Gastric Cancer. Front. Oncol. 2019, 9, 608. [Google Scholar] [CrossRef]
- Dinakaran, V.; Rathinavel, A.; Pushpanathan, M.; Sivakumar, R.; Gunasekaran, P. Elevated Levels of Circulating DNA in Cardiovascular Disease Patients: Metagenomic Profiling of Microbiome in the Circulation. PLoS ONE 2014, 9, 105221. [Google Scholar] [CrossRef]
- Huang, Y.F.; Chen, Y.J.; Fan, T.C.; Chang, N.C.; Chen, Y.J.; Midha, M.K.; Chen, T.H.; Yang, H.H.; Wang, Y.T.; Yu, A.L.; et al. Analysis of Microbial Sequences in Plasma Cell-Free DNA for Early-Onset Breast Cancer Patients and Healthy Females. BMC Med. Genom. 2018, 11, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Barrett, S.L.R.; Holmes, E.A.; Long, D.R.; Shean, R.C.; Bautista, G.E.; Ravishankar, S.; Peddu, V.; Cookson, B.T.; Singh, P.K.; Greninger, A.L.; et al. Cell Free DNA from Respiratory Pathogens Is Detectable in the Blood Plasma of Cystic Fibrosis Patients. Sci. Rep. 2020, 10, 6903. [Google Scholar] [CrossRef]
- Cho, E.J.; Leem, S.; Kim, S.A.; Yang, J.; Lee, Y.B.; Kim, S.S.; Cheong, J.Y.; Cho, S.W.; Kim, J.W.; Kim, S.M.; et al. Circulating Microbiota-Based Metagenomic Signature for Detection of Hepatocellular Carcinoma. Sci. Rep. 2019, 17, 7536. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome Analyses of Blood and Tissues Suggest Cancer Diagnostic Approach. Nature 2020, 579, 567. [Google Scholar] [CrossRef]
- Grumaz, S.; Grumaz, C.; Vainshtein, Y.; Stevens, P.; Glanz, K.; Decker, S.O.; Hofer, S.; Weigand, M.A.; Brenner, T.; Sohn, K. Enhanced Performance of Next-Generation Sequencing Diagnostics Compared With Standard of Care Microbiological Diagnostics in Patients Suffering From Septic Shock. Crit. Care Med. 2019, 47, e394. [Google Scholar] [CrossRef]
- Echeverria, A.P.; Cohn, I.S.; Danko, D.C.; Shanaj, S.; Blair, L.; Hollemon, D.; Carli, A.V.; Sculco, P.K.; Ho, C.; Meshulam-Simon, G.; et al. Sequencing of Circulating Microbial Cell-Free DNA Can Identify Pathogens in Periprosthetic Joint Infections. J. Bone Jt. Surg. 2021, 103, 1705–1712. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Wang, P.; Zhang, J.; Li, K.; Ye, B.; Li, G.; Zhou, J.; Tong, Z.; Ke, L.; Shi, S.; et al. Plasma Metagenomic Next-Generation Sequencing of Microbial Cell-Free DNA Detects Pathogens in Patients with Suspected Infected Pancreatic Necrosis. BMC Infect. Dis. 2021, 22, 675. [Google Scholar] [CrossRef]
- Kitsios, G.D.; Bain, W.; Al-Yousif, N.; Duttagupta, R.; Ahmed, A.A.; McVerry, B.J.; Morris, A. Plasma Microbial Cell-Free DNA Load Is Associated with Mortality in Patients with COVID-19. Respir. Res. 2021, 22, 24. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Degner, N.; Scott, E.R.; Ruffin, F.; Franzone, J.; Sharma-Kuinkel, B.; Shah, P.; Hong, D.; Dalai, S.C.; Blair, L.; et al. Microbial Cell-Free DNA Identifies the Causative Pathogen in Infective Endocarditis and Remains Detectable Longer Than Conventional Blood Culture in Patients with Prior Antibiotic Therapy. Clin. Infect. Dis. 2022, ciac426. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, E.M.; de Vries, C.R.; Ruffin, F.; Sharma-Kuinkel, B.; Park, L.; Hong, D.; Scott, E.R.; Blair, L.; Degner, N.; Hollemon, D.H.; et al. Microbial Cell-Free DNA Identifies Etiology of Bloodstream Infections, Persists Longer Than Conventional Blood Cultures, and Its Duration of Detection Is Associated With Metastatic Infection in Patients With Staphylococcus Aureus and Gram-Negative Bacteremia. Clin. Infect. Dis. 2022, 74, 2020. [Google Scholar] [CrossRef] [PubMed]
- Benamu, E.; Gajurel, K.; Anderson, J.N.; Lieb, T.; Gomez, C.A.; Seng, H.; Aquino, R.; Hollemon, D.; Hong, D.K.; Blauwkamp, T.A.; et al. Plasma Microbial Cell-Free DNA Next-Generation Sequencing in the Diagnosis and Management of Febrile Neutropenia. Clin. Infect. Dis. 2022, 74, 1659–1668. [Google Scholar] [CrossRef]
- Witt, R.G.; Blair, L.; Frascoli, M.; Rosen, M.J.; Nguyen, Q.-H.; Bercovici, S.; Zompi, S.; Romero, R.; Mackenzie, T.C. Detection of Microbial Cell-Free DNA in Maternal and Umbilical Cord Plasma in Patients with Chorioamnionitis Using next Generation Sequencing. PLoS ONE 2020, 15, e0231239. [Google Scholar] [CrossRef]
