Effects of a Novel Infant Formula on the Fecal Microbiota in the First Six Months of Life: The INNOVA 2020 Study
Abstract
1. Introduction
2. Results
2.1. Phylum Level
2.2. Genus Level
2.3. Species Levels
2.4. Rivera-Pinto Microbiome Balance
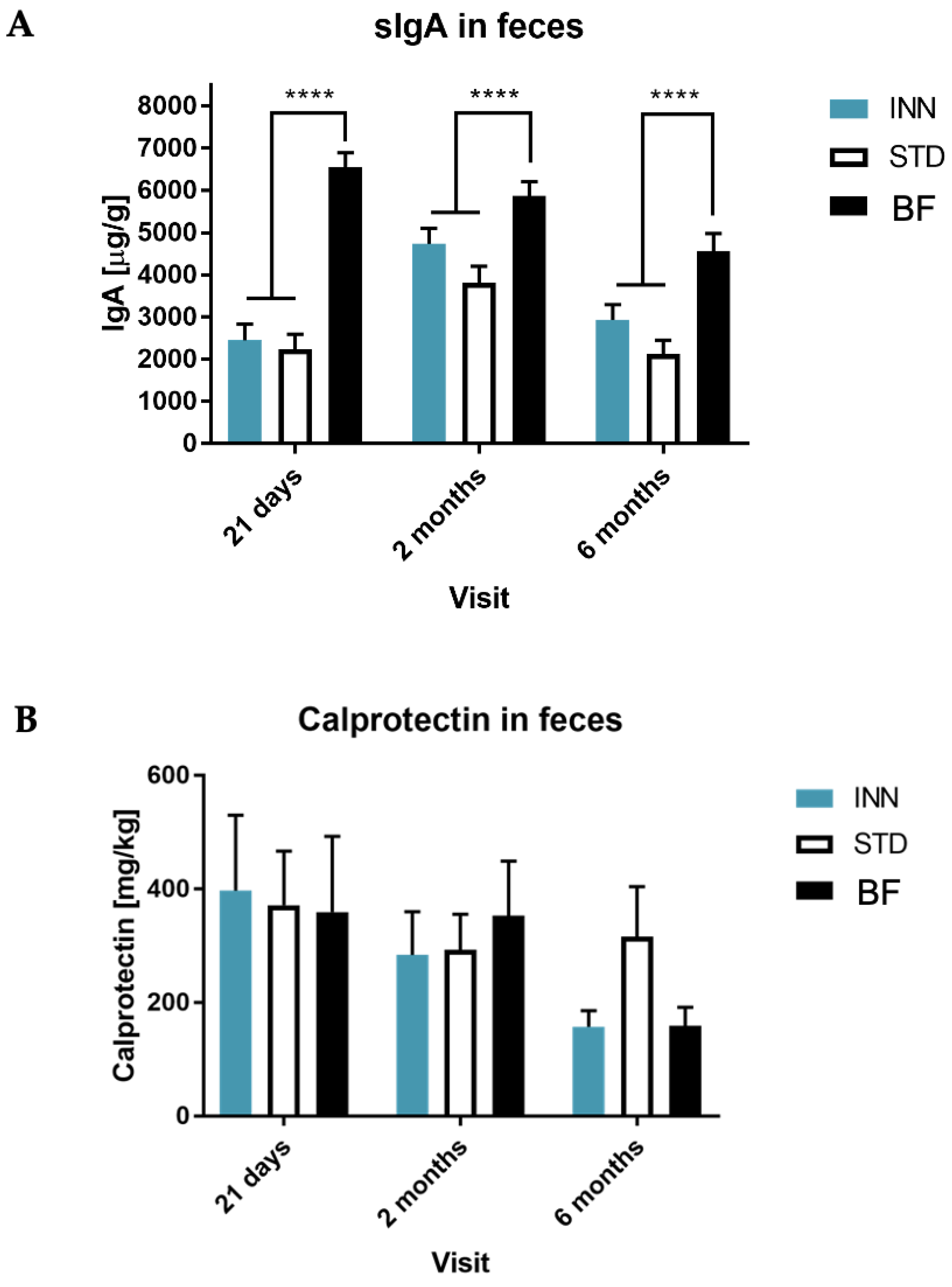
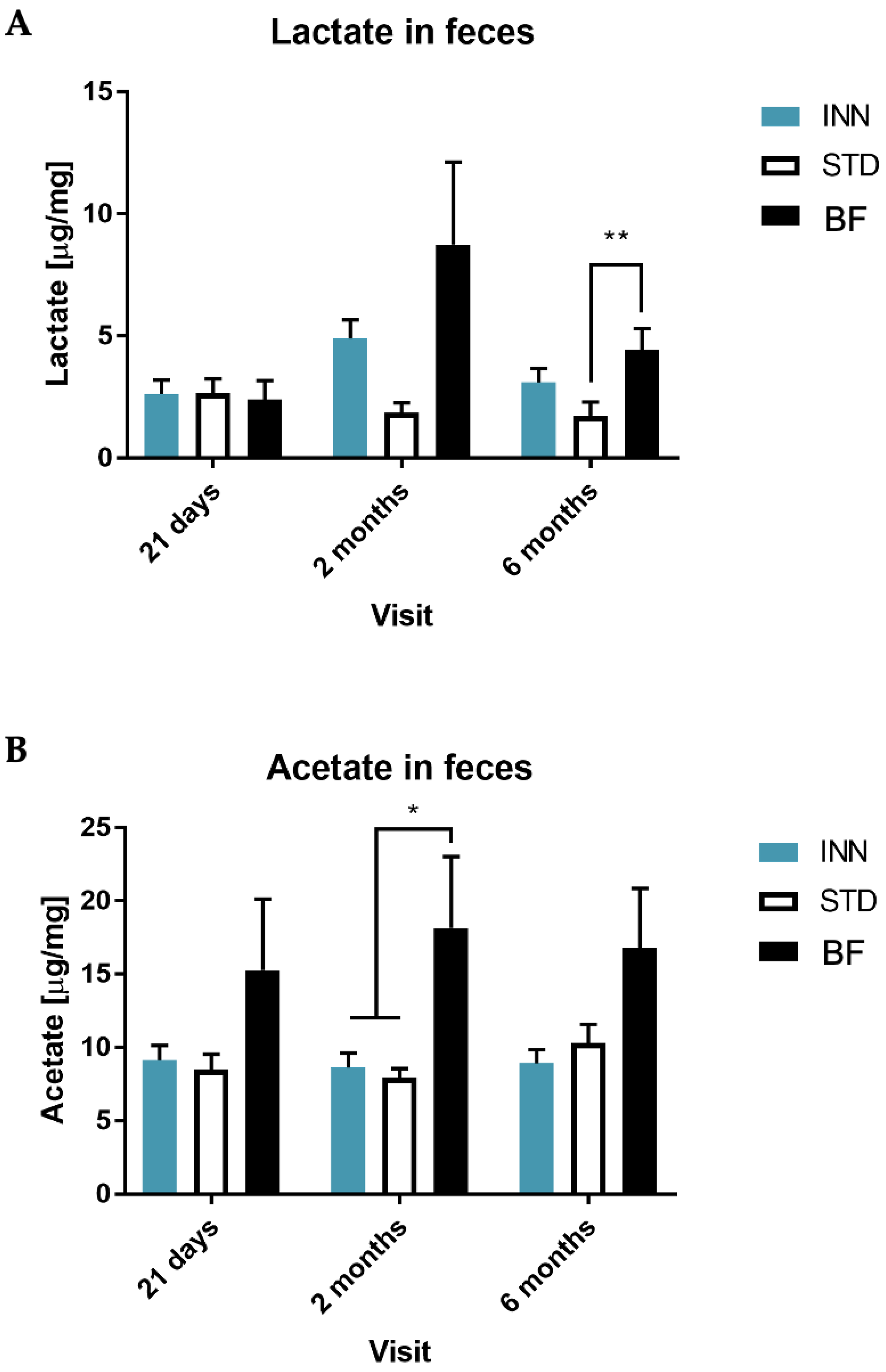
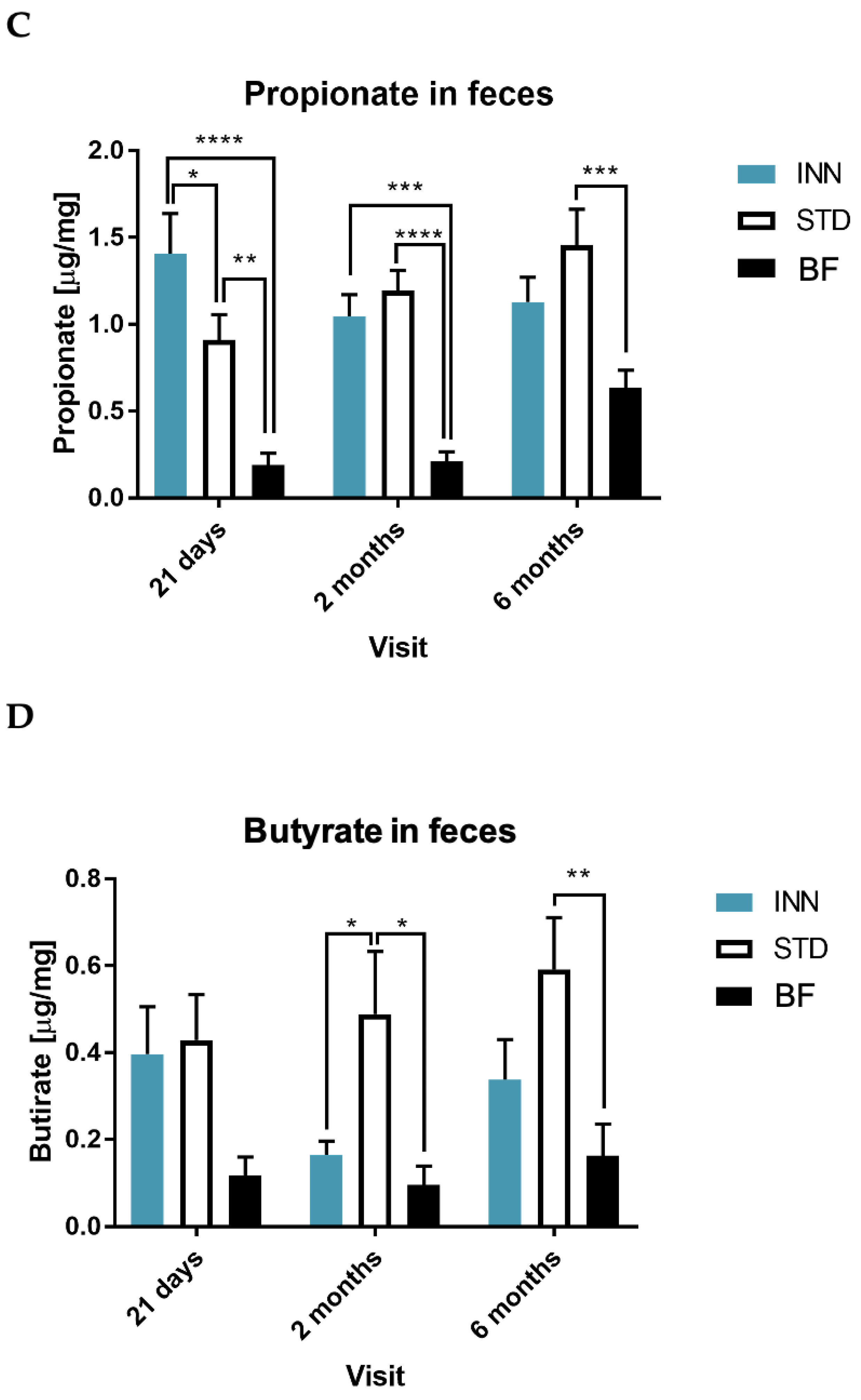
2.5. IgA, Calprotectin and Short-Chain Fatty Acids (SCFAs)
2.6. Correlations between Bacterial Diversity Indices, Bacterial Variables, SCFAs Levels, Metabolic Traits, and Clinical Outcomes
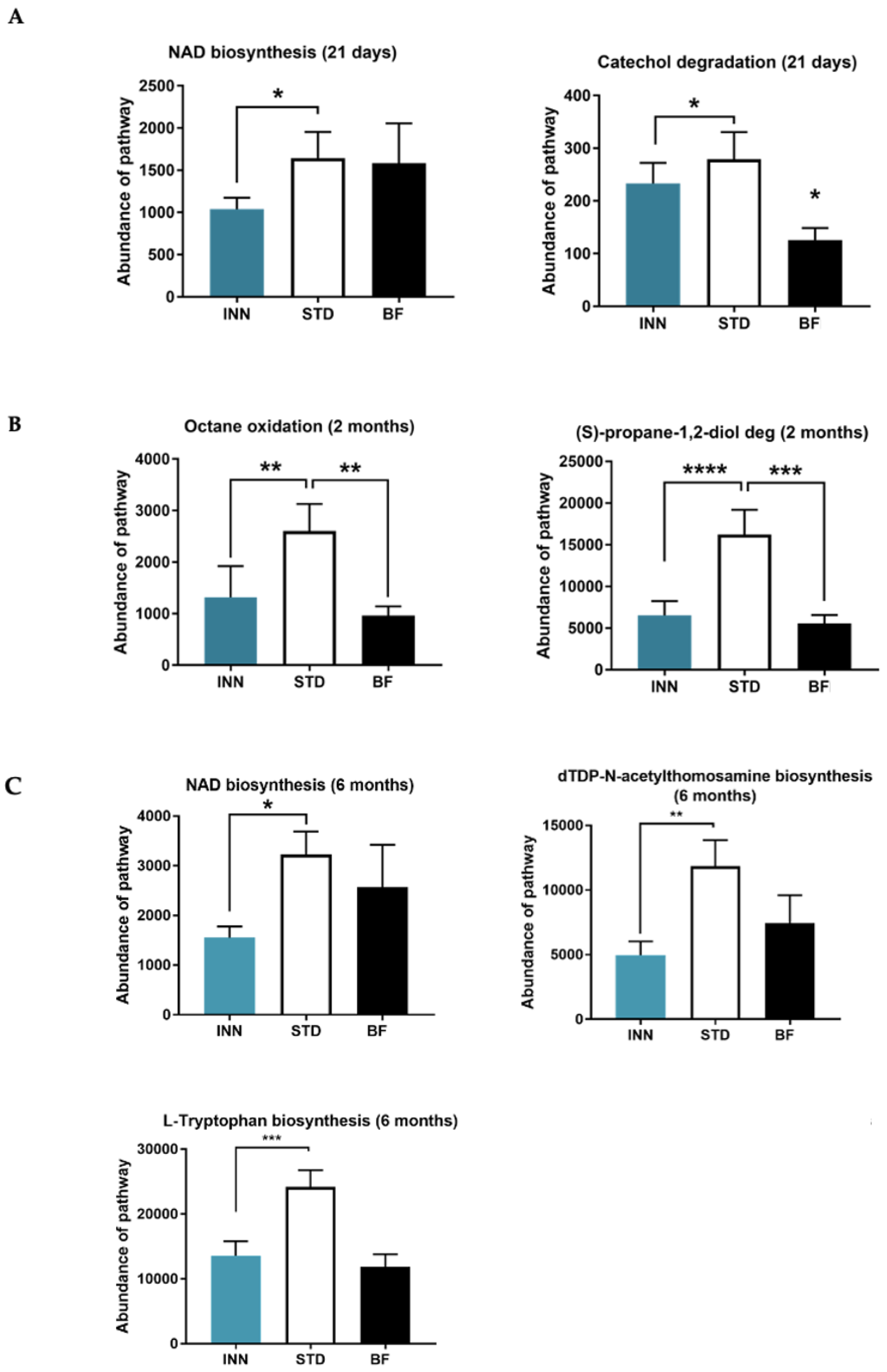
2.7. Major Bacteria Metabolic Pathways
3. Discussion
3.1. Effects on Richness and Diversity
3.2. Bifidobacterium and Other Genera
3.3. Secretory IgA
3.4. Short-Chain Fatty Acids
3.5. Metabolic Pathways Profile
3.6. Strengths, Limitations, and Suggestions
4. Materials and Methods
4.1. Ethics
4.2. Trial Design
4.3. Study Groups
- Group 1 (infant formula 1): Nutribén Innova® 1 (INN)
- Group 2 (infant formula 2): Nutribén® standard (STD)
- Group 3: Breastfeeding (external control exploratory analysis)
4.4. Inclusion and Exclusion Criteria
4.5. Sampling
4.6. DNA Extraction
4.7. Sequencing and Bioinformatic Analysis
4.8. Functional Profiles
4.9. Biochemical Analysis
