Microenvironmental Impact on InP/ZnS-Based Quantum Dots in In Vitro Models and in Living Cells: Spectrally- and Time-Resolved Luminescence Analysis
Abstract
1. Introduction
2. Results
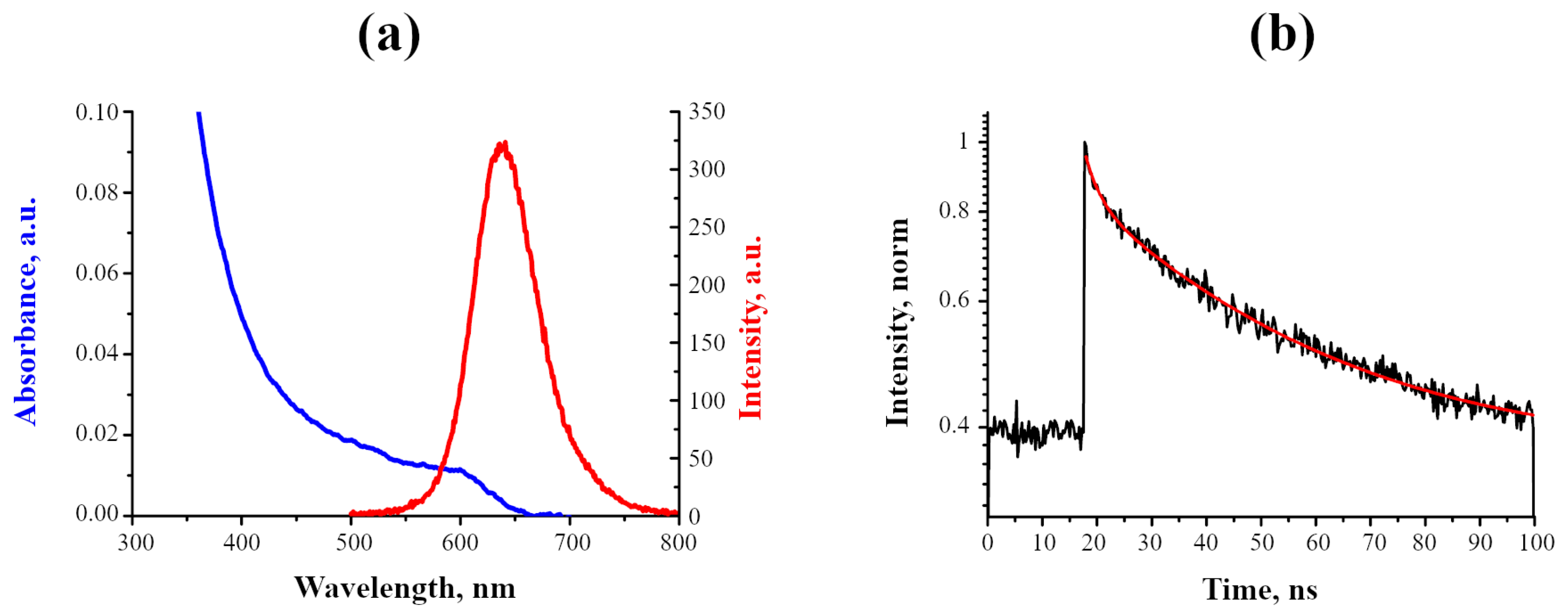