- Yang, H.; Haidar, G.; Al-Yousif, N.S.; Zia, H.; Kotok, D.; Ahmed, A.A.; Blair, L.; Dalai, S.; Bercovici, S.; Ho, C.; et al. Circulating Microbial Cell-Free DNA Is Associated with Inflammatory Host-Responses in Severe Pneumonia. Thorax 2021, 76, 1231–1235. [Google Scholar] [CrossRef]
- Velmurugan, G.; Dinakaran, V.; Rajendhran, J.; Swaminathan, K. Blood Microbiota and Circulating Microbial Metabolites in Diabetes and Cardiovascular Disease. Trends Endocrinol. Metab. 2020, 31, 835–847. [Google Scholar] [CrossRef]
- Zhang, L.; Morrison, M.; Nimmo, G.R.; Sriprakash, K.S.; Mondot, S.; Gowardman, J.R.; George, N.; Marsh, N.; Rickard, C.M. Molecular Investigation of Bacterial Communities on the Inner and Outer Surfaces of Peripheral Venous Catheters. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1083–1090. [Google Scholar] [CrossRef]
- Søby, J.H.; Watt, S.K.; Vogelsang, R.P.; Servant, F.; Lelouvier, B.; Raskov, H.; Knop, F.K.; Gögenur, I. Alterations in Blood Microbiota after Colonic Cancer Surgery. BJS Open. 2020, 4, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Tomás, I.; Diz, P.; Tobías, A.; Scully, C.; Donos, N. Periodontal Health Status and Bacteraemia from Daily Oral Activities: Systematic Review/Meta-Analysis. J. Clin. Periodontol. 2012, 39, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Wiest, R.; Lawson, M.; Geuking, M. Pathological Bacterial Translocation in Liver Cirrhosis. J. Hepatol. 2014, 60, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Gargari, G.; Mantegazza, G.; Taverniti, V.; del Bo, C.; Bernardi, S.; Andres-Lacueva, C.; González-Domínguez, R.; Kroon, P.A.; Winterbone, M.S.; Cherubini, A.; et al. Bacterial DNAemia Is Associated with Serum Zonulin Levels in Older Subjects. Sci. Rep. 2021, 11, 11054. [Google Scholar] [CrossRef] [PubMed]
- Rittirsch, D.; Flierl, M.A.; Nadeau, B.A.; Day, D.E.; Huber-Lang, M.S.; Grailer, J.J.; Zetoune, F.S.; Andjelkovic, A.V.; Fasano, A.; Ward, P.A. Zonulin as Prehaptoglobin2 Regulates Lung Permeability and Activates the Complement System. Am. J. Physiol. Cell. Mol. Physiol. 2013, 304, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The Microbiome of the Lung. Transl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef]
- Isler, B.; Kidd, T.J.; Stewart, A.G.; Harris, P.; Paterson, D.L. Achromobacter Infections and Treatment Options. Antimicrob. Agents Chemother. 2020, 64, e01025-20. [Google Scholar] [CrossRef]
- Michielan, A.; D’Incà, R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediat. Inflamm. 2015, 2015, 628157. [Google Scholar] [CrossRef]
- Martinez, R.M.; Wolk, D.M. Bloodstream Infections. Microbiol. Spectr. 2016, 4, 653–689. [Google Scholar] [CrossRef]
- Balajee, S.A.; Gribskov, J.L.; Hanley, E.; Nickle, D.; Marr, K.A. Aspergillus lentulus Sp. Nov., a New Sibling Species of A. fumigatus. Eukaryot Cell 2005, 4, 625–632. [Google Scholar] [CrossRef]
- MacIntyre, A.T.; Hirst, A.; Duttagupta, R.; Hollemon, D.; Hong, D.K.; Blauwkamp, T.A. Budget Impact of Microbial Cell-Free DNA Testing Using the Karius® Test as an Alternative to Invasive Procedures in Immunocompromised Patients with Suspected Invasive Fungal Infections. Appl. Health Econ. Health Policy 2021, 19, 231. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, Y.; Zhang, S.; Dong, L. Gut microbiota-mediated immunomodulation in tumor. J. Exp. Clin. Cancer Res. 2021, 40, 221. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Luong, M.K.; Shaw, H.; Nathan, P.; Bataille, V.; Spector, T.D. The Gut Microbiome: What the Oncologist Ought to Know. Br. J. Cancer 2021, 125, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Glyn, T.; Purcell, R. Circulating Bacterial DNA: A New Paradigm for Cancer Diagnostics. Front. Med. 2022, 9, 643. [Google Scholar] [CrossRef]
- Lee, R.A.; al Dhaheri, F.; Pollock, N.R.; Sharma, T.S. Assessment of the Clinical Utility of Plasma Metagenomic Next-Generation Sequencing in a Pediatric Hospital Population. J. Clin. Microbiol. 2020, 58, e00419-20. [Google Scholar] [CrossRef]
- Chiu, K.-P.; Yu, A.L. Application of Cell-Free DNA Sequencing in Characterization of Bloodborne Microbes and the Study of Microbe-Disease Interactions. PeerJ 2019, 7, e7426. [Google Scholar] [CrossRef]
