Identification of Key Biomarkers Associated with Immunogenic Cell Death and Their Regulatory Mechanisms in Severe Acute Pancreatitis Based on WGCNA and Machine Learning
Abstract
1. Introduction
2. Results
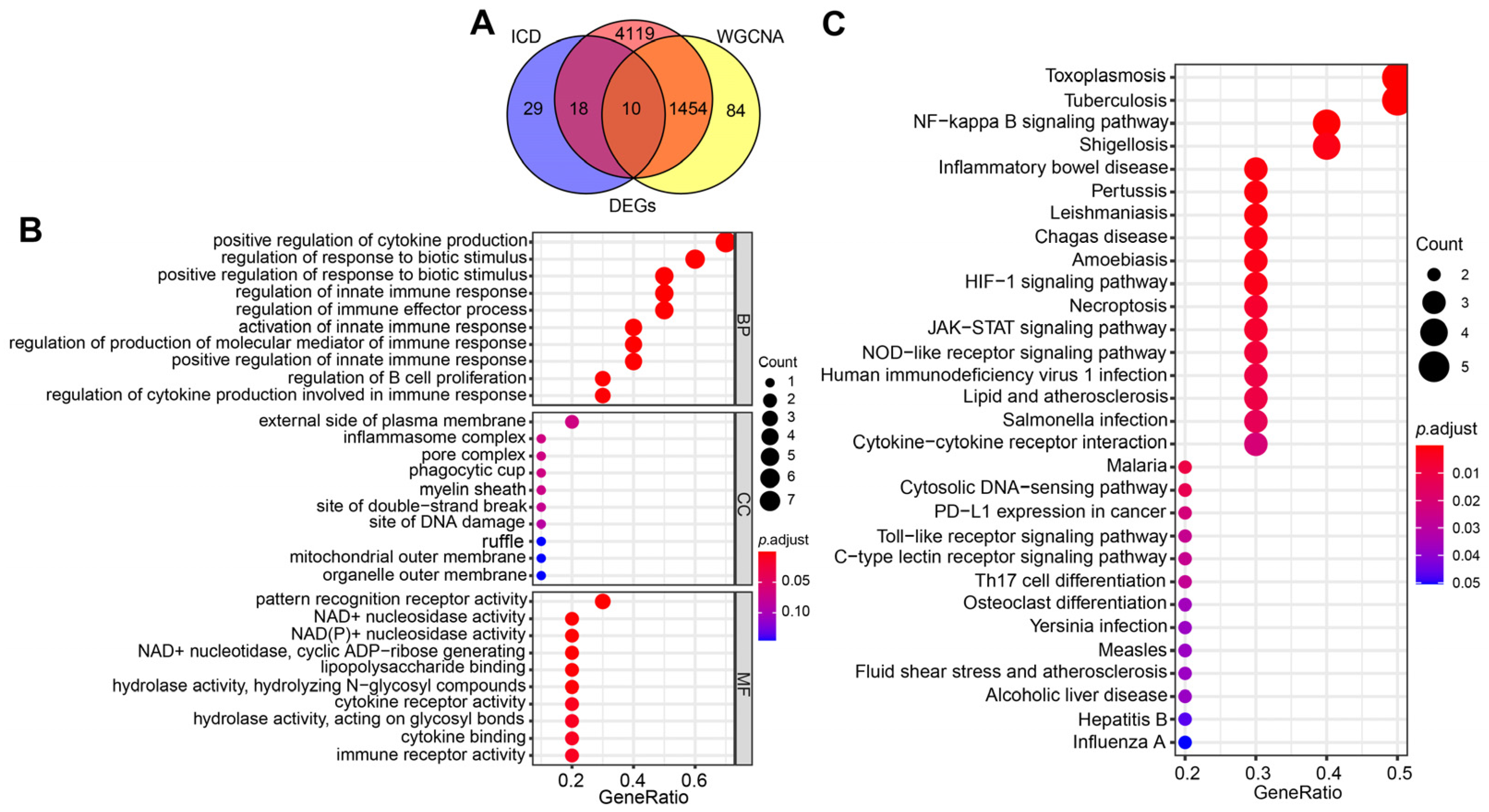
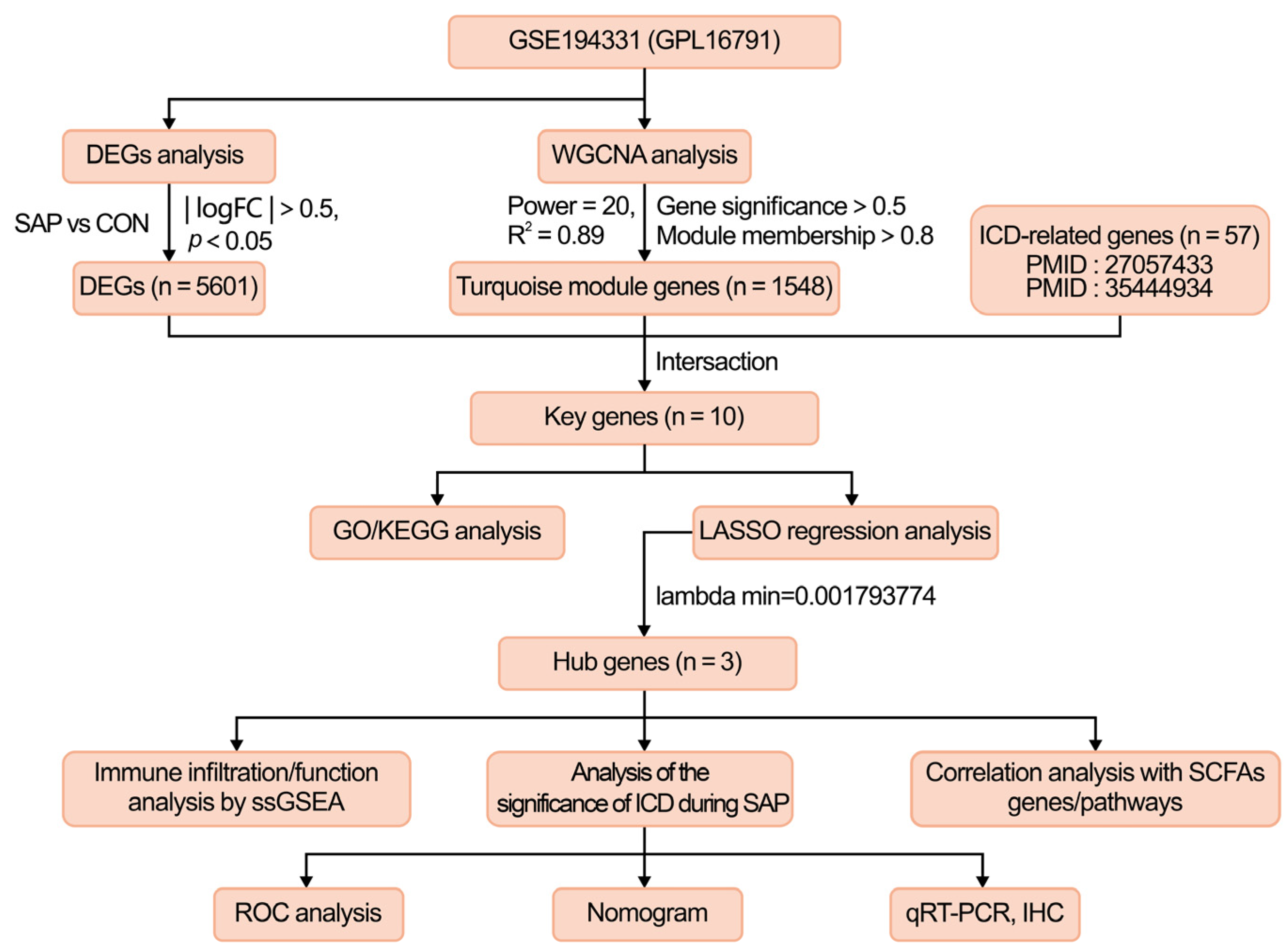
2.1. DEGs Screening
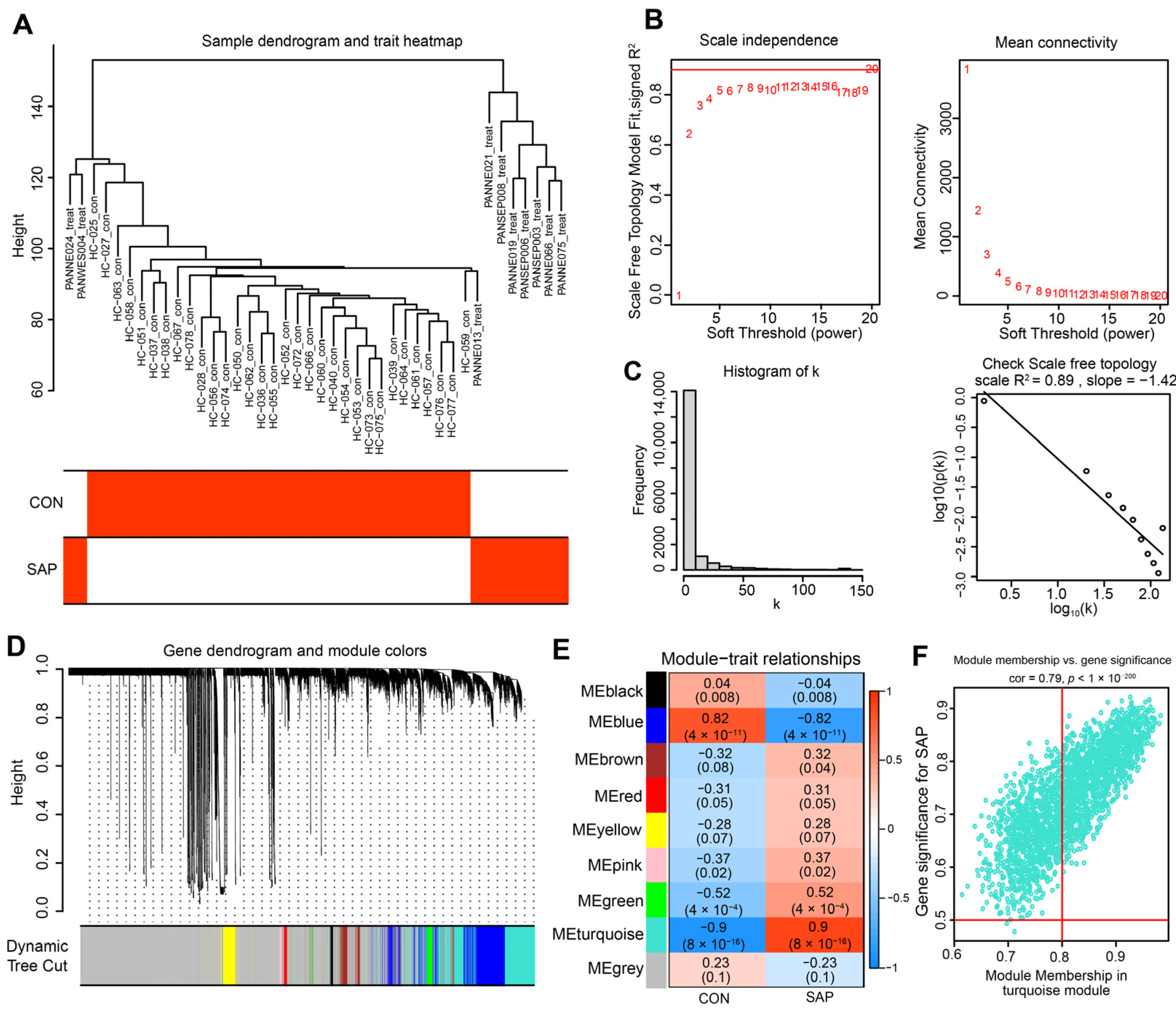
2.2. Construction of Co-Expression Network and Identification of SAP Core Genes
2.3. Selection and Functional Enrichment Analysis of ICD-Related Genes in SAP
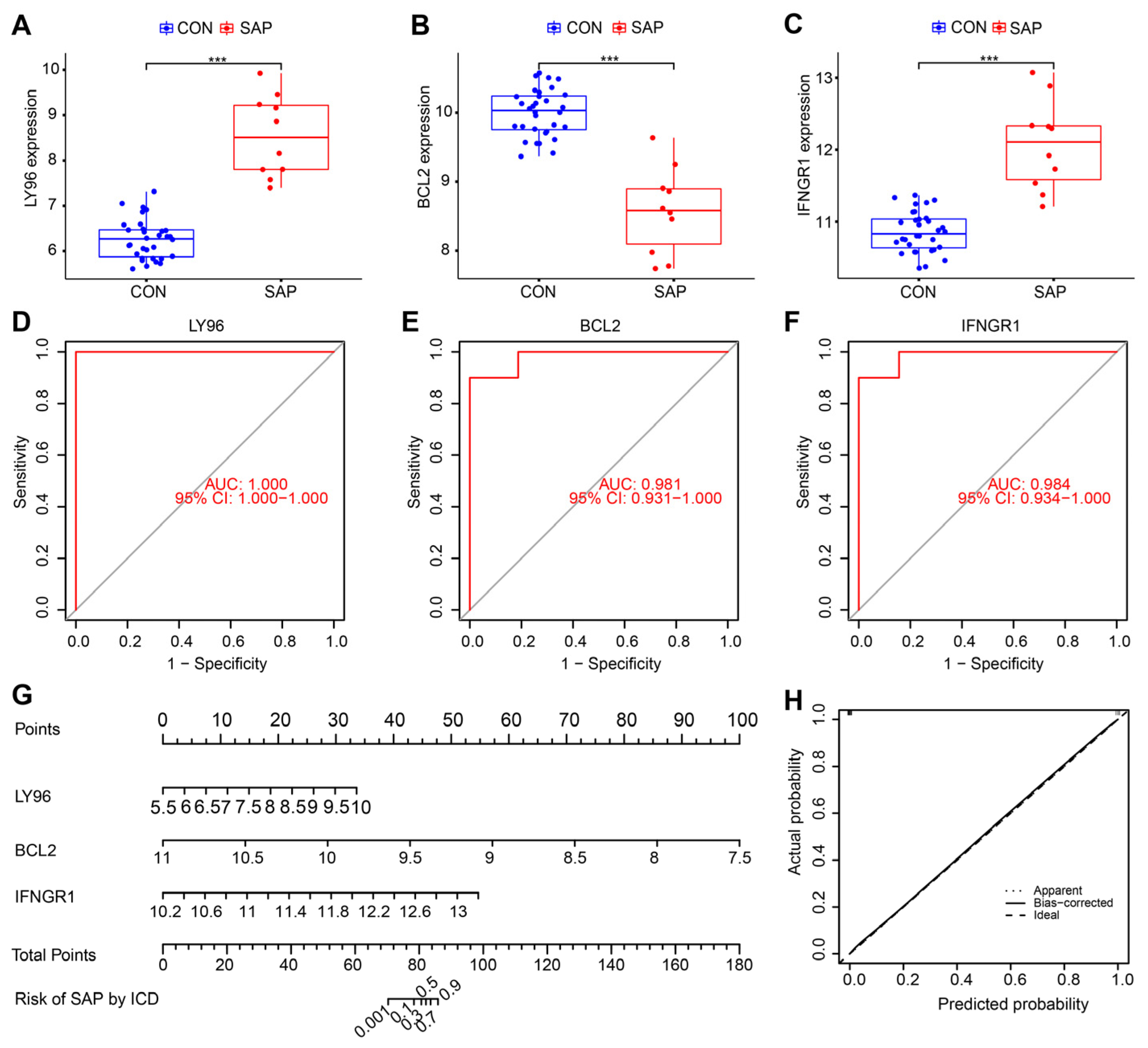
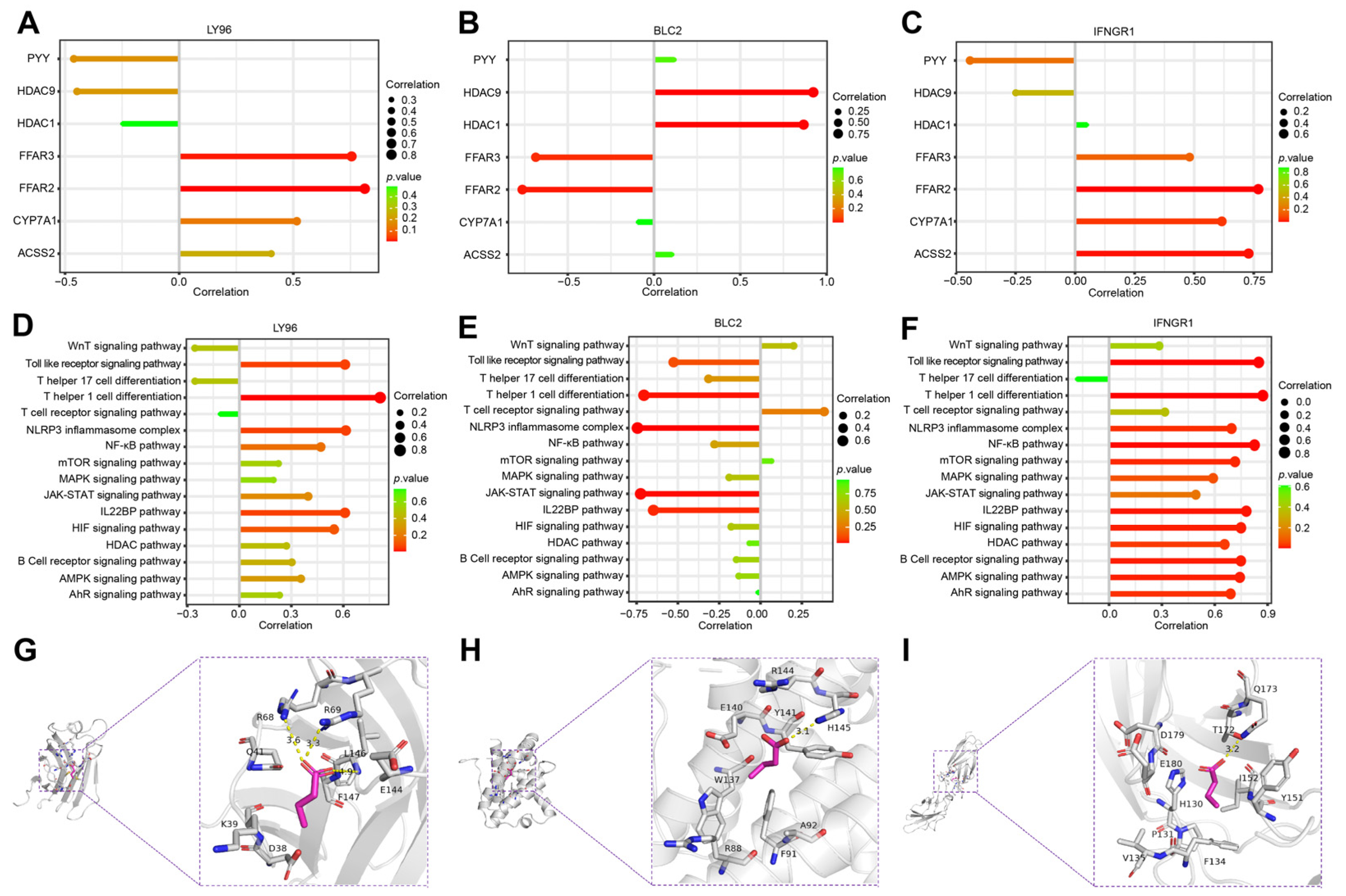
2.4. Identification of the Hub Genes Most Associated with ICD Occurrence in SAP
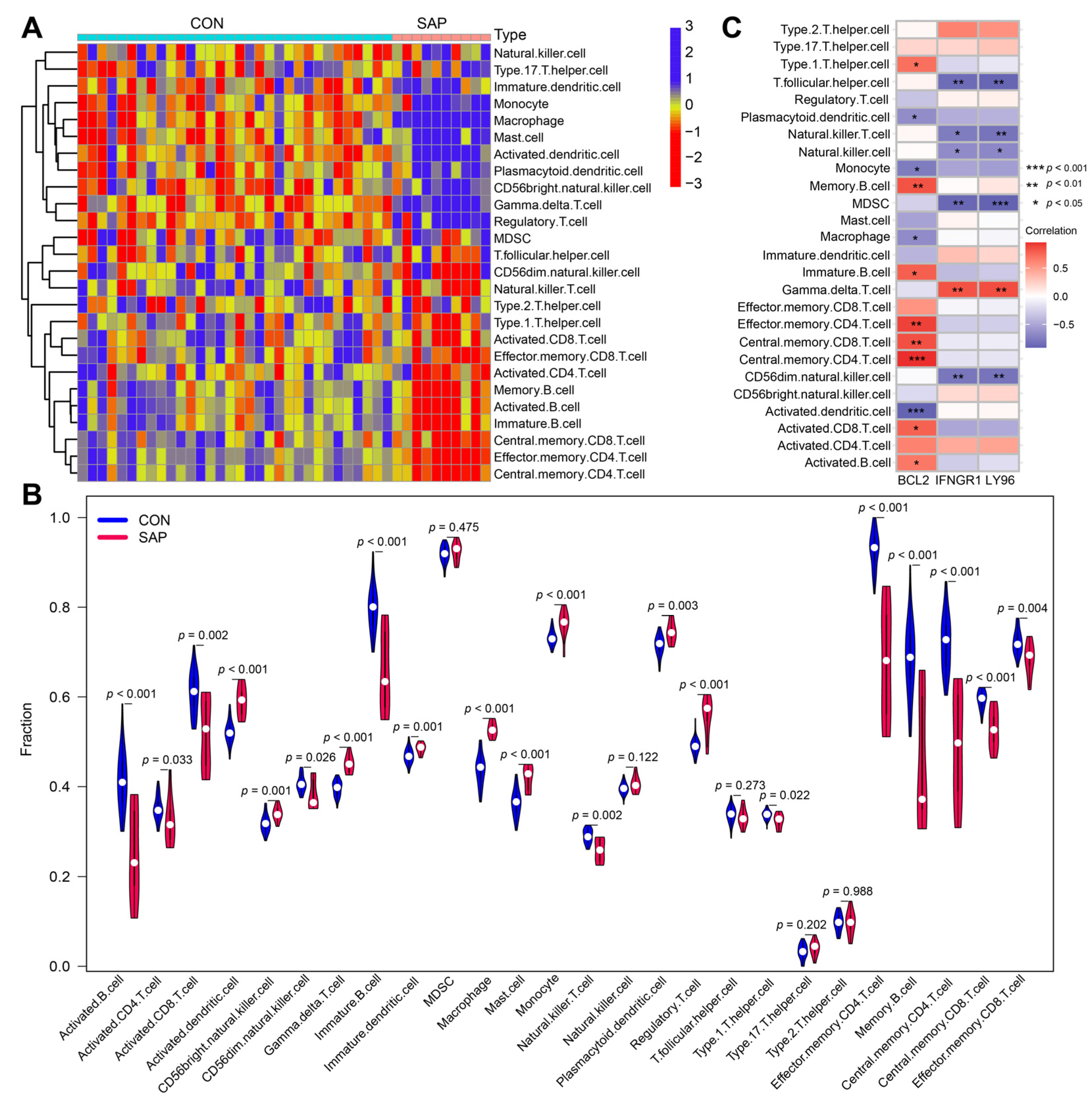
2.5. Immune Cell Infiltration/Functions and Its Association with Hub Genes
2.6. Correlation Analysis of Hub Genes and SCFAs
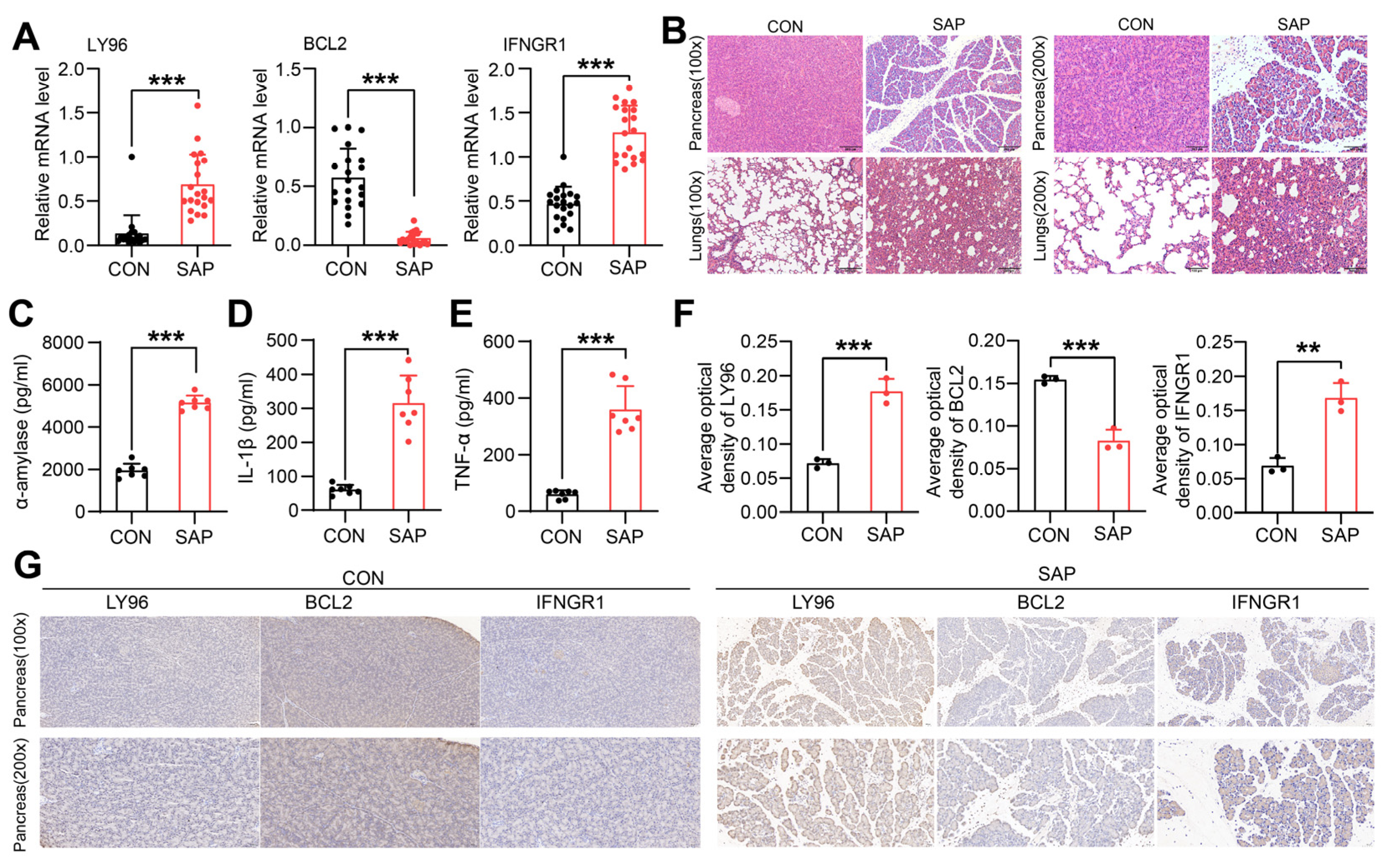
2.7. Clinical and Animal Validation of Hub Genes
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Processing
4.2. Identification and Visualization of Differentially Expressed Genes (DEGs)
4.3. WGCNA
4.4. LASSO Regression Screening of Central Genes
4.5. Gene Function Enrichment Analysis
4.6. ssGSEA
4.7. Differential Expression Analysis and Receiver Operating Characteristic (ROC) Curve Validation
4.8. Nomogram Construction and Verification
4.9. Peripheral Blood Mononuclear Cells (PBMCs) Collection and qRT-PCR
4.10. Establishment of Rat SAP Model
4.11. Histology and Immunohistochemistry
4.12. Serum Amylase and Inflammatory Factor Assay
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boxhoorn, L.; Voermans, R.P.; Bouwense, S.A.; Bruno, M.J.; Verdonk, R.C.; Boermeester, M.A.; van Santvoort, H.C.; Besselink, M.G. Acute pancreatitis. Lancet 2020, 396, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Papachristou, G.I. New insights into acute pancreatitis. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 479–496. [Google Scholar] [CrossRef] [PubMed]
- van Santvoort, H.C.; Bakker, O.J.; Bollen, T.L.; Besselink, M.G.; Ali, U.A.; Schrijver, A.M.; Boermeester, M.A.; van Goor, H.; Dejong, C.H.; van Eijck, C.H.; et al. A Conservative and Minimally Invasive Approach to Necrotizing Pancreatitis Improves Outcome. Gastroenterology 2011, 141, 1254–1263. [Google Scholar] [CrossRef]
- Leppäniemi, A.; Tolonen, M.; Tarasconi, A.; Lohse, H.A.S.; Gamberini, E.; Kirkpatrick, A.W.; Ball, C.G.; Parry, N.; Sartelli, M.; Wolbrink, D.R.J.; et al. 2019 WSES guidelines for the management of severe acute pancreatitis. World J. Emerg. Surg. 2019, 14, 27. [Google Scholar] [CrossRef]
- Duan, F.; Wang, X.; Wang, H.; Wang, Y.; Zhang, Y.; Chen, J.; Zhu, X.; Chen, B. GDF11 ameliorates severe acute pancreatitis through modulating macrophage M1 and M2 polarization by targeting the TGFβR1/SMAD-2 pathway. Int. Immunopharmacol. 2022, 108, 108777. [Google Scholar] [CrossRef]
