Discovery of Novel Thiazolidinedione-Derivatives with Multi-Modal Antidiabetic Activities In Vitro and In Silico
Abstract
1. Introduction
2. Results
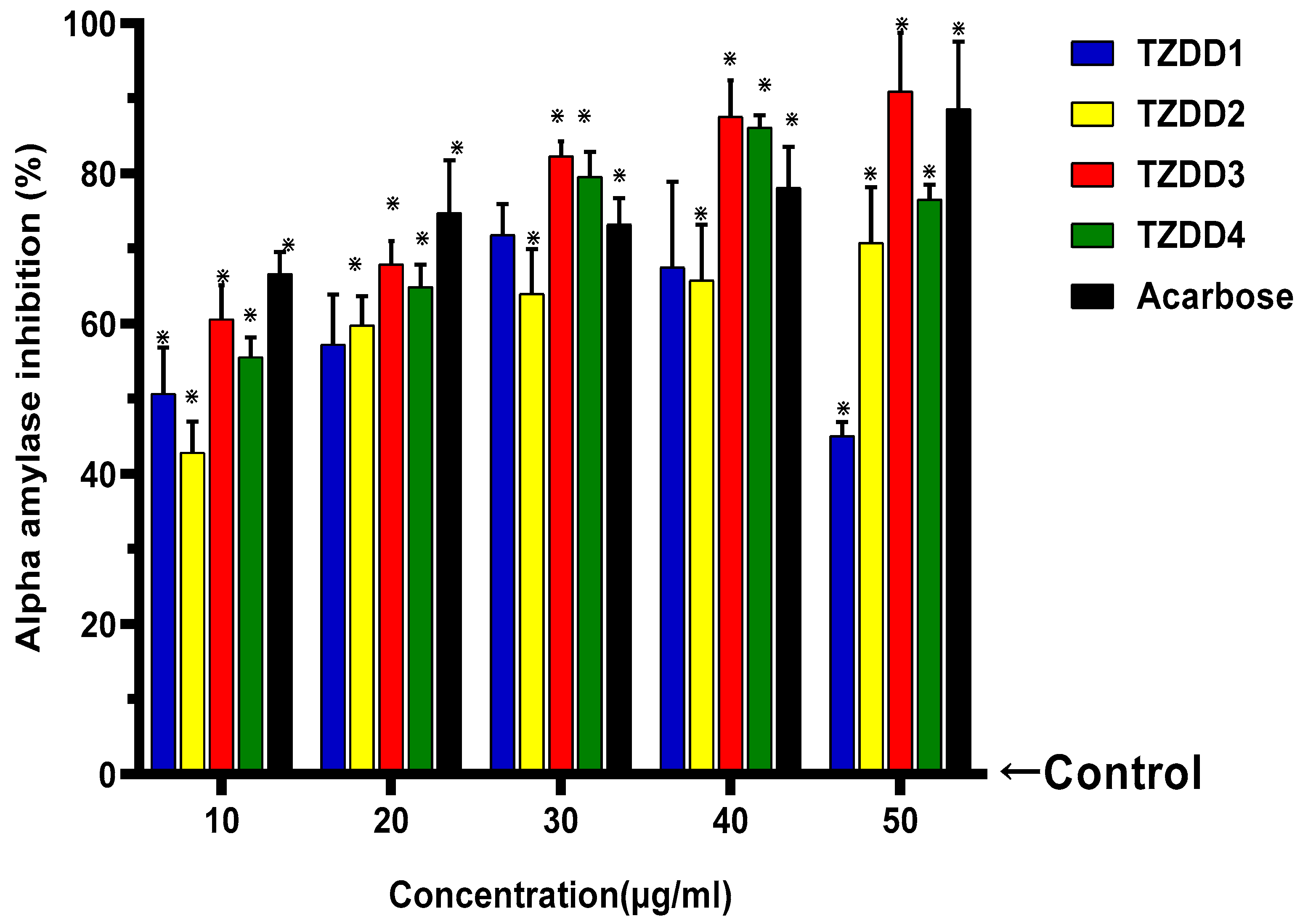
2.1. α-Amylase Activity
2.2. α-Glucosidase Activity
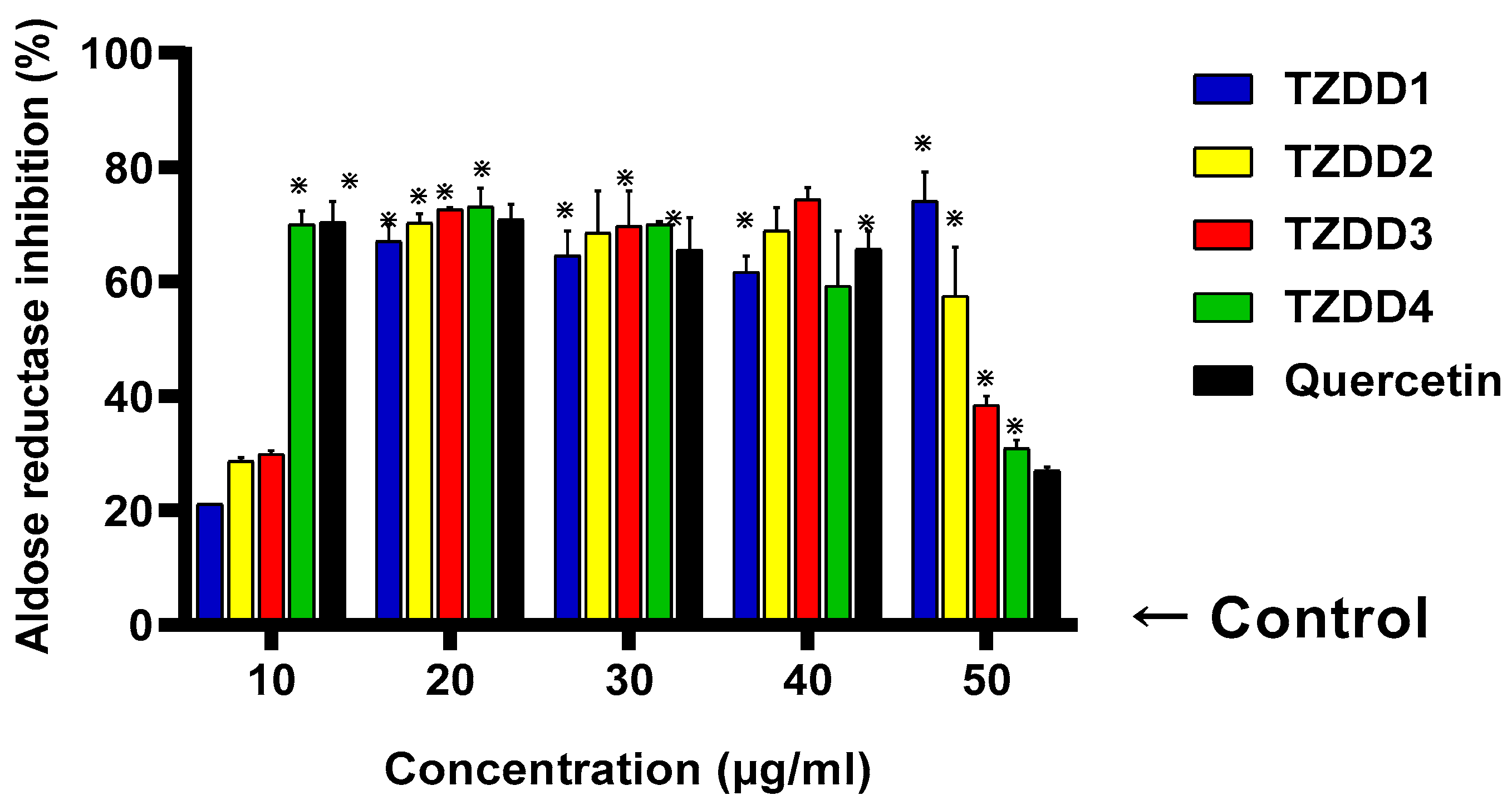
2.3. Aldose Reductase Activity
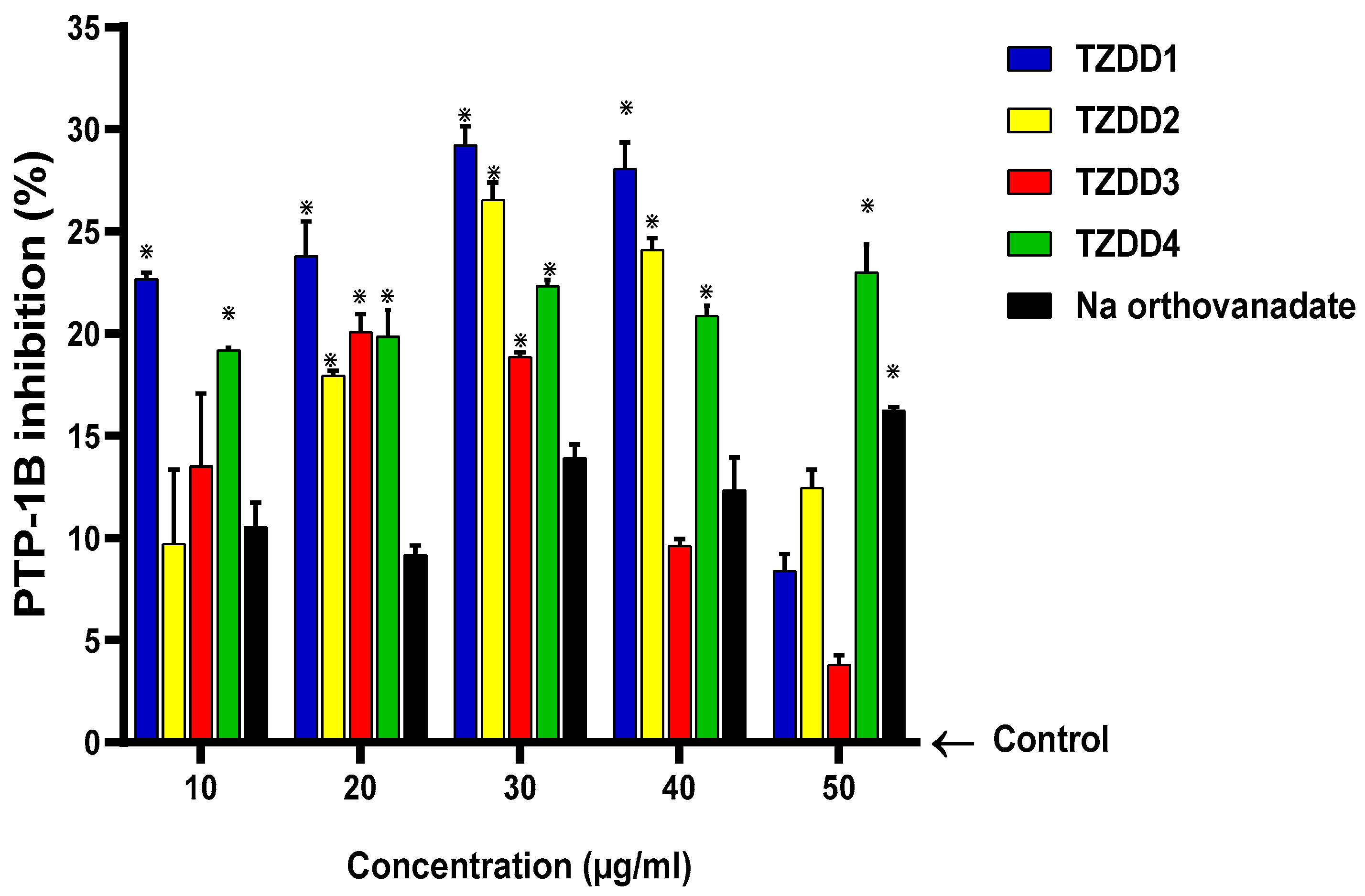
2.4. Protein Tyrosine Phosphatase 1B Activity
2.5. DPP-4 Activity
2.6. Cell Viability
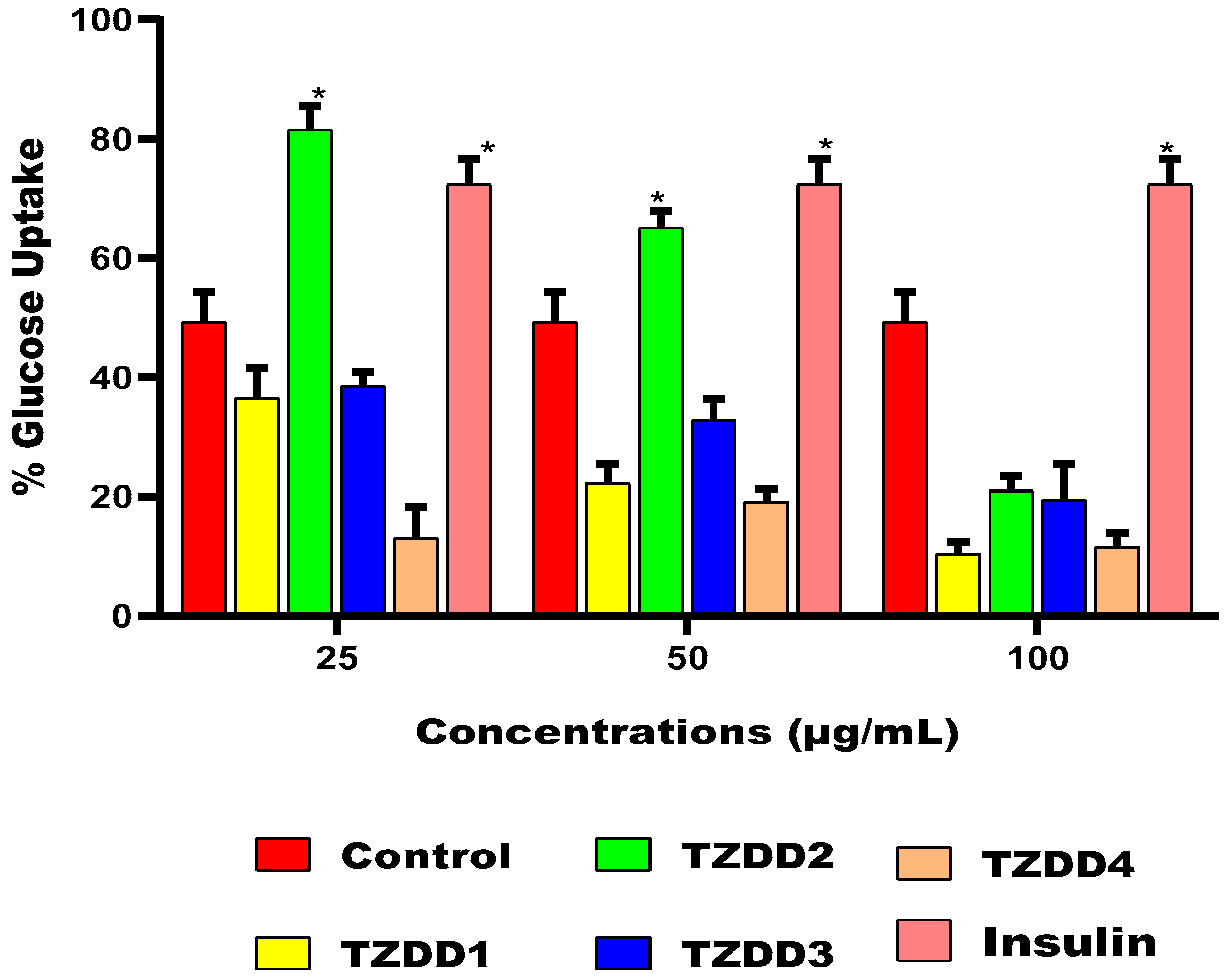
2.7. Glucose Uptake
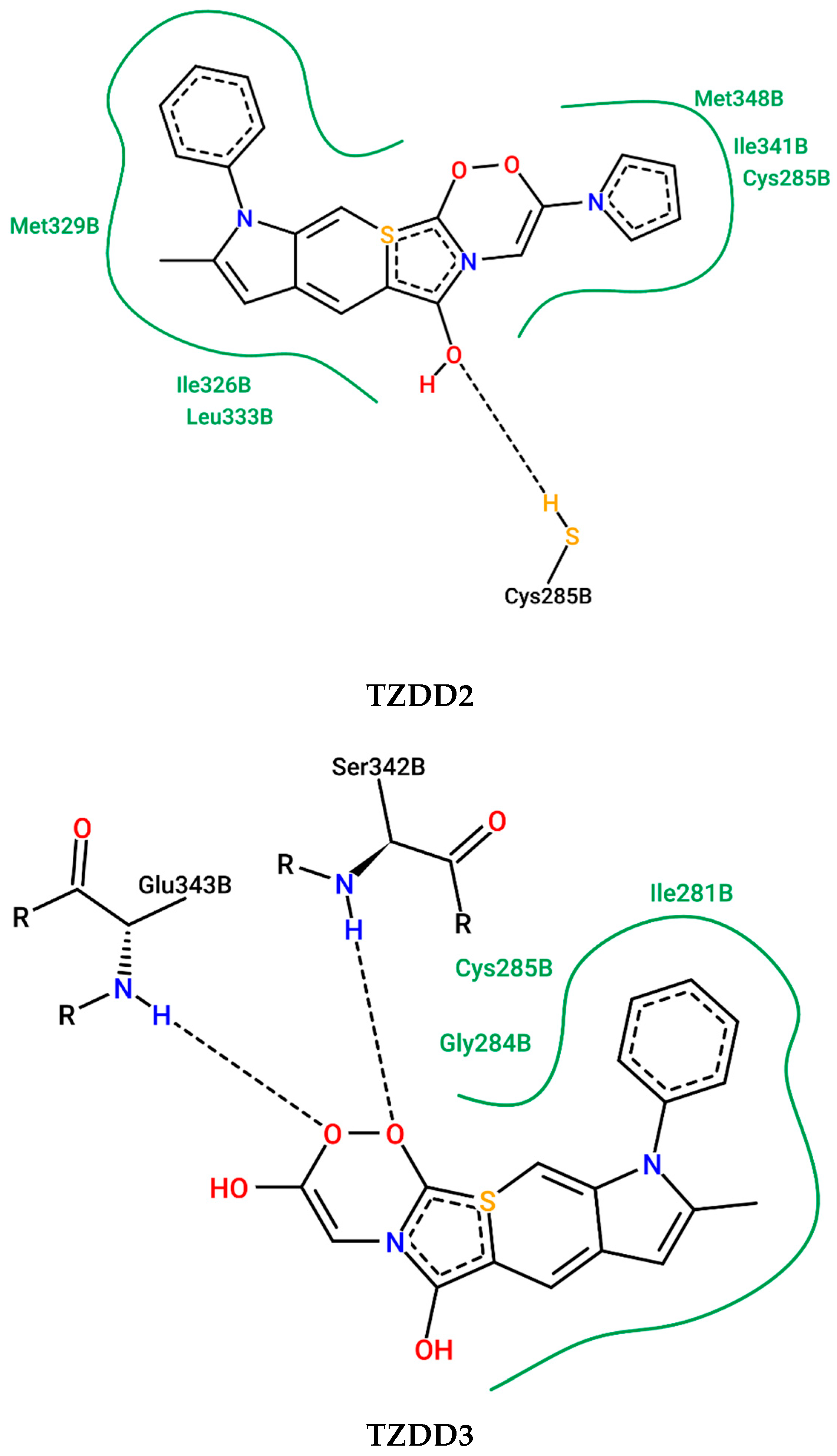
2.8. In Silico Determination of the PPAR-γ Activation
3. Discussion
4. Materials and Methods
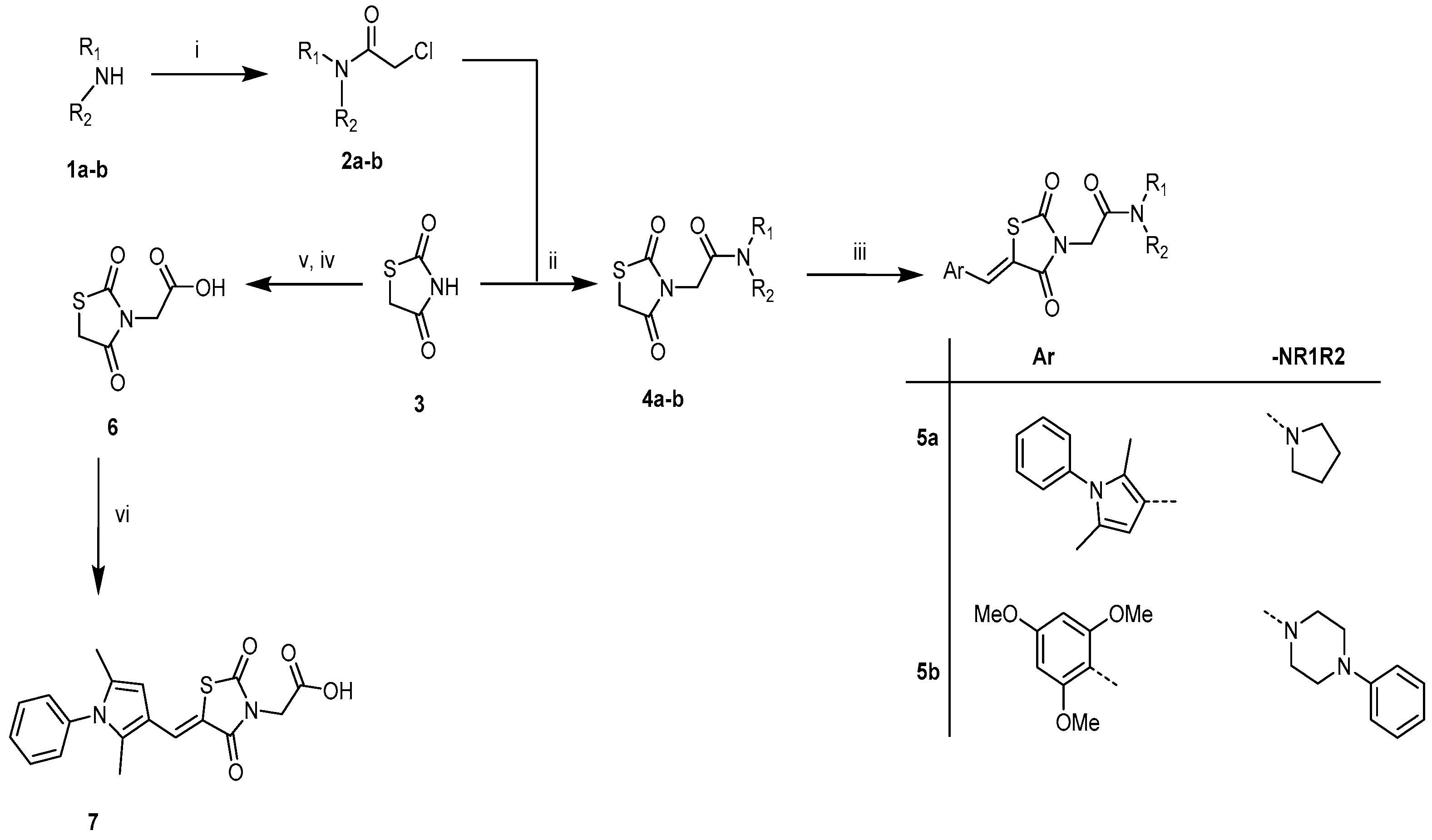
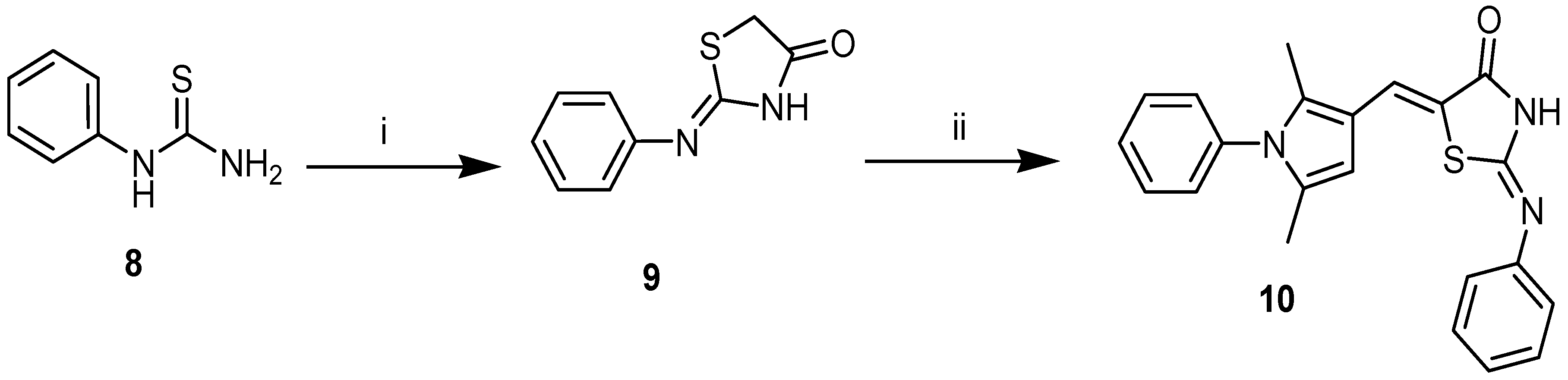
4.1. Synthesis of Compounds
4.1.1. Experimental Section
Materials and Physical Measurements
4.1.2. General Synthetic Procedure Target Compounds 5a (TZDD2) and 7 (TZDD3)
(Z)-5-((2,5-Dimethyl-1phenyl-1H-pyrrol-3-yl)methylene)-3-(2-oxo-2-(pyrrolidin-1-yl)ethyl)thiazolidine-2,4-dione, 5a (TZDD2)
(Z)-2-(5-((2,5-Dimethyl-1-phenyl-1H-pyrrol-3-yl)methylene)-2,4-dioxothiazolidin-3-yl)acetic Acid, 7 (TZDD3)
4.2. Biological Investigation
4.2.1. α-Amylase and Glucosidase Inhibition Assay
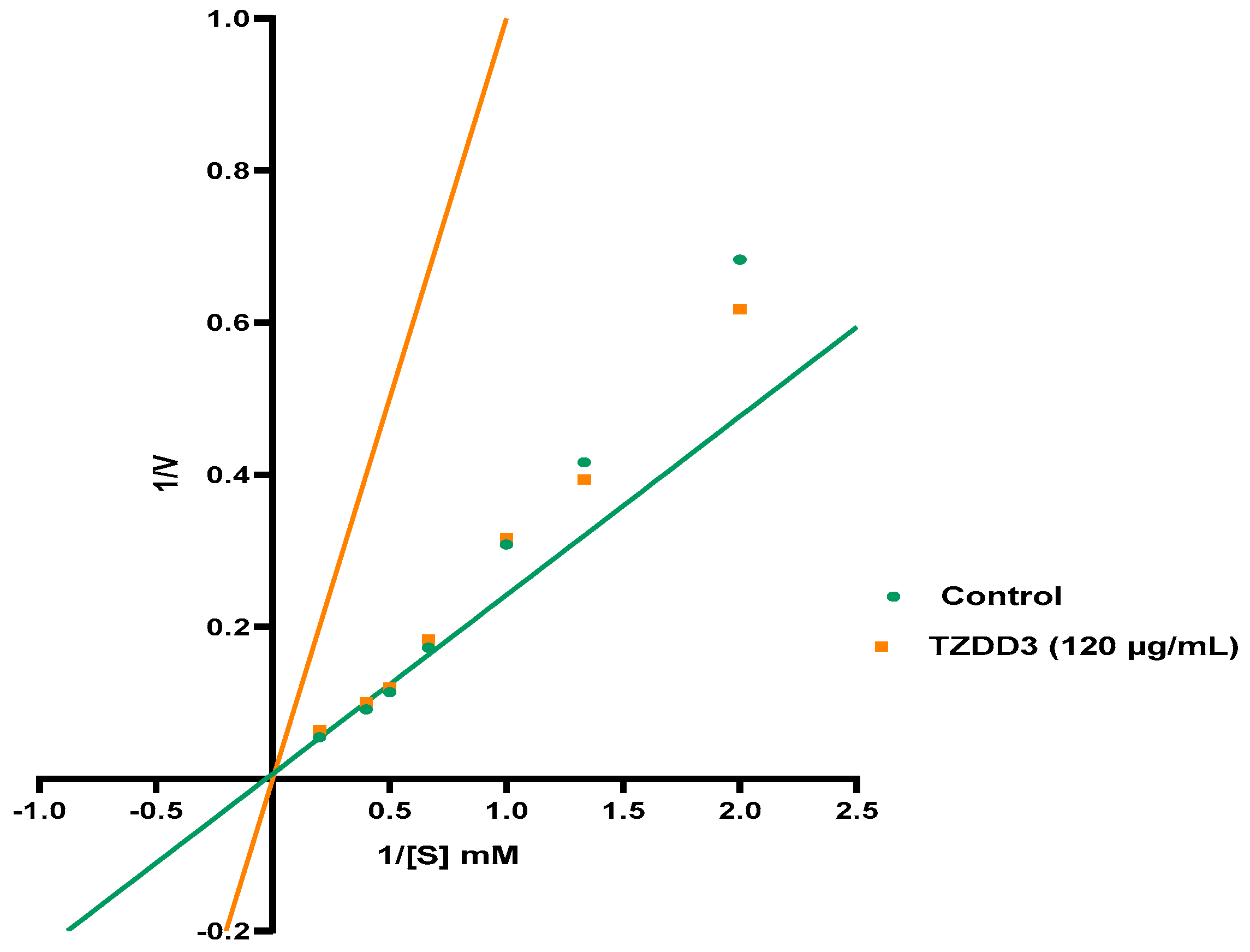
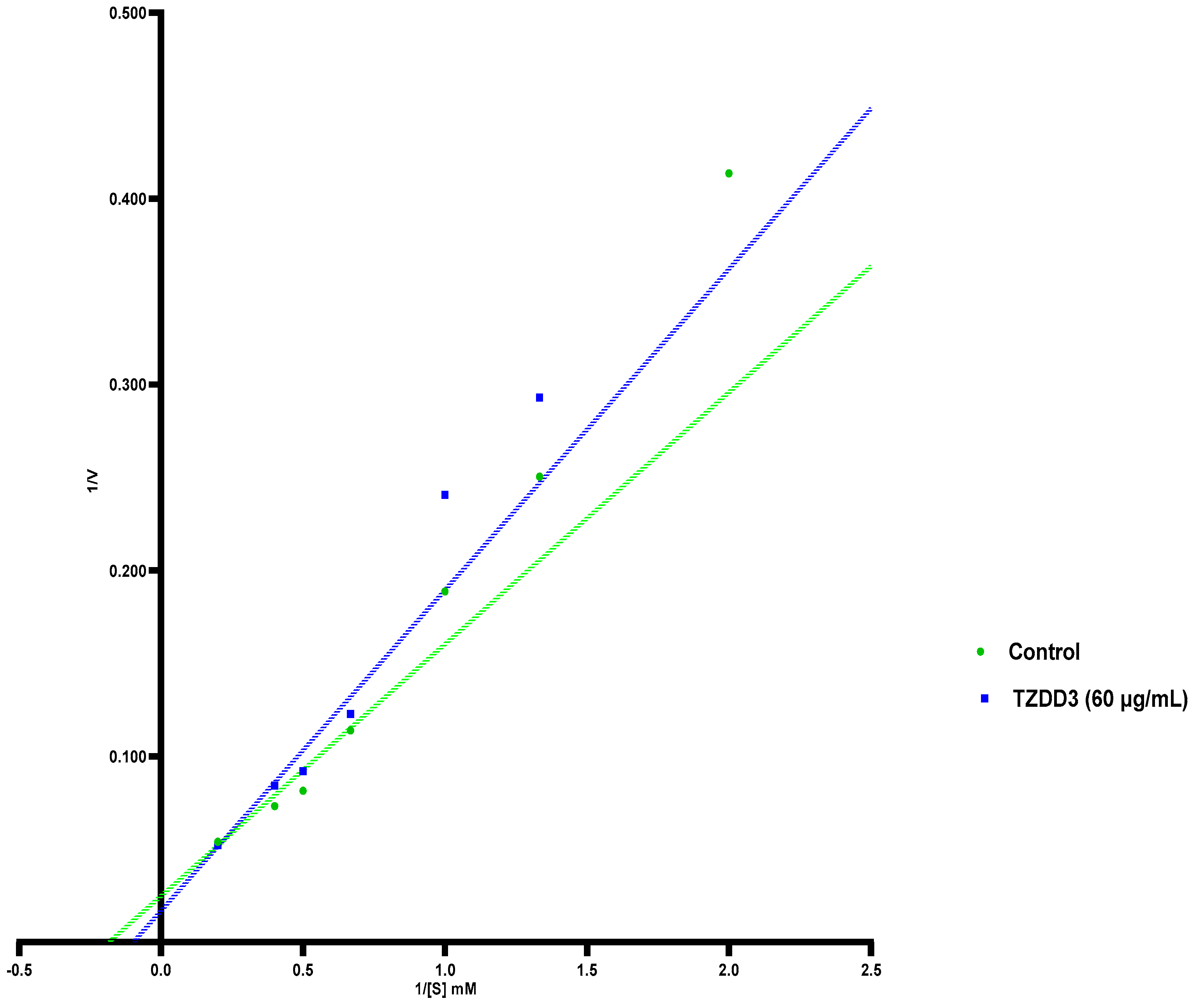
4.2.2. Determining Mode of Inhibition of Alpha Glucosidase
4.2.3. Aldose Reductase Inhibition Assay
4.2.4. Protein Tyrosine Phosphatase Inhibition Assay
4.2.5. Dipeptidyl Peptidase-4 Inhibition Assay
4.2.6. Glucose Uptake and Cell Viability
4.2.7. Peroxisome Proliferator–Activated Receptor-γ Docking In Silico
