Abstract
Taiwan has the highest incidence rate of oral cancer in the world. Although oral cancer is mostly an environmentally induced cancer, genetic factors also play an important role in its etiology. Genome-wide association studies (GWAS) have identified nine susceptibility regions for oral cancers in populations of European descent. In this study, we performed the first GWAS of oral cancer in Taiwan with 1529 cases and 44,572 controls. We confirmed two previously reported loci on the 6p21.33 (HLA-B) and 6p21.32 (HLA-DQ gene cluster) loci, highlighting the importance of the human leukocyte antigen and, hence, the immunologic mechanisms in oral carcinogenesis. The TERT-CLMPT1L locus on 5p15.33, the 4q23 ADH1B locus, and the LAMC3 locus on 9q34.12 were also consistent in the Taiwanese. We found two new independent loci on 6p21.32, rs401775 in SKIV2L gene and rs9267798 in TNXB gene. We also found two suggestive novel Taiwanese-specific loci near the TPRS1 gene on 8q23.3 and in the TMED3 gene on 15q25.1. This study identified both common and unique oral cancer susceptibility loci in the Taiwanese as compared to populations of European descent and shed significant light on the etiology of oral cancer in Taiwan.
1. Introduction
Oral cancer is the eighth most common cancer in men worldwide with an estimated 264,211 new cases in men in 2020 [1]. The age-standardized incidence rate was 2.6-fold higher in men (6.0 per 100,000) than in women (2.3 per 100,000) worldwide, although the rate varies widely across regions, with a 20-fold difference in men and 10-fold difference in women [1]. Taiwan has the highest incidence rate of oral cancer in men (27.01 per 100,000) in the world and the male predominance is even more pronounced with nearly 90% of oral cancers occurring in men, accounting for the third highest incidence rate and fourth highest cause of cancer deaths among all cancers among Taiwanese men [2]. The high incidence rate and striking male predominance of oral cancer in Taiwan are attributed to the high prevalence of major risk factors, including smoking, alcohol drinking, and particularly, betel quid chewing [2,3,4,5]. Betel quid chewing confers an approximately 8-fold increased risk of oral cancer, much higher that the risks conferred by smoking (3.6-fold) and drinking (2.2-fold) [6]. More significantly, there is a strong synergistic effect of smoking, drinking, and chewing in promoting oral carcinogenesis, resulting in an over 40-fold increased oral cancer risk for the smoking–drinking–chewing persons [6,7,8].
Although oral cancer is mostly an environmentally induced cancer, genetic factors and gene–environment interactions also play an important role in its etiology. Epidemiological studies have observed an elevated risk of oral cancer in individuals with a family history of head and neck cancer [9,10,11]. Numerous candidate gene studies have been performed to assess the associations of selected single nucleotide polymorphisms (SNPs) in genes involved in essential biological pathways such as carcinogen metabolism, DNA repair, cell-cycle control, and inflammatory response with the risks of head and neck cancer overall and/or oral cancer specifically [12,13,14,15,16,17,18,19,20,21,22]. The only consistent associations from numerous candidate gene studies are those related to alcohol dehydrogenase (ADH) genes: SNPs in several ADH genes were associated with the risks of upper aerodigestive cancers including oral cancer and the effects became more apparent with increasing alcohol consumption, indicating a gene–environment interaction [20,23]. The genome-wide association study (GWAS) has revolutionized genetic association research and identified hundreds of thousands of novel susceptibility loci for hundreds of genetic traits, including approximately 800 susceptibility loci for over 20 different cancers [24,25,26]. There were only three published GWAS of head and neck cancer or upper aerodigestive cancer that included the subtype of oral cancer, all in European populations [23,27,28]. These GWAS identified at least nine genetic susceptibility regions for oral cancers, including 2p23.3 (GPN1), 4q21 (HEL308 and FAM175A), 4q23 (ADH1B, ADH1C, ADH7), 5p15.33 (CLPTM1L), 6p21 (HLA), 6p22.1 (ZNRD1-AS1), 9p21.3 (CDKN2A–CDKN2B), 9q34.12 (LAMC3), and 12q24 (ALDH2). These loci only explain a small portion of the genetic heritability of oral cancer. More susceptibility loci remain to be discovered. Furthermore, these loci were found in European descendants, who have much a lower incidence rate than Taiwan and were not exposed to betel quid. No GWAS of oral cancer has been conducted in Taiwan. Given the common (smoking and alcohol drinking) and distinct environmental exposures (betel quid chewing) in Taiwan, and the diversity of genetic structure across different ethnicities, we hypothesize that there are common and unique genetic susceptibility loci of oral cancer in the Taiwanese as compared with the European population. We therefore performed the first GWAS of oral cancer in Taiwan.
2. Results
2.1. Demographics of Study Population
After strict quality control procedures, data from 1529 cases and 44,572 controls were included in the final analysis. Table 1 shows the selected characteristics of the cases and controls. The mean ages (standard deviation) of the cases and controls were 55 (11.5) and 54 (14.5) years, respectively. About 86.9% (N = 1328) of the patients and 77.7% (N = 34,616) of the controls were men.

Table 1.
Selected characteristics of the study population.