4.10. Rivera-Pinto Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. The Optimal Duration of Exclusive Breastfeeding: A Systematic Review; Department of Nutrition for Health and Development, Department of Child and Adolescent Health and Development: Geneva, Switzerland, 2002; Available online: https://apps.who.int/iris/bitstream/handle/10665/67208/WHO_NHD_01.08.pdf (accessed on 15 January 2023).
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; de Onis, M.; Ezzati, M.; Grantham-McGregor, S.; Katz, J.; Martorell, R.; et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Ip, S.; Chung, M.; Raman, G.; Trikalinos, T.A.; Lau, J. A summary of the Agency for Healthcare Research and Quality’s evidence report on breastfeeding in developed countries. Breastfeed. Med. 2009, 4 (Suppl. 1), S17–S30. [Google Scholar] [CrossRef] [PubMed]
- Ogbu, C.E.; Fongue, S.; Ogbu, S.C.; Kirby, R.S. Infant feeding practices and asthma in children aged 6 months to 5 years using a propensity score approach. Cent. Eur. J. Public Health 2021, 29, 284–289. [Google Scholar] [CrossRef]
- Rameez, R.M.; Sadana, D.; Kaur, S.; Ahmed, T.; Patel, J.; Khan, M.S.; Misbah, S.; Simonson, M.T.; Riaz, H.; Ahmed, H.M. Association of maternal lactation with diabetes and hypertension: A systematic review and meta-analysis. JAMA Netw. Open 2019, 2, e1913401. [Google Scholar] [CrossRef]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Christian, P.; Smith, E.R.; Lee, S.E.; Vargas, A.J.; Bremer, A.A.; Raiten, D.J. The need to study human milk as a biological system. Am. J. Clin. Nutr. 2021, 113, 1063–1072. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The role of microbiota in infant health: From early life to adulthood. Front. Immunol. 2021, 12, 1063–1072. [Google Scholar] [CrossRef]
- Forbes, J.D.; Azad, M.B.; Vehling, L.; Tun, H.M.; Konya, T.B.; Guttman, D.S.; Field, C.J.; Lefebvre, D.; Sears, M.R.; Becker, A.B.; et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018, 172, e181161. [Google Scholar] [CrossRef]
- Mesa, M.D.; Loureiro, B.; Iglesia, I.; Fernandez Gonzalez, S.; Llurba Olivé, E.; García Algar, O.; Solana, M.J.; Cabero Perez, M.J.; Sainz, T.; Martinez, L.; et al. The evolving microbiome from pregnancy to early infancy: A comprehensive review. Nutrients 2020, 12, 133. [Google Scholar] [CrossRef]
- Fabiano, V.; Indrio, F.; Verduci, E.; Calcaterra, V.; Pop, T.L.; Mari, A.; Zuccotti, G.V.; Cullu Cokugras, F.; Pettoello-Mantovani, M.; Goulet, O. Term infant formulas influencing gut microbiota: An overview. Nutrients 2021, 13, 4200. [Google Scholar] [CrossRef]
- World Health Organization. World Health Organization Breastfeeding Recommendations. 2022. Available online: https://www.who.int/health-topics/breastfeeding/ (accessed on 12 June 2022).