- Li, W.; Ou, L.; Fu, Y.; Chen, Y.; Yin, Q.; Song, H. Risk factors for concomitant infectious pancreatic necrosis in patients with severe acute pancreatitis: A systematic review and meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101901. [Google Scholar] [CrossRef]
- Kokosis, G.; Perez, A.; Pappas, T. Surgical management of necrotizing pancreatitis: An overview. World J. Gastroenterol. 2014, 20, 16106–16112. [Google Scholar] [CrossRef]
- Baron, T.H.; DiMaio, C.J.; Wang, A.Y.; Morgan, K.A. American Gastroenterological Association Clinical Practice Update: Management of Pancreatic Necrosis. Gastroenterology 2020, 158, 67–75. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; Liu, J.; Luo, Y.; Guo, H.; Yang, Q.; Xu, C.; Ma, S.; Chen, H. Intestinal Microbiota—An Unmissable Bridge to Severe Acute Pancreatitis-Associated Acute Lung Injury. Front. Immunol. 2022, 13, 913178. [Google Scholar] [CrossRef]
- Patel, B.K.; Patel, K.H.; Bhatia, M.; Iyer, S.G.; Madhavan, K.; Moochhala, S.M. Gut microbiome in acute pancreatitis: A review based on current literature. World J. Gastroenterol. 2021, 27, 5019–5036. [Google Scholar] [CrossRef]
- Li, X.-Y.; He, C.; Zhu, Y.; Lu, N.-H. Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J. Gastroenterol. 2020, 26, 2187–2193. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; He, C.; Li, X.; Cai, Y.; Hu, J.; Liao, Y.; Zhao, J.; Xia, L.; He, W.; Liu, L.; et al. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J. Gastroenterol. 2019, 54, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Si, C.; Deng, J.; Chen, X.; Kong, L.; Zhou, X.; Wang, W. Understanding the Effects of Metabolites on the Gut Microbiome and Severe Acute Pancreatitis. BioMed Res. Int. 2021, 2021, 1516855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, J.; Du, D.; Niu, N.; Liu, S.; Yang, X.; Lu, P.; Shen, X.; Shi, N.; Yao, L.; et al. Ketogenesis acts as an endogenous protective programme to restrain inflammatory macrophage activation during acute pancreatitis. Ebiomedicine 2022, 78, 103959. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, J.; Li, F.; Luo, Y.; Ge, P.; Zhang, Y.; Wen, H.; Yang, Q.; Ma, S.; Chen, H. The gut-lung axis in severe acute Pancreatitis-associated lung injury: The protection by the gut microbiota through short-chain fatty acids. Pharmacol. Res. 2022, 182, 106321. [Google Scholar] [CrossRef]
- Zhang, T.; Xia, M.; Zhan, Q.; Zhou, Q.; Lu, G.; An, F. Sodium Butyrate Reduces Organ Injuries in Mice with Severe Acute Pancreatitis Through Inhibiting HMGB1 Expression. Dig. Dis. Sci. 2015, 60, 1991–1999. [Google Scholar] [CrossRef]
- Lei, Y.; Tang, L.; Liu, S.; Hu, S.; Wu, L.; Liu, Y.; Yang, M.; Huang, S.; Tang, X.; Tang, T.; et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome 2021, 9, 115. [Google Scholar] [CrossRef]