2.2. Novel Variants Associated with Oral Cancer in the Taiwanese
A quantile–quantile plot of the observed versus expected χ2 test statistics did not show a large deviation from what was expected by chance (inflation factor λ = 1.023; Figure 1A). Fifteen variants on chromosome 6p21 were associated with oral cancer with genome-wide significance (p < 5 × 10−8) (Figure 1B, Table 2), twelve of which were highly linked and located in or near the HLA-B gene on 6p21.33 (Figure 2A). The other three, including one small insertion and two SNPs, were in the CFB, SKIV2L, and TNXB genes, respectively, on 6p21.32 (Figure 2B). The CFB and SIKV2L SNPs were in moderate linkage, and the TNXB SNP was not linked to the other two.
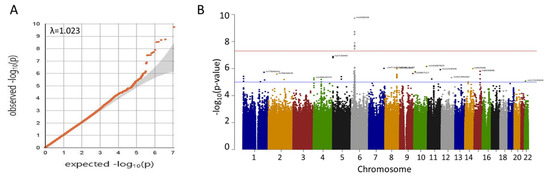
Figure 1.
(A) Quantile–quantile plot of observed p-values for associations. The data indicate only a small amount of population inflation (λ = 1.023). (B) Manhattan plot of genome-wide p-values for associations. Red line indicates p = 5 × 10−8. Only SNPs in 6p21 reach genome-wide significance.

Table 2.
Significant variants (p < 5 × 10−8) associated with oral cancer in the Taiwanese.
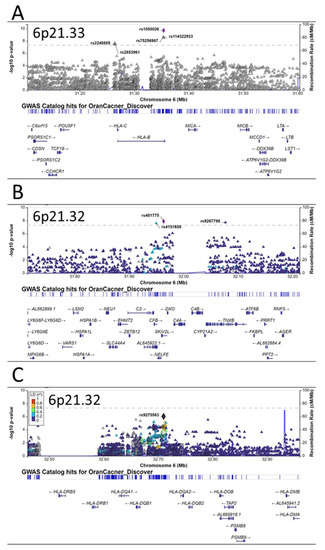
Figure 2.
Regional plots of associations in 6p21. Three independent susceptibility loci for oral cancer are observed in 6p21 region (A–C). The y-axis presents the −log10 p-values of the SNPs and the x-axis presents the corresponding chromosomal position of each SNP. The genomic locations of genes within the regions were annotated from the UCSC Genome Browser.
Besides these three independent loci on 6p21, there were several other promising loci that were close to having genome-wide significance, including another independent locus on 6p21.33 (HLA-DQ gene cluster, Figure 2C), the TERT-CLMPT1L locus on 5p15.33 (Figure 3A), a locus near the TRPS1 gene on 8q23.3 (Figure 3B), and the TMED3 gene on 15q25.1 (Figure 3C).
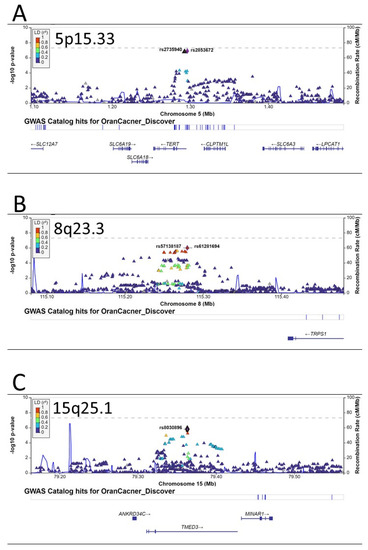
Figure 3.
Regional plots of associations for regions of 5p15.33 (A), 8q23.3 (B), and 15q25.1 (C). The y-axis presents the −log10 p-values of the SNPs and the x-axis presents the corresponding chromosomal position of each SNP. The genomic locations of genes within the regions were annotated from the UCSC Genome Browser.
2.3. Common Variants Associated with Oral Cancer Validated among Various Populations
Previous GWAS in populations of European descent identified nine susceptibility loci for oral cancer. We queried these loci in our population. The consistent loci included rs1229984 in the ADH1B gene on 4q23, rs928674 in the LAMC3 gene on 9q34.12, and several loci on 6p21.32 and 6p21.33 (Table 3). For those loci that were close to genome-wide significance in previous GWAS, the TERT-CLMPT1L locus on 5p15.33 was highly consistent and the lead SNP was rs7726159 in the TERT gene (OR = 1.19, p = 2.18 × 10−7 in previous GWAS and OR = 1.16, p = 6.40 × 10−5 in our population) (Table 4).

Table 3.
Replication of previous GWAS-identified oral cancer susceptibility loci in Taiwan population.

Table 4.
Borderline significant SNPs (5 × 10−8 < p < 1 × 10−7) in Europeans and their associations with oral cancer in Taiwanese.
3. Discussion
In this study, we performed the first GWAS of oral cancer in Taiwan. We confirmed two previously reported 6p21.33 and 6p21.32 HLA loci, and found two additional independent 6p21.33 loci for oral cancer in the Taiwanese. We also confirmed the previously reported oral cancer susceptibility loci at the TERT-CLMPT1L locus on 5p15.33, thet 4q23 ADH1B locus, and the LAMC3 locus on 9q34.12 in Taiwanese populations. We found suggestive novel susceptibility loci near the TPRS1 gene on 8q23.3 and in the TMED3 gene on 15q25.1.