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aynalem, S.B.; Zeleke, A.J. Prevalence of Diabetes Mellitus and Its Risk Factors among Individuals Aged 15 Years and Above in Mizan-Aman Town, Southwest Ethiopia, 2016: A Cross Sectional Study. Int. J. Endocrinol. 2018, 2018, 9317987. [Google Scholar] [CrossRef]
- Pheiffer, C.; Pillay-van Wyk, V.; Joubert, J.D.; Levitt, N.; Nglazi, M.D.; Bradshaw, D. The prevalence of type 2 diabetes in South Africa: A systematic review protocol. BMJ Open 2019, 8, e021029. [Google Scholar] [CrossRef]
- Bradshaw, D.; Norman, R.; Pieterse, D.; Levitt, N.S.; The South African Comparative Risk Assessment Group. Estimating the burden of disease attributable to diabetes South Africa in 2000. S. Afr. Med. J. 2007, 96, 700–705. [Google Scholar] [CrossRef]
- Erasmus, R.; Soita, D.; Hassan, M.; Blanco-Blanco, E.; Vergotine, Z.; Kengne, A.; Matsha, T. High prevalence of diabetes mellitus and metabolic syndrome in a South African coloured population: Baseline data of a study in Bellville, Cape Town. S. Afr. Med. J. 2012, 102, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Govender, R.D.; Gathiram, P.; Panajatovic, M. Poor control and management of type 2 diabetes mellitus at an under-resourced South African Hospital: Is it a case of clinical inertia? S. Afr. Fam. Pract. 2017, 59, 154–159. [Google Scholar] [CrossRef]
- Erzse, A.; Stacey, N.; Chola, L.; Tugendhaft, A.; Freeman, M.; Hofman, K. The direct medical cost of type 2 diabetes mellitus in South Africa: A cost of illness study. Glob. Health Action 2019, 12, 1636611. [Google Scholar] [CrossRef] [PubMed]
- Mohammed Iqbal, A.K.; Khan, A.Y.; Kalashetti, M.B.; Belavagi, N.S.; Gong, Y.; Khazi, I.A.M. Synthesis, hypoglycaemic and hypolipidemic activities of novel thiazolidinedione derivatives containing thiazole/triazole/oxadiazole ring. Eur. J. Med. Chem. 2012, 53, 308–315. [Google Scholar] [CrossRef]