- Carver, J.D. Advances in nutritional modifications of infant formulas. Am. J. Clin. Nutr. 2003, 77, 1550S–1554S. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.M.; Harris, B.J.; Lien, E.L.; Pramuk, K.; Trabulsi, J. Alpha-lactalbumin-rich infant formula fed to healthy term infants in a multicenter study: Plasma essential amino acids and gastrointestinal tolerance. Eur. J. Clin. Nutr. 2008, 62, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Bergmann, K.; Brenna, J.T.; Calder, P.C.; Campoy, C.; Clandinin, M.T.; Colombo, J.; Daly, M.; Decsi, T.; Demmelmair, H. Should formula for infants provide arachidonic acid along with DHA? A position paper of the European Academy of Paediatrics and the Child Health Foundation. Am. J. Clin. Nutr. 2020, 111, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Campoy, C.; Chisaguano Tonato, A.M.; de la Garza Puentes, A.; Sáenz de Pipaón, M.; Verduci, E.; Koletzko, B.; Casanova, G.; Larqué, E.; Valenzuela, R.; Moreno Villares, J.M.; et al. Controversy about the critical role of long-chain polyunsaturated fatty acids, arachidonic acid (ARA) and docosahexaenoic acid (DHA), during infancy. Nutr. Hosp. 2021, 38, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, F.; Enrique, M.; Llopis, S.; Barrena, M.; Navarro, V.; Alvarez, B.; Chenoll, E.; Ramon, D.; Tortajada, M.; Martorell, P. Lipoteichoic acid from Bifidobacterium animalis subsp. lactis BPL1: A novel postbiotic that reduces fat deposition via IGF-1 pathway. Microb. Biotechnol. 2022, 15, 805–816. [Google Scholar] [CrossRef]
- Carreras, N.L.; Martorell, P.; Chenoll, E.; Genoves, S.; Ramon, D.; Aleixandre, A. Anti-obesity properties of the strain Bifidobacterium animalis subsp. lactis CECT 8145 in Zucker fatty rats. Benef. Microbes 2018, 9, 629–641. [Google Scholar] [CrossRef]
- Pedret, A.; Valls, R.M.; Calderon-Perez, L.; Llaurado, E.; Companys, J.; Pla-Paga, L.; Moragas, A.; Martin-Lujan, F.; Ortega, Y.; Giralt, M.; et al. Effects of daily consumption of the probiotic Bifidobacterium animalis subsp. lactis CECT 8145 on anthropometric adiposity biomarkers in abdominally obese subjects: A randomized controlled trial. Int. J. Obes. 2019, 43, 1863–1868. [Google Scholar] [CrossRef]
- Silva, A.; Gonzalez, N.; Terren, A.; Garcia, A.; Martinez-Blanch, J.F.; Illescas, V.; Morales, J.; Maroto, M.; Genoves, S.; Ramon, D.; et al. An infant milk formula supplemented with heat-treated probiotic Bifidobacterium animalis subsp. lactis CECT 8145, reduces fat deposition in C. elegans and augments acetate and lactate in a fermented infant slurry. Foods 2020, 9, 652. [Google Scholar] [CrossRef]
- Wall, R.; Ross, R.P.; Ryan, C.A.; Hussey, S.; Murphy, B.; Fitzgerald, G.F.; Stanton, C. Role of gut microbiota in early infant development. Clin. Med. Pediatr. 2009, 3, 45–54. [Google Scholar] [CrossRef]
- Nermes, M.; Kantele, J.M.; Atosuo, T.J.; Salminen, S.; Isolauri, E. Interaction of orally administered Lactobacillus rhamnosus GG with skin and gut microbiota and humoral immunity in infants with atopic dermatitis. Clin. Exp. Allergy 2011, 41, 370–377. [Google Scholar] [CrossRef]
- Rivera-Pinto, J.; Egozcue, J.J.; Pawlowsky-Glahn, V.; Paredes, R.; Noguera-Julian, M.; Calle, M.L. Balances: A new perspective for microbiome analysis. MSystems 2018, 3, e00053-18. [Google Scholar] [CrossRef]
- Bezirtzoglou, E.; Tsiotsias, A.; Welling, G.W. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 2011, 17, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Brink, L.R.; Mercer, K.E.; Piccolo, B.D.; Chintapalli, S.V.; Elolimy, A.; Bowlin, A.K.; Matazel, K.S.; Pack, L.; Adams, S.H.; Shankar, K.; et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am. J. Clin. Nutr. 2020, 111, 1190–1202. [Google Scholar] [CrossRef]
- Lee, S.A.; Lim, J.Y.; Kim, B.S.; Cho, S.J.; Kim, N.Y.; Kim, O.B.; Kim, Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr. Res. Pract. 2015, 9, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.P.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T.; et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018, 9, 4169. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.F.; Brown, B.P.; Lennard, K.; Karaoz, U.; Havyarimana, E.; Passmore, J.S.; Hesseling, A.C.; Edlefsen, P.T.; Kuhn, L.; Mulder, N.; et al. Feeding-related gut microbial composition associates with peripheral T-cell activation and mucosal gene expression in African infants. Clin. Infect. Dis. 2018, 67, 1237–1246. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Chung, J.; Battaglia, T.; Henderson, N.; Jay, M.; Li, H.; A, D.L.; Wu, F.; Perez-Perez, G.I.; Chen, Y.; et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016, 8, 343ra82. [Google Scholar] [CrossRef]
- Ma, J.; Li, Z.; Zhang, W.; Zhang, C.; Zhang, Y.; Mei, H.; Zhuo, N.; Wang, H.; Wang, L.; Wu, D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci. Rep. 2020, 10, 15792. [Google Scholar] [CrossRef]
- Rhoads, J.M.; Collins, J.; Fatheree, N.Y.; Hashmi, S.S.; Taylor, C.M.; Luo, M.; Hoang, T.K.; Gleason, W.A.; Van Arsdall, M.R.; Navarro, F.; et al. Infant colic represents gut inflammation and dysbiosis. J. Pediatr. 2018, 203, 55–61. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Zijlmans, M.A.; Kuitunen, M.; Kukkonen, K.; Savilahti, E.; Salonen, A.; de Weerth, C.; de Vos, W.M. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 2017, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Taft, D.H.; Liu, J.; Maldonado-Gomez, M.X.; Akre, S.; Huda, M.N.; Ahmad, S.M.; Stephensen, C.B.; Mills, D.A. Bifidobacterial dominance of the gut in early life and acquisition of antimicrobial resistance. MSphere 2018, 3. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Lönnerdal, B.; Lien, E.L. Nutritional and physiologic significance of alpha-lactalbumin in infants. Nutr. Rev. 2003, 61, 295–305. [Google Scholar] [CrossRef]
- Bezkorovainy, A.; Topouzian, N. Bifidobacterium bifidus var. Pennsylvanicus growth promoting activity of human milk casein and its derivatives. Int. J. Biochem. 1981, 13, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Petschow, B.W.; Talbott, R.D. Response of bifidobacterium species to growth promoters in human and cow milk. Pediatr. Res. 1991, 29, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Tamana, S.K.; Tun, H.M.; Konya, T.; Chari, R.S.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Moraes, T.J.; Turvey, S.E.; Subbarao, P.; et al. Bacteroides-dominant gut microbiome of late infancy is associated with enhanced neurodevelopment. Gut Microbes 2021, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- York, A. Delivery of the gut microbiome. Nat. Rev. Microbiol. 2018, 16, 520–521. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Messina, N.; Mohn, W.W.; Finlay, B.B.; Curtis, N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: A systematic review. J. Allergy Clin. Immunol. 2019, 143, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dai, W.; Feng, X.; Zhou, Q.; Wang, H.; Yang, Y.; Li, S.; Zheng, Y. Microbiota composition in upper respiratory tracts of healthy children in Shenzhen, China, differed with respiratory sites and ages. Biomed. Res. Int. 2018, 2018, 6515670. [Google Scholar] [CrossRef] [PubMed]