The most notable finding of this study is the multiple independent oral cancer susceptibility loci spanning the 6p21.3 regions, which contain the human leukocyte antigen (HLA) gene clusters. The HLA system plays essential roles in innate and adaptive immune responses [29]. The HLA system consists of three regions: the class I region encodes HLA-A, -B, and -C; the class II region encodes HLA-DR, -DQ, and –DP; and the class III region genes code for proteins of the complement system and the TNF family members. The functions of class I and class II molecules are to bind intracellular and extracellular peptide antigens and present them to antigen-specific T lymphocytes. Peptide antigens associated with HLA class I molecules are recognized by CD8+ T cells and those associated with HLA class II molecules are recognized by CD4+ T cells [30]. The 6p21.3 is the most gene-dense region in the human genome and the HLA genes are the most polymorphic in the human genome [31]. Previous GWAS have reported genetic variants in various HLA genes as susceptibility loci for many human diseases, including a few cancers such as lung cancer [32], liver cancer [33], cervical cancer [34], colorectal cancer [35], leukemia [36], lymphoma [37,38], and different subtypes of head and neck cancers [23,27,28,39]. Specifically, for oral cancer, previous GWAS identified three loci in the HLA region, including rs3828805 (chromosome 6 position 32636120) [27] and rs3135001 (6:32670136) in HLA-DQB1 (6p21.32) [28] and rs1265081 (6:31111675) in CCHCR1 (6p21.33) [28]. Recently, Ji et al. found that two independent SNPs in the 6p21.33 regions were associated with altered oral cancer risks in a Chinese population: rs2524182 (6:31130593) in TRIM39-RPP21-HLA-E and rs3131018 (6:31143582) in PSORS1C3-TCF19 [40]. In our current study, we found many SNPs in 6p21 that reached genome-wide or borderline genome-wide significance in their associations with oral cancer in Taiwan, covering multiple HLA genes (Table 2, Figure 2), further supporting the important roles of the HLA system in oral cancer etiology.
For other GWAS-identified oral cancer susceptibility loci in European populations, we were able to replicate the loci on 5p15.33, the 4q23 ADH1B locus, and the LAMC3 locus on 9q34.12 (Table 3). The 5p15.33 region encompassing the TERT-CLPTM1L genes has been associated with the risks of at least 11 different cancers, including lung, prostate, breast, pancreatic, bladder, esophageal, endometrial, gastric, and head and neck cancers, glioma, and melanoma [28,32,41,42,43,44,45,46]. Multiple mechanisms, including telomere structure, epigenetic modification, transcriptional regulation, and apoptosis, have been suggested to explain the associations of the TERT-CLPTM1L locus and cancer susceptibility [47,48]. Chromosome 4q23 contains a cluster of alcohol dehydrogenase (ADH) genes and several SNPs in different ADH genes have been identified as susceptibility loci for oral cancer in European populations (Table 3) and other populations [49]. The best-studied SNP is rs1229984, a missense SNP at codon 47 (Arg47His) of the ADH1B. The A allele (coding for His) is about 40 times more active in metabolizing alcohol and is associated with a reduced risk of oral cancer. The frequency of the A (His) allele is only ~5% in European populations but reaches ~80% in East Asians. We used the predominant A (His) allele as the reference group, and those with the less active G (Arg) allele had 20% (OR = 1.20, 95% CI, 1.06–1.35, p = 0.003), consistent with literature reports in Europeans and Asians [49].
We also found potential novel susceptibility loci at 8q23.3 near the TRPS1 gene. TRPS1 is an atypical member of the GATA transcriptional factor family, exhibiting transcriptional repression by interacting with corepressors [50,51,52]. Recent studies have suggested that TRPS1 is overexpressed in several cancers and can act as an oncogenic driver through various mechanisms [53], such as driving heterochromatic origin refiring and genome amplifications [54], controlling the cell-cycle progression [55], promoting epithelial-to-mesenchymal transition [56], promoting angiogenesis [57], and causing epigenetic alterations (DNA methylation and histone acetylation) [50,58,59]. There has been no report of TRPS1 in oral cancer. Future studies are warranted to investigate the role of TRPS1 in oral carcinogenesis. Another novel oral cancer susceptibility locus is in the TMED3 gene on 15q25.1. TMED3 is a transmembrane protein and plays an important function in vesicular transport and innate immunity [60]. Several recent studies have shown an increased expression of TMED3 in a number of cancers and TMED3 promotes the carcinogenesis of liver, breast, colorectal, lung, and endometrial cancer as well as glioma and osteosarcoma [61,62,63,64,65,66,67]. Biologically, TMED3 can activate IL-11/STAT3 and Wnt/beta-catenin signaling pathways [61,62]. The role of TMED3 in oral carcinogenesis remains to be investigated.
Whether these oral susceptibility loci in the Taiwanese are consistent in other Asian populations are largely unknown. A recent study in China [40] used Human Exome BeadChip (~240K mostly nonsynonymous coding variants), but the final analyzable SNPs were only ~63K because most of the SNPs were monomorphic. They found two independent oral cancer susceptibility loci in the 6p21.33 regions: rs2524182 in TRIM39-RPP21-HLA-E and rs3131018 168 in PSORS1C3-TCF19 [40], consistent with our data. Other regions remain to be investigated. The data in India were more limited. Only one early pilot GWAS using human CNV370k BeadChip in only 55 cases and 92 controls was published [68]. Due to its small sample size, none of the reported European and Taiwanese oral cancer susceptibility loci were among the top hits in that study. The genetic susceptibility loci to oral cancer in India warrant further investigation.