- Bhattarai, B.R.; Kafle, B.; Hwang, J.; Ham, S.W.; Lee, K.; Park, H.; Han, I.; Cho, H. Novel thiazolidinedione derivatives with anti-obesity effects: Dual action as PTP1B inhibitors and PPAR-c activators. Bioorg. Med. Chem. Lett. 2010, 20, 6758–6763. [Google Scholar] [CrossRef]
- Maccari, R.; Ciurleo, R.; Giglio, M.; Cappiello, M.; Moschini, R.; Del Corso, A.; Mura, U.; Ottanà, R. Identification of new non-carboxylic acid containing inhibitors of aldose reductase. Bioorg. Med. Chem. 2010, 18, 4049–4055. [Google Scholar] [CrossRef]
- Hidalgo Figueroa, S.; Estrada-Soto, S.; Ramírez-Espinosa, J.; Paoli, P.; Lori, G.; Rivera, I.; Navarrete-Vázquez, G. Synthesis and evaluation of thiazolidine-2,4-dione/benzazole derivatives as inhibitors of protein tyrosine phosphatase 1B (PTP-1B): Antihyperglycemic activity with molecular docking study. Biomed. Pharmacother. 2018, 107, 1302–1310. [Google Scholar] [CrossRef]
- Sharma, R.K.; Younis, Y.; Mugumbate, G.; Njoroge, M.; Gut, J.; Rosenthal, P.J.; Chibale, K. Synthesis and structure-activity-relationship studies of thiazolidinediones as antiplasmodial inhibitors of the Plasmodium falciparum cysteine protease falcipain-2. Eur. J. Med. Chem. 2015, 27, 507–518. [Google Scholar] [CrossRef]
- Salamone, S.; Colin, C.; Grillier-Vuissoz, I.; Kuntz, S.; Mazerbourg, S.; Flament, S.; Martin, H.; Richert, L.; Chapleur, Y.; Boisbrun, M. Synthesis of new troglitazone derivatives: Anti-proliferative activity in breast cancer cell lines and preliminary toxicological study. Eur. J. Med. Chem. 2012, 51, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Stocks, P.A.; Raynes, K.J.; Bray, P.G.; Park, B.K.; O’Neill, P.M.; Ward, S.A. Novel Short Chain Chloroquine Analogues Retain Activity Against Chloroquine Resistant K1 Plasmodium falciparum. J. Med. Chem. 2002, 45, 4975–4983. [Google Scholar] [CrossRef]
- Matsuda, H.; Morikawa, T.; Yoshikawa, M. Antidiabetogenic constituents from several natural medicines. Pure Appl. Chem. 2002, 74, 1301–1308. [Google Scholar] [CrossRef]
- Shai, L.J.; Masoko, P.; Mokgotho, M.P.; Magano, S.R.; Mogale, A.M.; Boaduo, N.; Eloff, J.N. Yeast alpha glucosidase inhibitory and antioxidant activities of six medicinal plants collected in Phalaborwa, South Africa. SAJB 2010, 76, 465–470. [Google Scholar] [CrossRef]
- Amtul, Z.; Rahman, A.; Siddiqui, R.; Choudhary, M.I. Chemistry and Mechanism of Urease Inhibition. Curr. Med. Chem. 2002, 9, 1323–1348. [Google Scholar] [CrossRef]
- Park, H.; Hwang, K.Y.; Kim, Y.H.; Oh, K.H.; Lee, J.Y.; Kim, K. Discovery and biological evaluation of novel α-glucosidase inhibitors with in vivo antidiabetic effect. Bioorg. Med. Chem. Lett. 2008, 18, 3711–3715. [Google Scholar] [CrossRef]
- Li, N.; Wang, F.; Niu, S.; Cao, J.; Wu, K.; Li, Y.; Yin, Y. Discovery of novel inhibitors of Streptococcus pneumoniae based on the virtual screening with the homology-modeled structure of histidine kinase (VicK). BMC Microbiol. 2009, 9, 1–11. [Google Scholar] [CrossRef]
- Chen, J.; Wu, S.; Zhang, Q.; Yin, Z.; Zhang, L. α-Glucosidase inhibitory effect of anthocyanins from Cinnamomum camphora fruit: Inhibition kinetics and mechanistic insights through in vitro and in silico studies. Int. J. Biol. Macromol. 2002, 143, 696–703. [Google Scholar] [CrossRef]