- Bosheva, M.; Tokodi, I.; Krasnow, A.; Pedersen, H.K.; Lukjancenko, O.; Eklund, A.C.; Grathwohl, D.; Sprenger, N.; Berger, B.; Cercamondi, C.I. Infant formula with a specific blend of five human milk oligosaccharides drives the gut microbiota development and improves gut maturation markers: A randomized controlled trial. Front. Nutr. 2022, 9, 920362. [Google Scholar] [CrossRef]
- Shim, J.O. Clostridium difficile in children: To treat or not to treat? Pediatr. Gastroenterol. Hepatol. Nutr. 2014, 17, 80–84. [Google Scholar] [CrossRef]
- Xiao, S.; Jiang, S.; Qian, D.; Duan, J. Modulation of microbially derived short-chain fatty acids on intestinal homeostasis, metabolism, and neuropsychiatric disorder. Appl. Microbiol. Biotechnol. 2020, 104, 589–601. [Google Scholar] [CrossRef]
- Zhu, B.; Zheng, S.; Lin, K.; Xu, X.; Lv, L.; Zhao, Z.; Shao, J. Effects of infant formula supplemented with prebiotics and OPO on infancy fecal microbiota: A pilot randomized clinical trial. Front. Cell. Infect. Microbiol. 2021, 11, 650407. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Bosi, A.; Banfi, D.; Bistoletti, M.; Giaroni, C.; Baj, A. Tryptophan metabolites along the microbiota-gut-brain axis: An interkingdom communication system influencing the gut in health and disease. Int. J. Tryptophan Res. 2020, 13, 1178646920928984. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.M.A.; Sundekilde, U.K.; Andersen, H.J.; Nielsen, D.S.; Bertram, H.C. Lactose and bovine milk oligosaccharides synergistically stimulate B. longum subsp. longum growth in a simplified model of the infant gut microbiome. J. Proteome Res. 2019, 18, 3086–3098. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liu, B.; Li, J.; Dong, X.; Lin, M.; Zhang, M.; Zhao, J.; Dai, Y.; Chen, L. Association between sn-2 fatty acid profiles of breast milk and development of the infant intestinal microbiome. Food. Funct. 2018, 9, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Cerdó, T.; Diéguez, E.; Campoy, C. Infant growth, neurodevelopment and gut microbiota during infancy: Which nutrients are crucial? Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 434–441. [Google Scholar] [CrossRef]
- Lee, H.; Li, Z.; Christensen, B.; Peng, Y.; Li, X.; Hernell, O.; Lönnerdal, B.; Slupsky, C.M. Metabolic phenotype and microbiome of infants fed formula containing Lactobacillus paracasei strain F-19. Front. Pediatr. 2022, 10, 856951. [Google Scholar] [CrossRef]
- Bazanella, M.; Maier, T.V.; Clavel, T.; Lagkouvardos, I.; Lucio, M.; Maldonado-Gòmez, M.X.; Autran, C.; Walter, J.; Bode, L.; Schmitt-Kopplin, P.; et al. Randomized controlled trial on the impact of early-life intervention with bifidobacteria on the healthy infant fecal microbiota and metabolome. Am. J. Clin. Nutr. 2017, 106, 1274–1286. [Google Scholar] [CrossRef]
- Chi, C.; Xue, Y.; Liu, R.; Wang, Y.; Lv, N.; Zeng, H.; Buys, N.; Zhu, B.; Sun, J.; Yin, C. Effects of a formula with a probiotic Bifidobacterium lactis supplement on the gut microbiota of low birth weight infants. Eur. J. Nutr. 2020, 59, 1493–1503. [Google Scholar] [CrossRef]
- Simeoni, U.; Berger, B.; Junick, J.; Blaut, M.; Pecquet, S.; Rezzonico, E.; Grathwohl, D.; Sprenger, N.; Brüssow, H.; Szajewska, H.; et al. Gut microbiota analysis reveals a marked shift to bifidobacteria by a starter infant formula containing a synbiotic of bovine milk-derived oligosaccharides and Bifidobacterium animalis subsp. lactis CNCM I-3446. Environ. Microbiol. 2016, 18, 2185–2195. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Secretory IgA: Designed for anti-microbial defense. Front. Immunol. 2013, 4, 222. [Google Scholar] [CrossRef]
- Maruyama, K.; Hida, M.; Kohgo, T.; Fukunaga, Y. Changes in salivary and fecal secretory IgA in infants under different feeding regimens. Pediatr. Int. 2009, 51, 342–345. [Google Scholar] [CrossRef]
- Aadland, E.; Fagerhol, M.K. Faecal calprotectin: A marker of inflammation throughout the intestinal tract. Eur. J. Gastroenterol. Hepatol. 2002, 14, 823–825. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Ben, R.A.; Pons, S.; de Paolo, M.I.; Bustos Fernández, L. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J. Pediatr. Gastroenterol. Nutr. 1992, 15, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Shats, I.; Williams, J.G.; Liu, J.; Makarov, M.V.; Wu, X.; Lih, F.B.; Deterding, L.J.; Lim, C.; Xu, X.; Randall, T.A.; et al. Bacteria boost mammalian host NAD metabolism by engaging the deamidated biosynthesis pathway. Cell Metab. 2020, 31, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Janssen, D.B.; Witholt, B. Growth on octane alters the membrane lipid fatty acids of Pseudomonas oleovorans due to the induction of alkB and synthesis of octanol. J. Bacteriol. 1995, 177, 6894–6901. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Xue, J.; Murrieta, C.M.; Rule, D.C.; Miller, K.W. Exogenous or L-rhamnose-derived 1,2-propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 2008, 74, 7073–7079. [Google Scholar] [CrossRef]
- Palacios, S.; Starai, V.J.; Escalante-Semerena, J.C. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 2003, 185, 2802–2810. [Google Scholar] [CrossRef]
- Ruiz-Ojeda, F.J.; Plaza-Díaz, J.; Morales, J.; Cristina de la Torre, A.I.; García-García, A.; de Prado, C.N.; Coronel, C.; Crespo, C.; Ortega, E.; Martín-Pérez, E.; et al. A multicenter, randomized, blinded, controlled clinical trial investigating the effect of a novel infant formula on the body composition of infants: INNOVA 2020 study protocol. Adv. Pediatr. Res. 2022, 9. [Google Scholar] [CrossRef]
- Task Force on Clinical Testing of Infant Formulas Committee on Nutrition; American Academy of Pediatrics Committee on Nutrition. Clinical Testing of Infant Formulas with Respect to Nutritional Suitability for Term Infants. 1988. Available online: https://wayback.archive-it.org/7993/20170722090324/https://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/InfantFormula/ucm170649.htm (accessed on 24 June 2022).