The findings from our study not only shed significant insight into the biology of oral cancer etiology in Taiwan, but also have an important clinical and public health impact. The identified susceptibility genes may become potential preventive and/or therapeutic targets. For example, strategies that improve immune response, telomere maintenance, and alcohol metabolism may prevent oral cancer development given the importance of relevant genes in these pathways in oral cancer susceptibility. Another potential translational application is to use multiple genetic susceptibility loci to develop a polygenic risk score (PRS) for each person. This PRS can be integrated with environmental exposures such as smoking, drinking, and betel nut chewing to identify individuals at the highest risk of developing oral cancer, who then would be subjected to targeted cancer prevention and screening.
This is the first GWAS of oral cancer in Taiwan. We had the largest oral cancer cases and controls in any association studies of oral cancer to date. We found both common and unique oral cancer susceptibility loci in the Taiwanese as compared to European populations. Our large sample size enabled the unequivocal confirmation of HLA regions as the most prominent oral cancer susceptibility loci in Taiwan. We also confirmed the susceptibility loci at 5p15.33, the 4q23 ADH1B locus, and the LAMC3 locus on 9q34.12. We found novel suggestive susceptibility loci near the TPRS1 gene on 8q23.3 and in the TMED3 gene on 15q25.1.
4. Materials and Methods
4.1. Study Population and Data Collection
The study participants were part of the China Medical University Hospital (CMUH) Precision Medicine Project, a systemic effort initiated in 2018 to recruit subjects and collect biospecimens from all patients who come to CMUH for medical visits [69,70]. More than 170,000 subjects have been enrolled to date. The recruitment and sample collection procedures were approved by the ethical committees of CMUH (CMUH107-REC3-058 and CMUH110-REC3-005). Each participant signed an informed consent form and provided blood samples. Clinical information was abstracted from the electronic medical records (EMRs) of the CMUH. A total of 1529 oral cancer patients (ICD-10-CM Diagnosis Code C00 to C06) and 44,572 controls (without a history of any cancer) were included in the study.
4.2. Genotyping and Imputation
The whole genome SNP genotyping using the Affymetrix genome-wide human SNP array 6.0 chip was performed according to the manufacturer’s protocol [68]. We excluded samples and SNPs with genotyping call rates of <90%. We filtered out SNPs with a Hardy–Weinberg equilibrium p-value of <10−6, and a minor allele frequency (MAF) < 10−4. We excluded SNPs on sex chromosomes. Genotype imputation was performed as we recently described [69]. Briefly, we first constructed a population-specific reference panel by using whole genome sequencing data (1463 individuals) from the Taiwan Biobank (TWB). We used four algorithms (IMPUTE2, IMPUTE4, IMPUTE5, and Beagle5.2) and two reference panels (TWB and East Asian participants of the 1000 Genomes Project) to perform genotype imputation and found Beagle5.2 exhibited the fastest calculation speed, smallest storage space, highest specificity, and highest number of high-quality variants (15,277,414). The Beagle5.2 imputations were performed using its default parameters, except for the effective population size (20,000) and the buffer region (500,000 bases). The accuracy of the imputation result was measured using BCFtools gtcheck [71] to assess the concordance rate between the imputed genotypes and the WGS data. The imputation accuracy was 98.75% by Beagle5.2.
4.3. Statistical Analysis
For the participants’ characteristics, continuous data were presented as the means with standard deviation, and categorical data were presented as proportions. We used t-tests to compare the mean values of continuous variables and chi-squared tests to compare the frequencies of categorical variables between the cases and controls. The association of each SNP with the risk of oral cancer was analyzed using an additive model in the logistic regression analysis with PLINK V.1.90 [72]. To control for population structure, we performed principal component analysis (PCA) in EIGENSTRAT and adjusted significant principal components (PC) associated with the cancer status in unconditional logistic regression analysis, together with demographic variables including age and gender when estimating odds ratio (OR) and 95% confidence interval (CI). A genome-wide significance level was set at 5 × 10−8.
Author Contributions
Conceptualization, D.-T.B., C.-W.T. and W.-S.C.; data curation, D.-T.B., T.-Y.L. and J.G.; methodology, T.-Y.L., J.G. and W.-S.C.; statistics, C.-W.T., L.-C.S. and J.-S.Y.; project administration, D.-T.B. and F.-J.T.; supervision, D.-T.B. and F.-J.T.; writing—original draft, D.-T.B. and J.G.; writing—review and editing, D.-T.B., J.G. and F.-J.T. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are grateful to every subject who donated their samples to this study. We appreciate the support from China Medical University Hospital (DMR-112-130). The funders had no role in the study design, data collection, statistical analysis, or decision to publish or preparation of the manuscript.