- Arulmozhiraja, S.; Matsuo, N.; Ishitsubo, E.; Okazaki, S.; Shimano, H.; Tokiwa, H. Comparative Binding Analysis of Dipeptidyl Peptidase IV (DPP-4) with Antidiabetic Drugs—An Ab Initio Fragment Molecular Orbital Study. PLoS ONE 2016, 11, e0166275. [Google Scholar] [CrossRef]
- Lalloyer, F.; Staels, B. Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vascul. Biol. 2010, 30, 894–899. [Google Scholar] [CrossRef]
- Kroker, A.J.; Bruning, J.B. Review of the Structural and Dynamic Mechanisms of PPAR-γ Partial Agonism. PPAR Res. 2015, 2015, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Long, N.; Le Gresley, A.; Wren, S.P. Thiazolidinediones: An In–Depth Study of Their Synthesis and Application to Medicinal Chemistry in the Treatment of Diabetes Mellitus. ChemMedChem 2021, 16, 1717–1736. [Google Scholar] [CrossRef]
- Oyedemi, S.O.; Oyedemi, B.O.; Arowosegbe, S.; Afolayan, A.J. Phyto-chemicals analysis and medicinal potentials of hydroalcoholic extract from Curtisia dentata (Burm. F) Ca Sm stem bark. Int. J. Mol. Sci. 2012, 13, 6189–6203. [Google Scholar] [CrossRef]
- Akter, R.; Afrose, A.; Rahman, M.R.; Chowdhury, R.; Nirzhor, S.; Khan, R.I.; Kabir, M.T. A Comprehensive Analysis into the Therapeutic Application of Natural Products as SIRT6 Modulators in Alzheimer’s Disease, Aging, Cancer, Inflammation, and Diabetes. Int. J. Mol. Sci. 2021, 22, 4180. [Google Scholar] [CrossRef]
- Veeresham, C.; Rao, A.; Asres, K. Aldose Reductase Inhibitors of Plant Origin. Phytother. Res. 2014, 28, 317–333. [Google Scholar] [CrossRef]