- Pan, X.; Fang, X.; Wang, F.; Li, H.; Niu, W.; Liang, W.; Wu, C.; Li, J.; Tu, X.; Pan, L.; et al. Butyrate ameliorates caerulein-induced acute pancreatitis and associated intestinal injury by tissue-specific mechanisms. Br. J. Pharmacol. 2019, 176, 4446–4461. [Google Scholar] [CrossRef]
- Van der Berg, F.F.; van Dalen, D.; Hyoju, S.K.; van Santvoort, H.C.; Besselink, M.G.; Wiersinga, W.J.; Zaborina, O.; Boermeester, M.A.; Alverdy, J. Western-type diet influences mortality from necrotising pancreatitis and demonstrates a central role for butyrate. Gut 2021, 70, 915–927. [Google Scholar] [CrossRef]
- Xiao, S.; Jing, S.; Jiakui, S.; Lei, Z.; Ying, L.; Han, L.; Xinwei, M.; Weiqin, L. Butyrate Ameliorates Intestinal Epithelial Barrier Injury Via Enhancing Foxp3+ Regulatory T-Cell Function in Severe Acute Pancreatitis Model. Turk. J. Gastroenterol. 2022, 33, 710–719. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Wei, M.; Yang, B. Recent advances in nanomedicines for photodynamic therapy (PDT)-driven cancer immunotherapy. Theranostics 2022, 12, 434–458. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Chen, L.; Li, Y.; Hu, Z.; He, F. Ferroptosis, necroptosis, and pyroptosis in the tumor microenvironment: Perspectives for immunotherapy of SCLC. Semin. Cancer Biol. 2022, 86, 273–285. [Google Scholar] [CrossRef]
- Kroemer, G.; Galassi, C.; Zitvogel, L.; Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 2022, 23, 487–500. [Google Scholar] [CrossRef]
- Xu, M.; Lu, J.-H.; Zhong, Y.-Z.; Jiang, J.; Shen, Y.-Z.; Su, J.-Y.; Lin, S.-Y. Immunogenic Cell Death-Relevant Damage-Associated Molecular Patterns and Sensing Receptors in Triple-Negative Breast Cancer Molecular Subtypes and Implications for Immunotherapy. Front. Oncol. 2022, 12, 870914. [Google Scholar] [CrossRef]
- Mardi, A.; Shirokova, A.V.; Mohammed, R.N.; Keshavarz, A.; Zekiy, A.O.; Thangavelu, L.; Mohamad, T.A.M.; Marofi, F.; Shomali, N.; Zamani, A.; et al. Biological causes of immunogenic cancer cell death (ICD) and anti-tumor therapy; Combination of Oncolytic virus-based immunotherapy and CAR T-cell therapy for ICD induction. Cancer Cell Int. 2022, 22, 168. [Google Scholar] [CrossRef]
- Birmpilis, A.I.; Paschalis, A.; Mourkakis, A.; Christodoulou, P.; Kostopoulos, I.V.; Antimissari, E.; Terzoudi, G.; Georgakilas, A.G.; Armpilia, C.; Papageorgis, P.; et al. Immunogenic Cell Death, DAMPs and Prothymosin α as a Putative Anticancer Immune Response Biomarker. Cells 2022, 11, 1415. [Google Scholar] [CrossRef]
- Hoque, R.; Sohail, M.; Malik, A.; Sarwar, S.; Luo, Y.; Shah, A.; Barrat, F.; Flavell, R.; Gorelick, F.; Husain, S.; et al. TLR9 and the NLRP3 Inflammasome Link Acinar Cell Death With Inflammation in Acute Pancreatitis. Gastroenterology 2011, 141, 358–369. [Google Scholar] [CrossRef]