Institutional Review Board Statement
The recruitment and sample collection procedures were conducted according to the guidelines of the Declaration of Helsinki, and approved by the ethical committees of CMUH (CMUH107-REC3-058 and CMUH110-REC3-005).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Jhuang, J.-R.; Su, S.-Y.; Chiang, C.-J.; Yang, Y.-W.; Lin, L.-J.; Hsu, T.-H.; Lee, W.-C. Forecast of peak attainment and imminent decline after 2017 of oral cancer incidence in men in Taiwan. Sci. Rep. 2022, 12, 5726. [Google Scholar] [CrossRef]
- Chuang, S.-L.; Su, W.W.-Y.; Chen, S.L.-S.; Yen, A.M.-F.; Wang, C.-P.; Fann, J.C.-Y.; Chiu, S.Y.-H.; Lee, Y.-C.; Chiu, H.-M.; Chang, D.-C.; et al. Population-based screening program for reducing oral cancer mortality in 2,334,299 Taiwanese cigarette smokers and/or betel quid chewers. Cancer 2017, 123, 1597–1609. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Ho, P.-S.; Jou, Y.-Y.; Wu, C.-Y.; Wang, Y.-W.; Yang, Y.-H. Determining High Prevalence of Betel-Quid Chewing and Cigarette Smoking by Occupation Using the Taiwan National Health Interview Survey. Subst. Use Misuse 2020, 55, 1472–1482. [Google Scholar] [CrossRef]
- Wang, T.-H.; Hsia, S.-M.; Shih, Y.-H.; Shieh, T.-M. Association of Smoking, Alcohol Use, and Betel Quid Chewing with Epigenetic Aberrations in Cancers. Int. J. Mol. Sci. 2017, 18, 1210. [Google Scholar] [CrossRef]
- Petti, S.; Masood, M.; Scully, C. The Magnitude of Tobacco Smoking-Betel Quid Chewing-Alcohol Drinking Interaction Effect on Oral Cancer in South-East Asia. A Meta-Analysis of Observational Studies. PLoS ONE 2013, 8, e78999. [Google Scholar] [CrossRef]
- Lee, Y.A.; Li, S.; Chen, Y.; Li, Q.; Chen, C.; Hsu, W.; Lou, P.; Zhu, C.; Pan, J.; Shen, H.; et al. Tobacco smoking, alcohol drinking, betel quid chewing, and the risk of head and neck cancer in an East Asian population. Head Neck 2019, 41, 92–102. [Google Scholar] [CrossRef]
- Lin, W.-J.; Jiang, R.-S.; Wu, S.-H.; Chen, F.-J.; Liu, S.-A. Smoking, Alcohol, and Betel Quid and Oral Cancer: A Prospective Cohort Study. J. Oncol. 2011, 2011, 525976. [Google Scholar] [CrossRef]
- Toporcov, T.N.; Znaor, A.; Zhang, Z.-F.; Yu, G.-P.; Winn, D.M.; Wei, Q.; Vilensky, M.; Vaughan, T.; Thomson, P.; Talamini, R.; et al. Risk factors for head and neck cancer in young adults: A pooled analysis in the INHANCE consortium. Int. J. Epidemiol. 2015, 44, 169–185. [Google Scholar] [CrossRef]
- Garavello, W.; Foschi, R.; Talamini, R.; La Vecchia, C.; Rossi, M.; Maso, L.D.; Tavani, A.; Levi, F.; Barzan, L.; Ramazzotti, V.; et al. Family history and the risk of oral and pharyngeal cancer. Int. J. Cancer 2008, 122, 1827–1831. [Google Scholar] [CrossRef]
- Radoï, L.; Paget-Bailly, S.; Guida, F.; Cyr, D.; Menvielle, G.; Schmaus, A.; Carton, M.; Cenée, S.; Sanchez, M.; Guizard, A.-V.; et al. Family history of cancer, personal history of medical conditions and risk of oral cavity cancer in France: The ICARE study. BMC Cancer 2013, 13, 560. [Google Scholar] [CrossRef] [PubMed]
- Bau, D.-T.; Tsai, M.-H.; Huang, C.-Y.; Lee, C.-C.; Tseng, H.-C.; Lo, Y.-L.; Tsai, Y.; Tsai, F.-J. Relationship between polymorphisms of nucleotide excision repair genes and oral cancer risk in Taiwan: Evidence for modification of smoking habit. Chin. J. Physiol. 2007, 50, 294–300. [Google Scholar] [PubMed]
- Bau, D.T.; Tsai, M.H.; Lo, Y.L.; Hsu, C.M.; Tsai, Y.; Lee, C.C.; Tsai, F.J. Association of p53 and p21(CDKN1A/WAF1/CIP1) polymorphisms with oral cancer in Taiwan patients. Anticancer Res. 2007, 27, 1559–1564. [Google Scholar]
- Tsai, M.-H.; Tseng, H.-C.; Liu, C.-S.; Chang, C.-L.; Tsai, C.-W.; Tsou, Y.-A.; Wang, R.-F.; Lin, C.-C.; Wang, H.-C.; Chiu, C.-F.; et al. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 2009, 45, e90–e94. [Google Scholar] [CrossRef]
- Bau, D.-T.; Chang, C.-H.; Tsai, M.-H.; Chiu, C.-F.; Tsou, Y.-A.; Wang, R.-F.; Tsai, C.-W.; Tsai, R.-Y. Association between DNA repair gene ATM polymorphisms and oral cancer susceptibility. Laryngoscope 2010, 120, 2417–2422. [Google Scholar] [CrossRef]
- Tsai, M.-H.; Tsai, C.-W.; Tsou, Y.-A.; Hua, C.-H.; Hsu, C.-F.; Bau, D.-T. Significant association of cyclin D1 single nucleotide polymorphisms with oral cancer in taiwan. Anticancer Res. 2011, 31, 227–231. [Google Scholar]