- Williamson, A.E.; Ylioja, P.M.; Robertson, M.N.; Antonova-Koch, Y.; Avery, V.; Baell, J.B.; Batchu, H.; Batra, S.; Burrows, J.N.; Bhattacharyya, S.; et al. Open source drug discovery: Highly potent antimalarial compounds derived from the Tres Cantos arylpyrroles. ACS Cent. Sci. 2016, 2, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Worthington Biochemical Corp. Maltase-a-glucosidase. In Worthington Enzyme Manual; Freehold, 261; Worthington, V., Ed.; Worthington Biochemical Corporation: Lakewook, NJ, USA, 1993. [Google Scholar]
- Balan, K.; Ratha, P.; Prakash, G.; Viswanathamurthi, P.; Adisakwattana, S.; Palvannan, T. Evaluation of invitro α-amylase and α-glucosidase inhibitory potential of N2O2 schiff base Zn complex. Arab. J. Chem. 2017, 10, 732–738. [Google Scholar] [CrossRef]
- Bowden, A.C. A simple method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 1974, 137, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Fatmawati, S.; Ersam, T.; Shimizu, K. The inhibitory activity of aldose reductase in vitro by constituents of Garcinia mangostana Linn. Phytomedicine 2015, 22, 49–51. [Google Scholar] [CrossRef]
- Kazeem, M.I.; Adeyemi1, A.A.; Adenowo, A.F.; Akinsanya, M.A. Carica papaya Linn. Fruit extract inhibited the activities of aldose reductase and sorbitol dehydrogenase: Possible mechanism for amelioration of diabetic complications. Future J. Pharmaceut. Sci. 2020, 6, 96. [Google Scholar] [CrossRef]
- Ang, L.; Cao, J.; Duan, L.; Tang, Y.; Zhao, Y. Protein tyrosine phosphatase 1B (PTP1B) and a-glucosidase inhibitory activities of Schisandra chinensis (Turcz.) Baill. J. Function. Foods 2014, 9, 264–270. [Google Scholar]
- Song, Y.H.; Uddin, Z.; Jina, Y.M.; Lia, Z.; Curtis-Longb, M.J.; Kimc, K.D.; Choa, J.K.; Parka, K.H. Inhibition of protein tyrosine phosphatase (PTP1B) and a-glucosidase by geranylated flavonoids from Paulownia tomentosa. J. Enzy. Inhib. Med. Chem. 2017, 32, 1195–1202. [Google Scholar] [CrossRef]