| Clinical Context | Major Findings | References |
|---|---|---|
| Septic shock | >70% of the positivity rate for NGS-based pathogen identification over the whole study period. 96% of positive NGS results for acute sepsis time points were plausible. NGS results would have led to a change to a more adequate therapy in 53% of cases. | [30] |
| Relapsed or refractory pediatric cancers | 75% and 80% of predictive sensitivity of NGS for all BSIs and bacterial BSIs, respectively, in the 3 days before the onset of infection. 82% and 91% of the specificity of NGS, for any bacterial or fungal organism and any common BSI pathogen, respectively. | [17] |
| BSIs | 89.3% and 74.3% of the NGS sensitivity and specificity, respectively. NGS identified causative pathogens for a significantly longer interval than conventional blood cultures (median 15 days vs. 2 days; p < 0.0001). The odds of metastatic infection significantly increased with each additional day of circulating cfmDNA detection (odds ratio, 2.89; p = 0.0011). | [35] |
| Sepsis | NGS reached the sensitivity and specificity of 0.952 and 1, respectively, for the identification of bacterial infection, and allowed for the simultaneous detection of viral pathogens. NGS revealed differences in the composition of circulating cfmDNA between septic and non-septic patients and between survivors and non-survivors by 28-day mortality. Improved performance was achieved in identifying sepsis and the prediction of clinical outcomes for ICU patients with AUC of 0.992 and 0.802, respectively, by integrating the information from circulating human and microbial cfDNA into a machine learning model. | [7] |
| Infective endocarditis | NGS achieved a sensitivity of 87%. NGS identified causative pathogens for a significantly longer interval than conventional blood cultures (median duration of positivity from antibiotic treatment initiation was 38.1 days vs. 3.7 days; p = 0.02771). The level of cfmDNA significantly decreased after surgical source control. | [34] |
| Periprosthetic joint infections | NGS identified causative pathogens in 57% of cases, increasing pathogen detection to 94% (as an adjunct to tissue cultures).NGS improved the time-to-speciation (the median time was 3 days less than standard-of-care methods).After treatment, NGS did not detect circulating cfmDNA of the infectious pathogen or there were reduced levels of circulating cfmDNA. | [31] |
| Infected pancreatic necrosis | The positivity rate of NGS was 54.55%. 83.33% of NGS-positive cases were consistent with the culture results of infected pancreatic necrosis drains. The PPA and NPA of NGS were 80.0% and 89.47%, respectively. Patients in the NGS positive group had new-onset septic shock significantly more frequently (12 (50.0%) vs. 4 (20.0%), p = 0.039) than those in the negative group. | [32] |
| Febrile neutropenia | The PPA and NPA of NGS were 90% and 31%, respectively. NGS sensitivity and specificity were 85% and 100%, respectively. NGS improved the time to diagnosis. NGS results would have led to a change to a more adequate therapy in 47% of cases. | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietrzak, B.; Kawacka, I.; Olejnik-Schmidt, A.; Schmidt, M. Circulating Microbial Cell-Free DNA in Health and Disease. Int. J. Mol. Sci. 2023, 24, 3051. https://doi.org/10.3390/ijms24033051
Pietrzak B, Kawacka I, Olejnik-Schmidt A, Schmidt M. Circulating Microbial Cell-Free DNA in Health and Disease. International Journal of Molecular Sciences. 2023; 24(3):3051. https://doi.org/10.3390/ijms24033051
Chicago/Turabian StylePietrzak, Bernadeta, Iwona Kawacka, Agnieszka Olejnik-Schmidt, and Marcin Schmidt. 2023. "Circulating Microbial Cell-Free DNA in Health and Disease" International Journal of Molecular Sciences 24, no. 3: 3051. https://doi.org/10.3390/ijms24033051
APA StylePietrzak, B., Kawacka, I., Olejnik-Schmidt, A., & Schmidt, M. (2023). Circulating Microbial Cell-Free DNA in Health and Disease. International Journal of Molecular Sciences, 24(3), 3051. https://doi.org/10.3390/ijms24033051