- Tsai, C.-W.; Chang, W.-S.; Lin, K.-C.; Shih, L.-C.; Tsai, M.-H.; Hsiao, C.-L.; Yang, M.-D.; Lin, C.-C.; Bau, D.-T. Significant association of Interleukin-10 genotypes and oral cancer susceptibility in Taiwan. Anticancer Res. 2014, 34, 3731–3737. [Google Scholar]
- Tsai, C.-W.; Chang, W.-S.; Liu, J.-C.; Tsai, M.-H.; Lin, C.-C.; Bau, D.-T. Contribution of DNA double-strand break repair gene XRCC3 genotypes to oral cancer susceptibility in Taiwan. Anticancer Res. 2014, 34, 2951–2956. [Google Scholar]
- Hung, Y.-W.; Tsai, C.-W.; Wu, C.-N.; Shih, L.-C.; Chen, Y.-Y.; Liu, Y.-F.; Hung, H.-S.; Shen, M.-Y.; Chang, W.-S.; Bau, D.-T. The Contribution of Matrix Metalloproteinase-8 Promoter Polymorphism to Oral Cancer Susceptibility. Vivo 2017, 31, 585–590. [Google Scholar] [CrossRef]
- Hashibe, M.; McKay, J.D.; Curado, M.P.; Oliveira, J.C.; Koifman, S.; Koifman, R.; Zaridze, D.; Shangina, O.; Wünsch-Filho, V.; Eluf-Neto, J.; et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat. Genet. 2008, 40, 707–709. [Google Scholar] [CrossRef]
- Fan, J.; Liu, W.; Zhang, M.; Xing, C. A literature review and systematic meta-analysis on XRCC3 Thr241Met polymorphism associating with susceptibility of oral cancer. Oncol. Lett. 2019, 18, 3265–3273. [Google Scholar] [CrossRef]
- Cadoni, G.; Boccia, S.; Petrelli, L.; Di Giannantonio, P.; Arzani, D.; Giorgio, A.; De Feo, E.; Pandolfini, M.; Gallì, P.; Paludetti, G.; et al. A review of genetic epidemiology of head and neck cancer related to polymorphisms in metabolic genes, cell cycle control and alcohol metabolism. Acta Otorhinolaryngol. Ital. 2012, 32, 1–11. [Google Scholar]
- McKay, J.D.; Truong, T.; Gaborieau, V.; Chabrier, A.; Chuang, S.C.; Byrnes, G.; Zaridze, D.; Shangina, O.; Szeszenia-Dabrowska, N.; Lissowska, J.; et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011, 7, e1001333. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Park, S.L.; Cheng, I.; Haiman, C.A. Genome-Wide Association Studies of Cancer in Diverse Populations. Cancer Epidemiol. Biomark. Prev. 2018, 27, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Lesseur, C.; Diergaarde, B.; Olshan, A.F.; Wünsch-Filho, V.; Ness, A.R.; Liu, G.; Lacko, M.; Eluf-Neto, J.; Franceschi, S.; Lagiou, P.; et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat. Genet. 2016, 48, 1544–1550. [Google Scholar] [CrossRef]
- Shete, S.; Liu, H.; Wang, J.; Yu, R.; Sturgis, E.M.; Li, G.; Dahlstrom, K.R.; Liu, Z.; Amos, C.I.; Wei, Q. A Genome-Wide Association Study Identifies Two Novel Susceptible Regions for Squamous Cell Carcinoma of the Head and Neck. Cancer Res 2020, 80, 2451–2460. [Google Scholar] [CrossRef]
- Mosaad, Y.M. Clinical Role of Human Leukocyte Antigen in Health and Disease. Scand. J. Immunol. 2015, 82, 283–306. [Google Scholar] [CrossRef]
- Traherne, J.A. Human MHC architecture and evolution: Implications for disease association studies. Int. J. Immunogenet. 2008, 35, 179–192. [Google Scholar] [CrossRef]
- Robinson, J.; Soormally, A.R.; Hayhurst, J.D.; Marsh, S.G. The IPD-IMGT/HLA Database—New developments in reporting HLA variation. Hum. Immunol. 2016, 77, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Broderick, P.; Webb, E.; Wu, X.; Vijayakrishnan, J.; Matakidou, A.; Qureshi, M.; Dong, Q.; Gu, X.; Chen, W.V.; et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 2008, 40, 1407–1409. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.-K.; Sun, J.; Cao, G.; Liu, Y.; Lin, D.; Gao, Y.-Z.; Ren, W.-H.; Long, X.-D.; Zhang, H.; Ma, X.-P.; et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus–related hepatocellular carcinoma. Nat. Genet. 2013, 45, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.J.; Bodinier, B.; Kalliala, I.; Zuber, V.; Vuckovic, D.; Doulgeraki, T.; Whitaker, M.D.; Wielscher, M.; Cartwright, R.; Tsilidis, K.K.; et al. Genetic variation in cervical preinvasive and invasive disease: A genome-wide association study. Lancet Oncol. 2021, 22, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Kweon, S.-S.; Tanikawa, C.; Jia, W.-H.; Xiang, Y.-B.; Cai, Q.; Zeng, C.; Schmit, S.L.; Shin, A.; Matsuo, K.; et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated with Risk for Colorectal Cancer. Gastroenterology 2019, 156, 1455–1466. [Google Scholar] [CrossRef]