- Dixit, A.; Cheema, H.; George, J.; Iyer, S.; Dudeja, V.; Dawra, R.; Saluja, A.K. Extracellular release of ATP promotes systemic inflammation during acute pancreatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G463–G475. [Google Scholar] [CrossRef]
- Frossard, J.-L.; Saluja, A.; Bhagat, L.; Lee, H.S.; Bhatia, M.; Hofbauer, B.; Steer, M.L. The role of intercellular adhesion molecule 1 and neutrophils in acute pancreatitis and pancreatitis-associated lung injury. Gastroenterology 1999, 116, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, M.; Brady, M.; Shokuhi, S.; Christmas, S.; Neoptolemos, J.P.; Slavin, J. Inflammatory mediators in acute pancreatitis. J. Pathol. 2000, 190, 117–125. [Google Scholar] [CrossRef]
- Tsuji, Y.; Watanabe, T.; Kudo, M.; Arai, H.; Strober, W.; Chiba, T. Sensing of Commensal Organisms by the Intracellular Sensor NOD1 Mediates Experimental Pancreatitis. Immunity 2012, 37, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.V.; Garg, A.D.; Kaczmarek, A.; Krysko, O.; Agostinis, P.; Vandenabeele, P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 2012, 12, 860–875. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, J.; Kong, Z.; Sun, X.; He, Z.; Sun, B.; Luo, C.; Sun, J. Emerging photodynamic nanotherapeutics for inducing immunogenic cell death and potentiating cancer immunotherapy. Biomaterials 2022, 282, 121433. [Google Scholar] [CrossRef]
- Bao, X.; Xie, L. Targeting purinergic pathway to enhance radiotherapy-induced immunogenic cancer cell death. J. Exp. Clin. Cancer Res. 2022, 41, 222. [Google Scholar] [CrossRef]
- Kepp, O.; Zitvogel, L.; Kroemer, G. Lurbinectedin: An FDA-approved inducer of immunogenic cell death for the treatment of small-cell lung cancer. Oncoimmunology 2020, 9, 1795995. [Google Scholar] [CrossRef]
- De Oca, R.M.; Alavi, A.S.; Vitali, N.; Bhattacharya, S.; Blackwell, C.; Patel, K.; Seestaller-Wehr, L.; Kaczynski, H.; Shi, H.; Dobrzynski, E.; et al. Belantamab Mafodotin (GSK2857916) Drives Immunogenic Cell Death and Immune-mediated Antitumor Responses In Vivo. Mol. Cancer Ther. 2021, 20, 1941–1955. [Google Scholar] [CrossRef]
- Kashefizadeh, A.; Kazemizadeh, H. Immunogenic cell death (ICD)-inducers in non-small-cell lung carcinoma (NSCLC): Current knowledge and future perspective. Clin. Transl. Oncol. 2022, 25, 316–322. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Nomiri, S.; Karami, H.; Baradaran, B.; Javadrashid, D.; Derakhshani, A.; Nourbakhsh, N.S.; Shadbad, M.A.; Solimando, A.G.; Tabrizi, N.J.; Brunetti, O.; et al. Exploiting systems biology to investigate the gene modules and drugs in ovarian cancer: A hypothesis based on the weighted gene co-expression network analysis. Biomed. Pharmacother. 2022, 146, 112537. [Google Scholar] [CrossRef]