- Cruz-Bermúdez, A.; Laza-Briviesca, R.; Vicente-Blanco, R.J.; García-Grande, A.; Coronado, M.J.; Laine-Menéndez, S.; Palacios-Zambrano, S.; Moreno-Villa, M.R.; Ruiz-Valdepeñas, A.M.; Lendinez, C.; et al. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free Radic. Biol. Med. 2019, 135, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Henneberry, M.O.; Engel, G.; Grayhack, J.T. Acid phosphatase. Urol. Clin. N. A 1979, 6, 629–641. [Google Scholar] [CrossRef]
- Liberato, M.V.; Nascimento, A.S.; Polikarpov, I. Human peroxisome proliferator-activated receptor gamma in complex with rosiglitazone. PLoS ONE 2012, 7, e36297. [Google Scholar]
- Dasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. Protein-Ligand Interaction Profiler (PLIP) 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucl. Acids Res. 2021, 49, W530–W534. [Google Scholar]




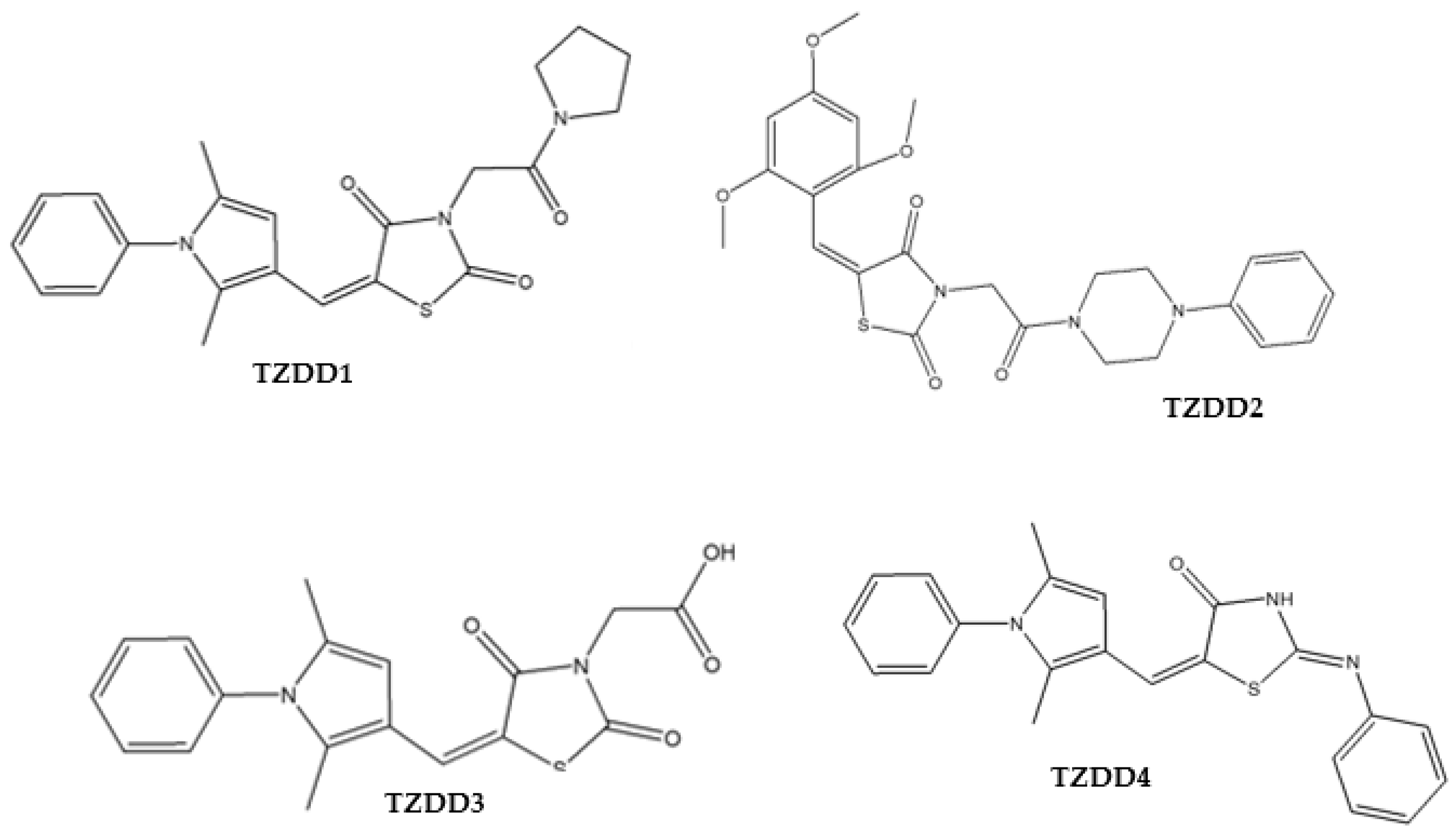
| IC50 for the Inhibitory Activity Assays (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Compounds Name | Structure | α-Amylase | α-Glucosidase | AR | PTP-1B | DPP-4 |
| TZDD1 | 24.85 ± 2.31 | 158.15 ± 11.59 | 27.54 ± 5.34 | unstable | 73.04 ± 5.59 | |
| TZDD2 | 18.24 ± 0.10 | 174.39 ± 4.35 | 82.27 ± 7.96 | 136.80 ± 4.06 | 31.93 ± 3.26 | |
| TZDD3 | 27.97 ± 0.76 | 56.82 ± 0.40 | 57.43 ± 7.80 | unstable | 75.82 ± 4.37 | |
| TZDD4 | 24.06 ± 2.28 | 698.92 ± 2.47 | 36.77 ± 5.10 | 258.78 ± 1.49 | 71.83 ± 2.34 | |
| Acarbose | 45.00 ± 6.90 | 44.84 ± 7.46 | n/a | n/a | n/a | |
| Quercetin | n/a | n/a | 36.71 ± 11.15 | n/a | n/a | |
| Na3VO4 | n/a | n/a | n/a | Unstable | n/a | |