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Morales, J.; de la Torre, A.I.C.; García-García, A.; de Prado, C.N.; Coronel-Rodríguez, C.; Crespo, C.; Ortega, E.; Martín-Pérez, E.; et al. Effects of a novel infant formula on weight gain, body composition, safety and tolerability to infants: The INNOVA 2020 Study. Nutrients 2022, 15, 147. [Google Scholar] [CrossRef]
- Yuan, S.; Cohen, D.B.; Ravel, J.; Abdo, Z.; Forney, L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 2012, 7, e33865. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Campeotto, F.; Elie, C.; Rousseau, C.; Giuseppi, A.; Hachem, T.; Gobalakichenane, P.; Le Touzey, M.; de Stefano, M.; Butel, M.J.; Kapel, N. Faecal calprotectin and gut microbiota do not predict enteropathy in very preterm infants. Acta Paediatr. 2021, 110, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.; Ye, K.; Li, M.; Ying, J.; Wang, H.; Han, J.; Shi, L.; Xiao, J.; Shen, Y.; Feng, X.; et al. Xylitol enhances synthesis of propionate in the colon via cross-feeding of gut microbiota. Microbiome 2021, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wei, T.S.V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J.; Freidank, M.; Cai, J.; Protivinsky, T. Package ‘Corrplot’. 2022. Available online: https://github.com/taiyun/corrplot (accessed on 15 January 2023).


| Phylum | 21 Days | 2 Months | 6 Months | p-Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BF (n = 68) | STD (n = 70) | INN (n = 75) | BF (n = 64) | STD (n = 64) | INN (n = 63) | BF (n = 52) | STD (n = 55) | INN (n = 52) | Treatment | Visit | Treatment × Visit | |
| Fisher | 5.1 ± 0.3 | 6.2 ± 0.3 | 6.2 ± 0.3 | 5.5 ± 0.3 | 5.6 ± 0.3 | 5.6 ± 0.3 | 4.9 ± 0.4 | 5.4 ± 0.3 | 4.8 ± 0.3 | 0.083 | 0.015 | 0.235 |
| Shannon index | 0.8 ± 0.07 | 1.1 ± 0.07 | 1.2 ± 0.06 | 0.9 ± 0.07 | 1.1 ± 0.07 | 0.7 ± 0.07 | 0.9 ± 0.08 | 1.2 ± 0.08 | 0.8 ± 0.08 | <0.001 | 0.082 | <0.001 |
| Inverse Simpson | 1.9 ± 0.2 | 2.3 ± 0.2 | 2.4 ± 0.1 | 1.8 ± 0.2 | 2.4 ± 0.2 | 1.6 ± 0.2 | 1.9 ± 0.2 | 2.4 ± 0.2 | 1.6 ± 0.2 | <0.001 | 0.061 | 0.004 |
| Pielou’s evenness | 0.23 ± 0.02 | 0.28 ± 0.02 | 0.31 ± 0.02 | 0.23 ± 0.02 | 0.31 ± 0.02 | 0.19 ± 0.02 | 0.25 ± 0.02 | 0.32 ± 0.02 | 0.22 ± 0.02 | <0.001 | 0.108 | <0.001 |
| Species richness | 37.7 ± 1.9 | 45.4 ± 1.9 | 44.9 ± 1.8 | 41.0 ± 2.0 | 41.4 ± 2.0 | 41.2 ± 2.0 | 37.2 ± 2.3 | 40.7 ± 2.1 | 36.4 ± 2.2 | 0.058 | 0.023 | 0.213 |
| Simpson | 0.33 ± 0.03 | 0.41 ± 0.03 | 0.47 ± 0.03 | 0.35 ± 0.03 | 0.47 ± 0.03 | 0.28 ± 0.03 | 0.37 ± 0.03 | 0.48 ± 0.03 | 0.32 ± 0.03 | <0.001 | 0.251 | <0.001 |
| Actinobacteria | 90.5 (1.2–99.4) | 76.4 (2.9–99.4) | 65.3 (3.2–98.9) | 89.2 (1.2–99.2) | 76.6 (2.7–98.9) | 92.9 (10.8–99.1) | 88.4 (4.3–99.1) | 80.5 (1.5–98.6) | 92.5 (6.0–98.0) | 0.599 | 0.205 | 0.247 |
| Firmicutes | 8 (0.4–98.5) | 21.6 (0.4–96.6) | 32.9 (0.8–96.3) | 10.1 (0.6–98.5) | 19.6 (1.0–96.9) | 5.8 (0.6–62.6) | 10.2 (0.8–95.6) | 15.8 (1.2–98.3) | 6.9 (1.3–93.8) | 0.838 | 0.168 | 0.235 |
| Verrucomicrobia | 0.08 (0–3.3) | 0.1 (0–71.9) | 0.08 (0–55.8) | 0.06 (0–1.6) | 0.06 (0–62.5) | 0.06 (0–42.0) | 0.05 (0–10.7) | 0.05 (0–66.6) | 0.04 (0–35.3) | 0.198 | 0.641 | 0.625 |
| Proteobacteria | 0.22 (0–1.9) | 0.4 (0.01–2.1) | 0.4 (0–2.6) | 0.2 (0–1.1) | 0.4 (0.04–2.7) | 0.5 (0.04–1.2) | 0.2 (0–1.1) | 0.2 (0.003–0.8) | 0.3 (0.01–1.2) | 0.008 | <0.001 | 0.98 |
| Bacteroidetes | 0.04 (0–0.6) | 0.02 (0–1.0) | 0.04 (0–0.7) | 0.04 (0–1.5) | 0.07 (0–1.1) | 0.03 (0–0.7) | 0.02 (0–1.0) | 0.1 (0–0.7) | 0.08 (0–0.8) | 0.828 | 0.242 | 0.261 |
| Fusobacteriota | 0 (0–1.0) | 0 (0–4.9) | 0 (0–0.6) | 0 (0–1.9) | 0 (0–0.1) | 0 (0–0.3) | 0.0008 (0–4.2) | 0 (0–1.2) | 0.004 (0–0.4) | 0.58 | 0.407 | 0.538 |
| Patescibacteria | 0 (0–0.9) | 0 (0–0.02) | 0 (0–0.02) | 0 (0–0.1) | 0 (0–0.3) | 0 (0–0.2) | 0 (0–3.2) | 0 (0–0.01) | 0 (0–0) | 0.558 | 0.076 | 0.931 |
| Synergistetes | 0 (0–0.3) | 0 (0–0.3) | 0 (0–0.09) | 0 (0–0.3) | 0 (0–0.07) | 0 (0–0.04) | 0 (0–0.05) | 0 (0–0.3) | 0 (0–0.2) | 0.156 | 0.447 | 0.366 |