- Zheng, P.-F.; Chen, L.-Z.; Liu, P.; Pan, H.W.; Fan, W.-J.; Liu, Z.-Y. Identification of immune-related key genes in the peripheral blood of ischaemic stroke patients using a weighted gene coexpression network analysis and machine learning. J. Transl. Med. 2022, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.H.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.M.; Hultman, P.; Toomey, C.B.; Cauvi, D.M.; Hoffman, H.M.; Hamel, J.C.; Kono, D.H. Definition of IFN-γ-related pathways critical for chemically-induced systemic autoimmunity. J. Autoimmun. 2012, 39, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.M.; Noon-Song, E.N.; Kemppainen, K.; Ahmed, C.M. Steroid-like signalling by interferons: Making sense of specific gene activation by cytokines. Biochem. J. 2012, 443, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.; Zhou, J.; Mao, W.; Chen, T.; Zhu, Y.; Pan, X.; Mei, H.; Singh, V.; Buxbaum, J.; Doig, G.; et al. Immune enhancement in patients with predicted severe acute necrotising pancreatitis: A multicentre double-blind randomised controlled trial. Intensiv. Care Med. 2022, 48, 899–909. [Google Scholar] [CrossRef]
- Nesvaderani, M.M.; Dhillon, B.K.; Chew, T.; Tang, B.F.; Baghela, A.; Hancock, R.E.O.; Eslick, G.D.F.; Cox, M.F. Gene Expression Profiling: Identification of Novel Pathways and Potential Biomarkers in Severe Acute Pancreatitis. J. Am. Coll. Surg. 2022, 234, 803–815. [Google Scholar] [CrossRef]
- Xue, L.; Wu, J.; Chen, J.; Yang, Y. Change in the pituitary stalk deviation angle after transsphenoidal surgery can predict the development of diabetes insipidus for pituitary adenomas. Endocr. Connect. 2022, 11, 11. [Google Scholar] [CrossRef]
- Chen, W.; Wang, S.; Wu, Y.; Shen, X.; Guo, Z.; Li, Q.; Xing, D. Immunogenic cell death: A link between gut microbiota and anticancer effects. Microb. Pathog. 2020, 141, 103983. [Google Scholar] [CrossRef]
- Zhang, R.; Kang, R.; Tang, D. The STING1 network regulates autophagy and cell death. Signal Transduct. Target. Ther. 2021, 6, 208. [Google Scholar] [CrossRef]
- Lu, W.; Rao, Y.; Li, Y.; Dai, Y.; Chen, K. The Landscape of Cell Death Processes with Associated Immunogenic and Fibrogenic Effects in Arrhythmogenic Cardiomyopathy. J. Cardiovasc. Dev. Dis. 2022, 9, 301. [Google Scholar] [CrossRef]