| Sitagliptin | n/a | n/a | n/a | n/a | 3.31 ± 0.45 | |
| Control | TZDD3 (60 µg/mL) | TZDD3 (120 µg/mL) | |
|---|---|---|---|
| Vmax | 40.28 | 59.43 | 32.76 |
| Km | 5.457 | 10.26 | 4.556 |
| Compound | LBE (kcal/mol) | EIC, ki (nM) | RMSD Values |
|---|---|---|---|
| TZDD1 | −12.02 | 1.55 | 21.67A |
| TZDD2 | −10.40 | 23.77 | 25.519A |
| TZDD3 | −8.84 | 332.32 | 23.664A |
| TZDD4 | −9.91 | 54.39 | 24.199A |
| Rosiglitazone | −8.63 | 472.78 | 26.194A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arineitwe, C.; Oderinlo, O.; Tukulula, M.; Khanye, S.; Khathi, A.; Sibiya, N. Discovery of Novel Thiazolidinedione-Derivatives with Multi-Modal Antidiabetic Activities In Vitro and In Silico. Int. J. Mol. Sci. 2023, 24, 3024. https://doi.org/10.3390/ijms24033024
Arineitwe C, Oderinlo O, Tukulula M, Khanye S, Khathi A, Sibiya N. Discovery of Novel Thiazolidinedione-Derivatives with Multi-Modal Antidiabetic Activities In Vitro and In Silico. International Journal of Molecular Sciences. 2023; 24(3):3024. https://doi.org/10.3390/ijms24033024
Chicago/Turabian StyleArineitwe, Charles, Ogunyemi Oderinlo, Matshawandile Tukulula, Setshaba Khanye, Andile Khathi, and Ntethelelo Sibiya. 2023. "Discovery of Novel Thiazolidinedione-Derivatives with Multi-Modal Antidiabetic Activities In Vitro and In Silico" International Journal of Molecular Sciences 24, no. 3: 3024. https://doi.org/10.3390/ijms24033024
APA StyleArineitwe, C., Oderinlo, O., Tukulula, M., Khanye, S., Khathi, A., & Sibiya, N. (2023). Discovery of Novel Thiazolidinedione-Derivatives with Multi-Modal Antidiabetic Activities In Vitro and In Silico. International Journal of Molecular Sciences, 24(3), 3024. https://doi.org/10.3390/ijms24033024