| Cyanobacteria | 0 (0–0.1) | 0 (0–0.1) | 0 (0–0.07) | 0 (0–0) | 0 (0–0.04) | 0 (0–0.02) | 0 (0–0.1) | 0 (0–0.4) | 0 (0–0.09) | 0.599 | 0.677 | 0.902 |
| Genus | 21 Days | 2 Months | 6 Months | p-Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BF (n = 68) | STD (n = 70) | INN (n = 75) | BF (n = 64) | STD (n = 64) | INN (n = 63) | BF (n = 52) | STD (n = 55) | INN (n = 52) | Treatment | Visit | Treatment × Visit | |
| Bifidobacterium | 79.9 (0.7–99.4) | 70.1 (2.3–98.6) | 50.3 (3.2–97.1) | 70.4 (0.9–98.9) | 67.2 (2.1–94.9) | 87.4 (7.7–98.9) | 78.7 (3.6–98.9) | 68.5 (0.8–98.3) | 82.7 (4.8–95.7) | 0.002 | <0.001 | <0.001 |
| Clostridium sensu stricto 1 | 1.1 (0–92.5) | 1.8 (0.06–82.9) | 5.8 (0.1–93.8) | 1.0 (0–94.2) | 2.5 (0.2–62.2) | 1.2 (0.02–37.0) | 0.5 (0–72.4) | 0.5 (0–88.5) | 1.4 (0.01–16.9) | 0.104 | <0.001 | 0.039 |
| Collinsella | 0.3 (0–30.1) | 0.3 (0–33.4) | 0.4 (0–44.0) | 0.3 (0–88.6) | 0.3 (0.01–23.8) | 0.4 (0–53.4) | 0.4 (0.03–50.9) | 0.3 (0.01–54.9) | 0.6 (0–63.7) | 0.331 | 0.001 | 0.845 |
| Blautia | 0.08 (0–30.6) | 0.1 (0–94.2) | 0.1 (0–2.4) | 0.1 (0–18.0) | 0.09 (0–89.7) | 0.06 (0–10.5) | 0.06 (0–12.3) | 0.1 (0–41.4) | 0.05 (0–13.3) | 0.016 | 0.973 | 1 |
| Ruminococcus gnavus group | 0.1 (0–95.6) | 0.2 (0–43.3) | 0.2 (0–15.0) | 0.2 (0–90.7) | 0.2 (0–73.9) | 0.08 (0–10.1) | 0.2 (0.01–41.0) | 1.6 (0–46.7) | 0.2 (0–73.9) | 0.009 | 0.362 | 0.827 |
| Clostridioides | 0.08 (0–2.9) | 0.05 (0–70.0) | 0.1 (0–32.2) | 0.04 (0–47.2) | 0.07 (0–25.0) | 0.03 (0–8.5) | 0.04 (0–5.8) | 0.3 (0–24.3) | 0.03 (0–39.7) | 0.086 | 0.967 | 0.535 |
| Akkermansia | 0.06 (0–3.3) | 0.09 (0–70.9) | 0.08 (0–55.2) | 0.05 (0–1.6) | 0.07 (0–62.0) | 0.05 (0–42.4) | 0.06 (0–10.9) | 0.05 (0–66.1) | 0.04 (0–35.6) | <0.001 | 0.472 | 0.848 |
| Eggerthella | 0.1 (0–18.2) | 0.1 (0–43.5) | 0.2 (0–35.2) | 0.09 (0–6.8) | 0.1 (0–38.4) | 0.05 (0–23.8) | 0.2 (0–5.2) | 0.5 (0–15.5) | 0.09 (0–5.7) | 0.041 | 0.905 | 0.15 |
| Terrisporobacter | 0 (0–1.2) | 0 (0–28.9) | 0.01 (0–77.8) | 0 (0–0.8) | 0.01 (0–23.6) | 0 (0–2.7) | 0 (0–0.3) | 0 (0–0.5) | 0 (0–1.0) | 0.32 | 0.273 | 0.268 |
| Flavonifractor | 0.03 (0–2.1) | 0.05 (0–6.3) | 0.06 (0–44.1) | 0.04 (0–2.0) | 0.1 (0–59.3) | 0.03 (0–3.8) | 0.04 (0–13.4) | 0.2 (0–9.6) | 0.03 (0–3.2) | 0.059 | 0.465 | 0.002 |
| Cutibacterium | 0.05 (0–15.7) | 0.03 (0–11.8) | 0.02 (0–1.9) | 0.03 (0–70.9) | 0.01 (0–0.2) | 0.02 (0–1.9) | 0 (0–0.5) | 0 (0–0.2) | 0 (0–1.6) | 0.021 | 0.238 | 0.452 |
| Subdoligranulum | 0.01 (0–13.6) | 0.02 (0–7.4) | 0.03 (0–51.8) | 0.02 (0–19.9) | 0.01 (0–0.8) | 0 (0–0.8) | 0.02 (0–19.5) | 0.02 (0–13.6) | 0 (0–0.2) | 0.928 | 0.535 | 0.431 |
| Intestinibacter | 0 (0–0) | 0 (0–0) | 0 (0–0.1) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0.52 | 0.499 | 0.581 |
| Rothia | 0.06 (0–7.1) | 0.1 (0–6.4) | 0.1 (0–3.7) | 0.08 (0–13.2) | 0.09 (0–2.1) | 0.1 (0–1.2) | 0.04 (0–1.2) | 0.02 (0–0.3) | 0.06 (0–0.6) | 0.087 | 0.002 | 0.129 |
| Faecalibacterium | 0 (0–7.3) | 0.02 (0–5.5) | 0.03 (0–23.3) | 0.04 (0–1.6) | 0 (0–1.8) | 0.02 (0–8.8) | 0.04 (0–5.5) | 0.07 (0–27.1) | 0.05 (0–3.6) | 0.716 | 0.345 | 0.556 |
| Lachnoclostridium | 0 (0–7.2) | 0.02 (0–15.9) | 0.02 (0–18.0) | 0.005 (0–0.9) | 0.01 (0–15.7) | 0 (0–3.1) | 0 (0–17.3) | 0.06 (0–4.1) | 0 (0–3.5) | 0.199 | 0.977 | 0.489 |
| Corynebacterium | 0 (0–0) | 0 (0–0.3) | 0 (0–0.2) | 0 (0–0.2) | 0 (0–0.3) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0.553 | 0.302 | 0.427 |
| UBA1819 | 0 (0–8.6) | 0 (0–0.8) | 0 (0–24.6) | 0 (0–6.4) | 0 (0–5.8) | 0 (0–3.3) | 0 (0–12.5) | 0 (0–16.6) | 0 (0–2.5) | 0.43 | 0.082 | 0.005 |
| Tyzzerella | 0 (0–0) | 0 (0–0.2) | 0 (0–0.1) | 0 (0–0.3) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0 (0–0.2) | 0 (0–0) | 0.304 | 0.853 | 0.138 |
| Paeniclostridium | 0 (0–0.4) | 0 (0–10.5) | 0 (0–35.7) | 0 (0–0.1) | 0 (0–3.5) | 0 (0–1.6) | 0 (0–0.1) | 0 (0–0.7) | 0 (0–2.2) | 0.458 | 0.257 | 0.686 |
| Peptoniphilus | 0 (0–22.2) | 0.01 (0–1.6) | 0.01 (0–7.9) | 0 (0–0.3) | 0.03 (0–2.0) | 0.04 (0–1.1) | 0 (0–3.5) | 0.01 (0–0.1) | 0 (0–0.6) | 0.812 | 0.091 | 0.858 |