- Feng, D.; Park, O.; Radaeva, S.; Wang, H.; Yin, S.; Kong, X.; Zheng, M.; Zakhari, S.; Kolls, J.K.; Gao, B. Interleukin-22 Ameliorates Cerulein-Induced Pancreatitis in Mice by Inhibiting the Autophagic Pathway. Int. J. Biol. Sci. 2012, 8, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Choong, C.-J.; Okuno, T.; Ikenaka, K.; Baba, K.; Hayakawa, H.; Koike, M.; Yokota, M.; Doi, J.; Kakuda, K.; Takeuchi, T.; et al. Alternative mitochondrial quality control mediated by extracellular release. Autophagy 2021, 17, 2962–2974. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rivera, F.J.; Ryan, J.; Soto-Feliciano, Y.M.; Beytagh, M.C.; Xuan, L.; Feldser, D.M.; Hemann, M.T.; Zamudio, J.; Dimitrova, N.; Letai, A.; et al. Mitochondrial apoptotic priming is a key determinant of cell fate upon p53 restoration. Proc. Natl. Acad. Sci. USA 2021, 118, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-L.; Zhang, W.-X.; Yan, J.-Q.; Tang, Y.-L.; Jia, W.-J.; Xu, Z.-W.; Xu, M.-J.; Chattipakorn, N.; Wang, Y.; Feng, J.-P.; et al. Chalcone derivatives ameliorate lipopolysaccharide-induced acute lung injury and inflammation by targeting MD2. Acta Pharmacol. Sin. 2022, 43, 76–85. [Google Scholar] [CrossRef]
- Wu, S.; Lin, C.; Zhang, T.; Zhang, B.; Jin, Y.; Wang, H.; Li, H.; Wang, Y.; Wang, X. Pentamidine Alleviates Inflammation and Lipopolysaccharide-Induced Sepsis by Inhibiting TLR4 Activation via Targeting MD2. Front. Pharmacol. 2022, 13, 835081. [Google Scholar] [CrossRef]
- Jin, W.; Fang, Q.; Jiang, D.; Li, T.; Wei, B.; Sun, J.; Zhang, W.; Zhang, Z.; Zhang, F.; Linhardt, R.J.; et al. Structural characteristics and anti-complement activities of polysaccharides from Sargassum hemiphyllum. Glycoconj. J. 2020, 37, 553–563. [Google Scholar] [CrossRef]
- Stinson, W.A.; Miner, C.A.; Zhao, F.R.; Lundgren, A.J.; Poddar, S.; Miner, J.J. The IFN-γ receptor promotes immune dysregulation and disease in STING gain-of-function mice. J. Clin. Investig. 2022, 7, e155250. [Google Scholar] [CrossRef]
- Cossetti, C.; Iraci, N.; Mercer, T.R.; Leonardi, T.; Alpi, E.; Drago, D.; Alfaro-Cervello, C.; Saini, H.K.; Davis, M.P.; Schaeffer, J.; et al. Extracellular Vesicles from Neural Stem Cells Transfer IFN-γ via Ifngr1 to Activate Stat1 Signaling in Target Cells. Mol. Cell 2014, 56, 193–204. [Google Scholar] [CrossRef]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B.; Galla, S.; Chakraborty, S.; Cheng, X.; Yeo, J.; Mell, B.; Zhang, H.; et al. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Padoan, A.; Plebani, M.; Basso, D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int. J. Mol. Sci. 2019, 20, 676. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Li, Z.; Yu, X. The Role of Pancreatic Infiltrating Innate Immune Cells in Acute Pancreatitis. Int. J. Med. Sci. 2021, 18, 534–545. [Google Scholar] [CrossRef]
- Sendler, M.; van den Brandt, C.; Glaubitz, J.; Wilden, A.; Golchert, J.; Weiss, F.U.; Homuth, G.; Chama, L.L.D.F.; Mishra, N.; Mahajan, U.M.; et al. NLRP3 Inflammasome Regulates Development of Systemic Inflammatory Response and Compensatory Anti-Inflammatory Response Syndromes in Mice With Acute Pancreatitis. Gastroenterology 2020, 158, 253–269. [Google Scholar] [CrossRef]