- Lin, W.-Y.; Fordham, S.E.; Hungate, E.; Sunter, N.J.; Elstob, C.; Xu, Y.; Park, C.; Quante, A.; Strauch, K.; Gieger, C.; et al. Genome-wide association study identifies susceptibility loci for acute myeloid leukemia. Nat. Commun. 2021, 12, 6233. [Google Scholar] [CrossRef]
- Lin, G.-W.; Xu, C.; Chen, K.; Huang, H.-Q.; Chen, J.; Song, B.; Chan, J.K.C.; Li, W.; Liu, W.; Shih, L.-Y.; et al. Genetic risk of extranodal natural killer T-cell lymphoma: A genome-wide association study in multiple populations. Lancet Oncol. 2016, 21, 306–316. [Google Scholar] [CrossRef]
- Sud, A.; Thomsen, H.; Law, P.J.; Försti, A.; Filho, M.I.D.S.; Holroyd, A.; Broderick, P.; Orlando, G.; Lenive, O.; Wright, L.; et al. Genome-wide association study of classical Hodgkin lymphoma identifies key regulators of disease susceptibility. Nat. Commun. 2017, 8, 1892. [Google Scholar] [CrossRef]
- Wei, Q.; Yu, D.; Liu, M.; Wang, M.; Zhao, M.; Liu, M.; Jia, W.; Ma, H.; Fang, J.; Xu, W.; et al. Genome-wide association study identifies three susceptibility loci for laryngeal squamous cell carcinoma in the Chinese population. Nat. Genet. 2014, 46, 1110–1114. [Google Scholar] [CrossRef]
- Ji, P.; Chang, J.; Wei, X.; Song, X.; Yuan, H.; Gong, L.; Li, Y.; Ding, D.; Zhang, E.; Yan, C.; et al. Genetic variants associated with expression of TCF19 contribute to the risk of head and neck cancer in Chinese population. J. Med. Genet. 2022, 59, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Rafnar, T.; Sulem, P.; Stacey, S.N.; Geller, F.; Gudmundsson, J.; Sigurdsson, A.; Jakobsdottir, M.; Helgadottir, H.; Thorlacius, S.; Aben, K.K.H.; et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009, 41, 221–227. [Google Scholar] [CrossRef]
- Shete, S.; Hosking, F.J.; Robertson, L.B.; Dobbins, S.E.; Sanson, M.; Malmer, B.; Simon, M.; Marie, Y.; Boisselier, B.; Delattre, J.-Y.; et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009, 41, 899–904. [Google Scholar] [CrossRef]
- Petersen, G.M.; Amundadottir, L.; Fuchs, C.S.; Kraft, P.; Stolzenberg-Solomon, R.Z.; Jacobs, K.B.; Arslan, A.A.; Bueno-De-Mesquita, H.B.; Gallinger, S.; Gross, M.; et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010, 42, 224–228. [Google Scholar] [CrossRef]
- Haiman, C.; Chen, G.K.; Vachon, C.M.; Canzian, F.; Dunning, A.M.; Millikan, R.C.; Wang, X.; Ademuyiwa, F.; Ahmed, S. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 2011, 43, 1210–1214. [Google Scholar] [CrossRef]
- Chen, H.; Majumdar, A.; Wang, L.; Kar, S.; Brown, K.M.; Feng, H.; Turman, C.; Dennis, J.; Easton, D.; Michailidou, K.; et al. Large-scale cross-cancer fine-mapping of the 5p15.33 region reveals multiple independent signals. Hum. Genet. Genom. Adv. 2021, 2, 100041. [Google Scholar] [CrossRef]
- Tian, J.; Wang, Y.; Dong, Y.; Chang, J.; Wu, Y.; Chang, S.; Che, G. Cumulative Evidence for Relationships Between Multiple Variants in the TERT and CLPTM1L Region and Risk of Cancer and Non-Cancer Disease. Front. Oncol. 2022, 12, 946039. [Google Scholar] [CrossRef]
- Scherf, D.B.; Sarkisyan, N.; Jacobsson, H.; Claus, R.; Bermejo, J.L.; Peil, B.; Gu, L.; Muley, T.; Meister, M.; Dienemann, H.; et al. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene 2013, 32, 3329–3338. [Google Scholar] [CrossRef]
- Helbig, S.; Wockner, L.; Bouendeu, A.; Hille-Betz, U.; McCue, K.; French, J.D.; Edwards, S.L.; Pickett, H.A.; Reddel, R.R.; Chenevix-Trench, G.; et al. Functional dissection of breast cancer risk-associated TERT promoter variants. Oncotarget 2017, 8, 67203–67217. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, N.; Miao, L.; Yuan, H.; Wang, R.; Jiang, H. Alcohol dehydrogenase-1B Arg47His polymorphism is associated with head and neck cancer risk in Asian: A meta-analysis. Tumor Biol. 2015, 36, 1023–1027. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, L.; Wang, W.; Zhang, L.; Li, Z.; Zhao, Q.; Xing, K.; Feng, Z.; Peng, X. Genomic Analysis of Microbulbifer sp. Strain A4B-17 and the Characterization of Its Metabolic Pathways for 4-Hydroxybenzoic Acid Synthesis. Front. Microbiol. 2018, 9, 3115. [Google Scholar] [CrossRef]
- Witwicki, R.M.; Ekram, M.B.; Qiu, X.; Janiszewska, M.; Shu, S.; Kwon, M.; Trinh, A.; Frias, E.; Ramadan, N.; Hoffman, G.; et al. TRPS1 Is a Lineage-Specific Transcriptional Dependency in Breast Cancer. Cell Rep. 2018, 25, 1255–1267.e5. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, X.; Gong, X.; Wu, L.; Zhang, J.; Liu, W.; Li, J.; Chen, L. Atypical GATA transcription factor TRPS1 represses gene expression by recruiting CHD4/NuRD(MTA2) and suppresses cell migration and invasion by repressing TP63 expression. Oncogenesis 2018, 7, 96. [Google Scholar] [CrossRef]