| Anaerostipes | 0.02 (0–4.0) | 0.04 (0–16.8) | 0.06 (0–1.9) | 0.04 (0–5.4) | 0.02 (0–23.2) | 0.02 (0–7.4) | 0.06 (0–6.2) | 0.08 (0–6.6) | 0.03 (0–3.0) | 0.013 | 0.599 | 0.963 |
| Escherichia-Shigella | 0.1 (0–2.0) | 0.2 (0–1.2) | 0.1 (0–1.6) | 0.01 (0–1.1) | 0.3 (0–1.1) | 0.3 (0–1.1) | 0.2 (0–0.9) | 0.1 (0–0.7) | 0.3 (0–1.0) | 0.012 | 0.608 | 0.06 |
| Anaerococcus | 0 (0–22.5) | 0.02 (0–2.8) | 0.02 (0–5.2) | 0 (0–0.5) | 0.02 (0–2.7) | 0.03 (0–1.8) | 0 (0–5.2) | 0 (0–0.5) | 0.01 (0–0.5) | 0.715 | 0.125 | 0.871 |
| Finegoldia | 0 (0–5.8) | 0.03 (0–1.6) | 0.03 (0–7.8) | 0.1 (0–0.4) | 0.04 (0–4.7) | 0.03 (0–4.2) | 0 (0–5.1) | 0.01 (0–0.1) | 0.02 (0–0.4) | 0.263 | 0.138 | 0.158 |
| Romboutsia | 0 (0–0.3) | 0 (0–0.4) | 0 (0–0.2) | 0 (0–0) | 0 (0–0.5) | 0 (0–0) | 0 (0–0) | 0 (0–0.02) | 0 (0–0.03) | 0.246 | 0.406 | 0.881 |
| Streptococcus | 0.2 (0.01–1.9) | 0.2 (0–1.1) | 0.2 (0–1.1) | 0.1 (0–1.4) | 0.05 (0–0.6) | 0.2 (0–1.2) | 0.09 (0–1.4) | 0.01 (0–0.9) | 0.05 (0–0.7) | <0.001 | <0.001 | 0.195 |
| Ruminococcus | 0 (0–5.3) | 0 (0–5.3) | 0 (0–0.4) | 0 (0–1.1) | 0 (0–1.8) | 0 (0–0.3) | 0 (0–0.2) | 0 (0–0.7) | 0 (0–0.2) | 0.738 | <0.001 | 0.706 |
| Eubacterium hallii group | 0 (0–2.4) | 0 (0–1.6) | 0.01 (0–22.8) | 0 (0–0.6) | 0 (0–29.5) | 0 (0–0.5) | 0 (0–9.7) | 0.01 (0–6.0) | 0.01 (0–1.2) | 0.689 | 0.003 | 0.397 |
| Paraclostridium | 0 (0–0.3) | 0 (0–15.3) | 0 (0–9.2) | 0 (0–0.9) | 0 (0–1.3) | 0 (0–20.8) | 0 (0–0.1) | 0 (0–0.1) | 0 (0–1.2) | 0.308 | 0.996 | 0.687 |
| Ruminococcus torques group | 0 (0–62.9) | 0.03 (0–13.3) | 0.03 (0–11.3) | 0.04 (0–23.0) | 0.01 (0–5.4) | 0 (0–0.4) | 0.01 (0–1.1) | 0.03 (0–19.7) | 0.01 (0–9.9) | 0.548 | 0.47 | 0.534 |
| Veillonella | 0.03 (0–1.0) | 0.05 (0–0.8) | 0.2 (0–0.9) | 0.03 (0–1.3) | 0.2 (0–1.6) | 0.1 (0–1.9) | 0.1 (0–0.7) | 0.1 (0–0.8) | 0.2 (0–0.8) | <0.001 | 0.615 | 0.465 |
| Roseburia | 0 (0–0.4) | 0 (0–0.5) | 0 (0–0.3) | 0 (0–0.3) | 0 (0–0.1) | 0 (0–0.5) | 0 (0–0.07) | 0 (0–0.09) | 0 (0–0.08) | 0.636 | 0.097 | 0.067 |
| Bacteroides | 0.04 (0–0.6) | 0.03 (0–0.8) | 0.04 (0–0.7) | 0.06 (0–1.5) | 0.06 (0–0.8) | 0.02 (0–0.7) | 0.01 (0–0.9) | 0.07 (0–0.6) | 0.1 (0–0.9) | 0.944 | 0.023 | 0.075 |
| Enterococcus | 0.01 (0–0.6) | 0.05 (0–1.4) | 0.04 (0–1.2) | 0.01 (0–0.5) | 0.05 (0–0.5) | 0.03 (0–0.7) | 0.01 (0–0.4) | 0.03 (0–1.0) | 0.06 (0–0.8) | 0.006 | 0.141 | 0.723 |
| Eubacterium | 0 (0–0.3) | 0 (0–2.7) | 0 (0–8.3) | 0 (0–9.9) | 0 (0–15.3) | 0 (0–1.1) | 0 (0–5.9) | 0 (0–2.5) | 0 (0–1.6) | 0.705 | 0.379 | 0.573 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Ojeda, F.J.; Plaza-Diaz, J.; Morales, J.; Álvarez-Calatayud, G.; Climent, E.; Silva, Á.; Martinez-Blanch, J.F.; Enrique, M.; Tortajada, M.; Ramon, D.; et al. Effects of a Novel Infant Formula on the Fecal Microbiota in the First Six Months of Life: The INNOVA 2020 Study. Int. J. Mol. Sci. 2023, 24, 3034. https://doi.org/10.3390/ijms24033034
Ruiz-Ojeda FJ, Plaza-Diaz J, Morales J, Álvarez-Calatayud G, Climent E, Silva Á, Martinez-Blanch JF, Enrique M, Tortajada M, Ramon D, et al. Effects of a Novel Infant Formula on the Fecal Microbiota in the First Six Months of Life: The INNOVA 2020 Study. International Journal of Molecular Sciences. 2023; 24(3):3034. https://doi.org/10.3390/ijms24033034
Chicago/Turabian StyleRuiz-Ojeda, Francisco Javier, Julio Plaza-Diaz, Javier Morales, Guillermo Álvarez-Calatayud, Eric Climent, Ángela Silva, Juan F. Martinez-Blanch, María Enrique, Marta Tortajada, Daniel Ramon, and et al. 2023. "Effects of a Novel Infant Formula on the Fecal Microbiota in the First Six Months of Life: The INNOVA 2020 Study" International Journal of Molecular Sciences 24, no. 3: 3034. https://doi.org/10.3390/ijms24033034
APA StyleRuiz-Ojeda, F. J., Plaza-Diaz, J., Morales, J., Álvarez-Calatayud, G., Climent, E., Silva, Á., Martinez-Blanch, J. F., Enrique, M., Tortajada, M., Ramon, D., Alvarez, B., Chenoll, E., & Gil, Á. (2023). Effects of a Novel Infant Formula on the Fecal Microbiota in the First Six Months of Life: The INNOVA 2020 Study. International Journal of Molecular Sciences, 24(3), 3034. https://doi.org/10.3390/ijms24033034