- Zheng, L.; Xue, J.; Jaffee, E.M.; Habtezion, A. Role of Immune Cells and Immune-Based Therapies in Pancreatitis and Pancreatic Ductal Adenocarcinoma. Gastroenterology 2013, 144, 1230–1240. [Google Scholar] [CrossRef]
- Ribot, J.C.; Lopes, N.; Silva-Santos, B. γδ T cells in tissue physiology and surveillance. Nat. Rev. Immunol. 2021, 21, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Shiromizu, C.M.; Jancic, C.C. γδ T Lymphocytes: An Effector Cell in Autoimmunity and Infection. Front. Immunol. 2018, 9, 2389. [Google Scholar] [CrossRef]
- Papadopoulou, M.; Sanchez, G.S.; Vermijlen, D. Innate and adaptive γδ T cells: How, when, and why. Immunol. Rev. 2020, 298, 99–116. [Google Scholar] [CrossRef]
- Hayday, A.C. γδ T Cell Update: Adaptate Orchestrators of Immune Surveillance. J. Immunol. 2019, 203, 311–320. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Y.; Jing, H.; Chang, Q.; Wu, X.; Zhang, Z. RORγt inhibitor SR1001 alleviates acute pancreatitis by suppressing pancreatic IL-17-producing Th17 and γδ-T cells in mice with ceruletide-induced pancreatitis. Basic Clin. Pharmacol. Toxicol. 2021, 129, 357–368. [Google Scholar] [CrossRef]
- Garg, A.D.; De Ruysscher, D.; Agostinis, P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: A large-scale meta-analysis. Oncoimmunology 2016, 5, e1069938. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; He, S.; Yu, X.; Lai, X.; Tang, S.; El Akkawi Mariya, M.; Wang, M.; Yan, H.; Huang, X.; Zeng, S.; et al. Analysis and Experimental Validation of Rheumatoid Arthritis Innate Immunity Gene CYFIP2 and Pan-Cancer. Front. Immunol. 2022, 13, 954848. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Capitanio, U.; Jeldres, C.; Karakiewicz, P.I. Can nomograms be superior to other prediction tools? BJU Int. 2009, 103, 492–497. [Google Scholar] [CrossRef]
- Banks, P.A.; Bollen, T.L.; Dervenis, C.; Gooszen, H.G.; Johnson, C.D.; Sarr, M.G.; Tsiotos, G.G.; Vege, S.S.; Acute Pancreatitis Classification Working Group. Classification of acute pancreatitis—2012: Revision of the Atlanta classification and definitions by international consensus. Gut 2013, 62, 102–111. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, M.; Guo, H.; Liu, H.; Zhang, G.; Xu, C.; Chen, H. Emodin Alleviates Severe Acute Pancreatitis-Associated Acute Lung Injury by Inhibiting the Cold-Inducible RNA-Binding Protein (CIRP)-Mediated Activation of the NLRP3/IL-1β/CXCL1 Signaling. Front. Pharmacol. 2021, 12, 655372. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, G.; Chen, H.; Liu, G.; Owusu, L.; Wang, Y.; Wang, G.; Xu, C. Therapeutic effect of Qingyi decoction in severe acute pancreatitis-induced intestinal barrier injury. World J. Gastroenterol. 2015, 21, 3537–3546. [Google Scholar] [CrossRef]