- Yang, L.; Gong, X.; Wang, J.; Fan, Q.; Yuan, J.; Yang, X.; Sun, X.; Li, Y.; Wang, Y. Functional mechanisms of TRPS1 in disease progression and its potential role in personalized medicine. Pathol. Res. Pract. 2022, 237, 154022. [Google Scholar] [CrossRef]
- Yang, J.; Liu, X.; Huang, Y.; He, L.; Zhang, W.; Ren, J.; Wang, Y.; Wu, J.; Wu, X.; Shan, L.; et al. TRPS1 drives heterochromatic origin refiring and cancer genome evolution. Cell Rep. 2021, 34, 108814. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Y.; Liu, Y.; Yu, S.; Xie, H.; Shi, X.; Qin, S.; Ma, F.; Tan, T.Z.; Thiery, J.P.; et al. A central role for TRPS1 in the control of cell cycle and cancer development. Oncotarget 2014, 5, 7677–7690. [Google Scholar] [CrossRef] [PubMed]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal 2011, 4, ra41. [Google Scholar] [CrossRef]
- Hu, J.; Su, P.; Jia, M.; Wu, X.; Zhang, H.; Li, W.; Zhou, G. TRPS1 expression promotes angiogenesis and affects VEGFA expression in breast cancer. Exp. Biol. Med. 2014, 239, 423–429. [Google Scholar] [CrossRef]
- Liu, H.; Liao, Y.; Tang, M.; Wu, T.; Tan, D.; Zhang, S.; Wang, H. Trps1 is associated with the multidrug resistance of lung cancer cell by regulating MGMT gene expression. Cancer Med. 2018, 7, 1921–1932. [Google Scholar] [CrossRef]
- Serandour, A.A.; Mohammed, H.; Miremadi, A.; Mulder, K.W.; Carroll, J.S. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene 2018, 37, 5281–5291. [Google Scholar] [CrossRef]
- Strating, J.R.; Hafmans, T.G.; Martens, G.J. Functional diversity among p24 subfamily members. Biol. Cell 2009, 101, 207–220. [Google Scholar] [CrossRef]
- Zheng, H.; Yang, Y.; Han, J.; Jiang, W.-H.; Chen, C.; Wang, M.-C.; Gao, R.; Li, S.; Tian, T.; Wang, J.; et al. TMED3 promotes hepatocellular carcinoma progression via IL-11/STAT3 signaling. Sci. Rep. 2016, 6, 37070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, Y.; Li, Q. TMED3 Promotes Proliferation and Migration in Breast Cancer Cells by Activating Wnt/beta-Catenin Signaling. Onco. Targets Ther. 2020, 13, 5819–5830. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, Y.; Ye, X.; Ji, Y.; Chen, Y.; Zhang, X.; Li, Z. TMED3/RPS15A Axis promotes the development and progression of osteosarcoma. Cancer Cell Int. 2021, 21, 630. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, L.; Zhang, J. TMED3 exerts a protumor function in non-small cell lung cancer by enhancing the Wnt/beta-catenin pathway via regulation of AKT. Toxicol. Appl. Pharmacol. 2021, 433, 115793. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Zhang, M.; Wang, C. Analysis and Validation of TMED3 correlates with poor prognosis and tumor immune infiltration of glioma. J. Cancer Res. Clin. Oncol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qi, Y. Depleting TMED3 alleviates the development of endometrial carcinoma. Cancer Cell Int. 2022, 22, 231. [Google Scholar] [CrossRef]
- Wang, R.-F.; Hong, Y.-G.; Hao, L.-Q.; Yu, H.-T. Expression of TMED3 is independently associated with colorectal cancer prognosis. Exp. Ther. Med. 2022, 23, 286. [Google Scholar] [CrossRef]
- Bhatnagar, R.; Dabholkar, J.; Saranath, D. Genome-wide disease association study in chewing tobacco associated oral cancers. Oral Oncol. 2012, 48, 831–835. [Google Scholar] [CrossRef]
- Liu, T.-Y.; Lin, C.-F.; Wu, H.-T.; Wu, Y.-L.; Chen, Y.-C.; Liao, C.-C.; Chou, Y.-P.; Chao, D.; Lu, H.-F.; Chang, Y.-S.; et al. Comparison of Multiple Imputation Algorithms and Verification Using Whole-Genome Sequencing in the CMUH Genetic Biobank. Biomedicine 2021, 11, 57–65. [Google Scholar] [CrossRef]
- Liao, W.-L.; Liu, T.-Y.; Cheng, C.-F.; Chou, Y.-P.; Wang, T.-Y.; Chang, Y.-W.; Chen, S.-Y.; Tsai, F.-J. Analysis of HLA Variants and Graves’ Disease and Its Comorbidities Using a High Resolution Imputation System to Examine Electronic Medical Health Records. Front. Endocrinol. 2022, 13, 842673. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.; Whitwham, A.; Keane, T.; McCarthy, S.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).