| Characteristic | CON (n = 20) | SAP (n = 20) | p-Value |
|---|---|---|---|
| Gender (F/M) | 10/10 | 7/13 | 0.337 * N.S |
| Age (years) | 14.652 | 11.989 | 0.240 * N.S |
| PaO2 (mmHg) | 5.128 | 10.732 | 0.014 ** |
| FiO2 (%) | 0.000 | 7.816 | 0.001 *** |
| Oxygenation index (mmHg) | 24.420 | 107.183 | 0.002 *** |
| SpO2 (%) | 1.889 | 7.708 | 0.000 *** |
| WBC (109/L) | 1.147 | 5.120 | 0.000 *** |
| PCT (ng/mL) | 0.110 | 3.088 | 0.016 ** |
| CK-MB (ug/L) | 1.015 | 2.723 | 0.018 ** |
| Hs-Tnl (ng/mL) | 0.047 | 0.594 | 0.102 * N.S |
| ALT (U/L) | 10.516 | 217.789 | 0.009 *** |
| AST (U/L) | 5.261 | 323.926 | 0.038 ** |
| TBIL (umol/L) | 3.817 | 65.475 | 0.022 ** |
| DBIL (umol/L) | 1.293 | 44.053 | 0.021 ** |
| γ-GGT (U/L) | 12.672 | 315.916 | 0.030 ** |
| Creatinine (umol/L) | 17.243 | 79.348 | 0.031 ** |
| Urea (mmol/L) | 1.063 | 13.521 | 0.159 * N.S |
| Amylase (U/L) | 17.255 | 1581.329 | 0.007 *** |
| Lipase (U/L) | 55.457 | 1690.193 | 0.000 *** |
| Serum calcium (mmol/L) | 0.113 | 0.413 | 0.001 *** |
| Genes | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| GAPDH | CCTCGTCCCGTAGACAAAATG | TGAGGTCAATGAAGGGGTCGT |
| BCL2 | GGAAGCTTGTCATCAATGGAAATC | TGATGACCCTTTTGGCTCCC |
| LY96 | CATGAATCTTCCAAAGCGCAA | CTCCCAGAAATAGCTTCAACAACA |
| IFNGR1 | CAGAAAGGAGGAGAAGCAAATCA | ATCTCACTTCCGTTCATTCTCACAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, J.; Wang, Y.; Guo, H.; Li, F.; Cao, Y.; Zhao, L.; Chen, H. Identification of Key Biomarkers Associated with Immunogenic Cell Death and Their Regulatory Mechanisms in Severe Acute Pancreatitis Based on WGCNA and Machine Learning. Int. J. Mol. Sci. 2023, 24, 3033. https://doi.org/10.3390/ijms24033033
Wang Z, Liu J, Wang Y, Guo H, Li F, Cao Y, Zhao L, Chen H. Identification of Key Biomarkers Associated with Immunogenic Cell Death and Their Regulatory Mechanisms in Severe Acute Pancreatitis Based on WGCNA and Machine Learning. International Journal of Molecular Sciences. 2023; 24(3):3033. https://doi.org/10.3390/ijms24033033
Chicago/Turabian StyleWang, Zhengjian, Jin Liu, Yuting Wang, Hui Guo, Fan Li, Yinan Cao, Liang Zhao, and Hailong Chen. 2023. "Identification of Key Biomarkers Associated with Immunogenic Cell Death and Their Regulatory Mechanisms in Severe Acute Pancreatitis Based on WGCNA and Machine Learning" International Journal of Molecular Sciences 24, no. 3: 3033. https://doi.org/10.3390/ijms24033033
APA StyleWang, Z., Liu, J., Wang, Y., Guo, H., Li, F., Cao, Y., Zhao, L., & Chen, H. (2023). Identification of Key Biomarkers Associated with Immunogenic Cell Death and Their Regulatory Mechanisms in Severe Acute Pancreatitis Based on WGCNA and Machine Learning. International Journal of Molecular Sciences, 24(3), 3033. https://doi.org/10.3390/ijms24033033