2.1. Photophysical Characteristics of InP-QDs
2.1.1. Properties of InP-QDs in
2.1.2. Effect of Components of the Physiological Environment on the Photophysical Characteristics of InP-QDs In Vitro
2.1.3. Effect of pH on the Photophysical Characteristics of InP-QDs In Vitro
2.2. InP-QD Interactions with Cells
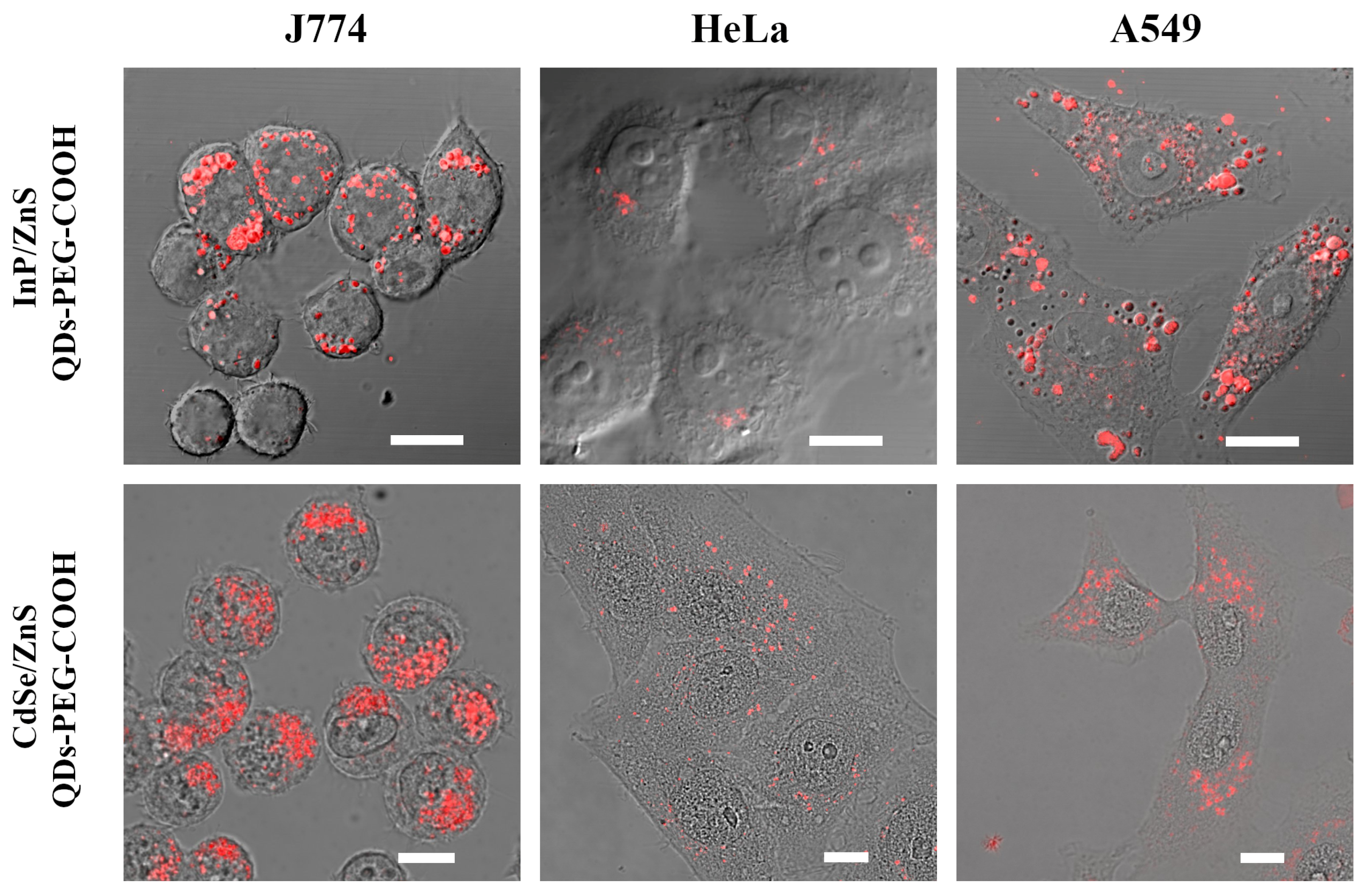
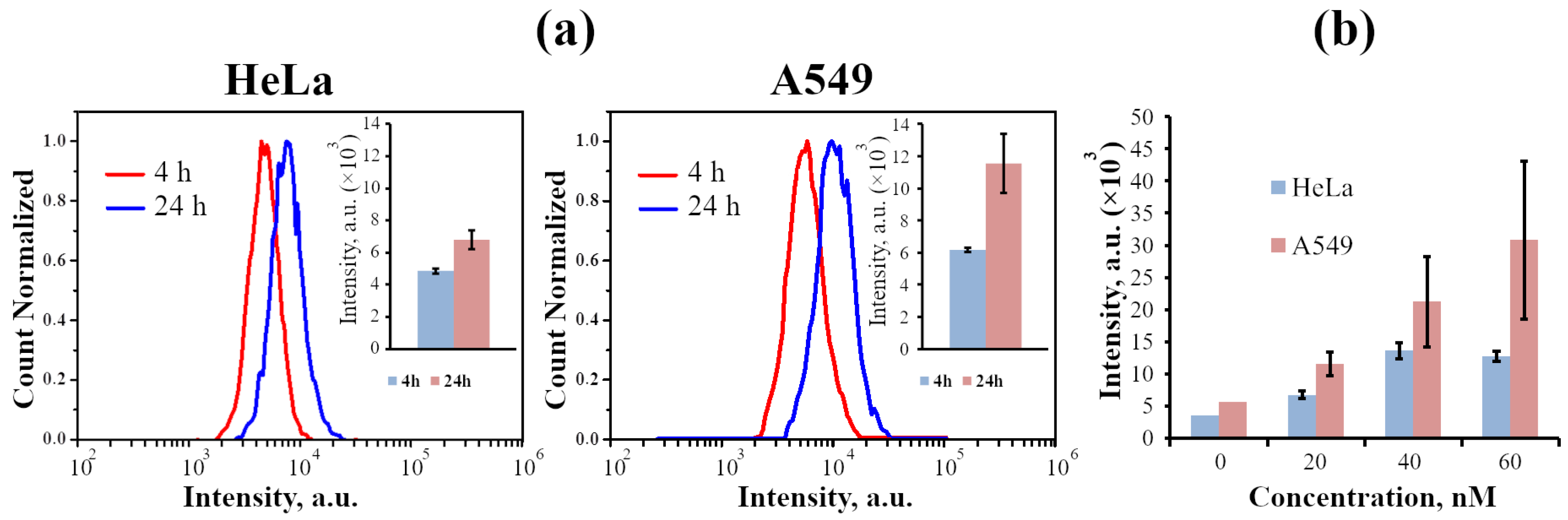
2.2.1. InP-QD Uptake by Cultured Cells of Different Types
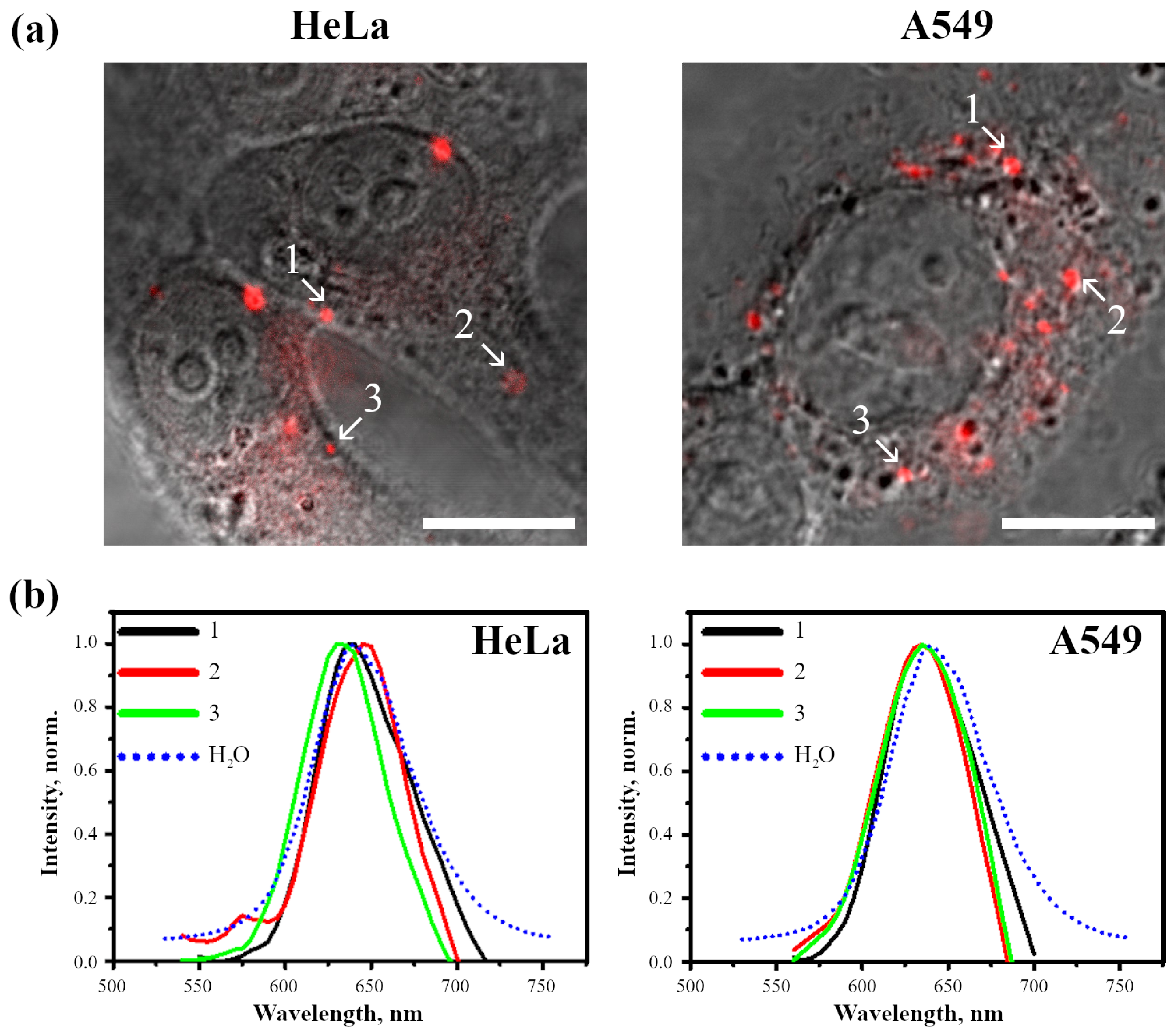
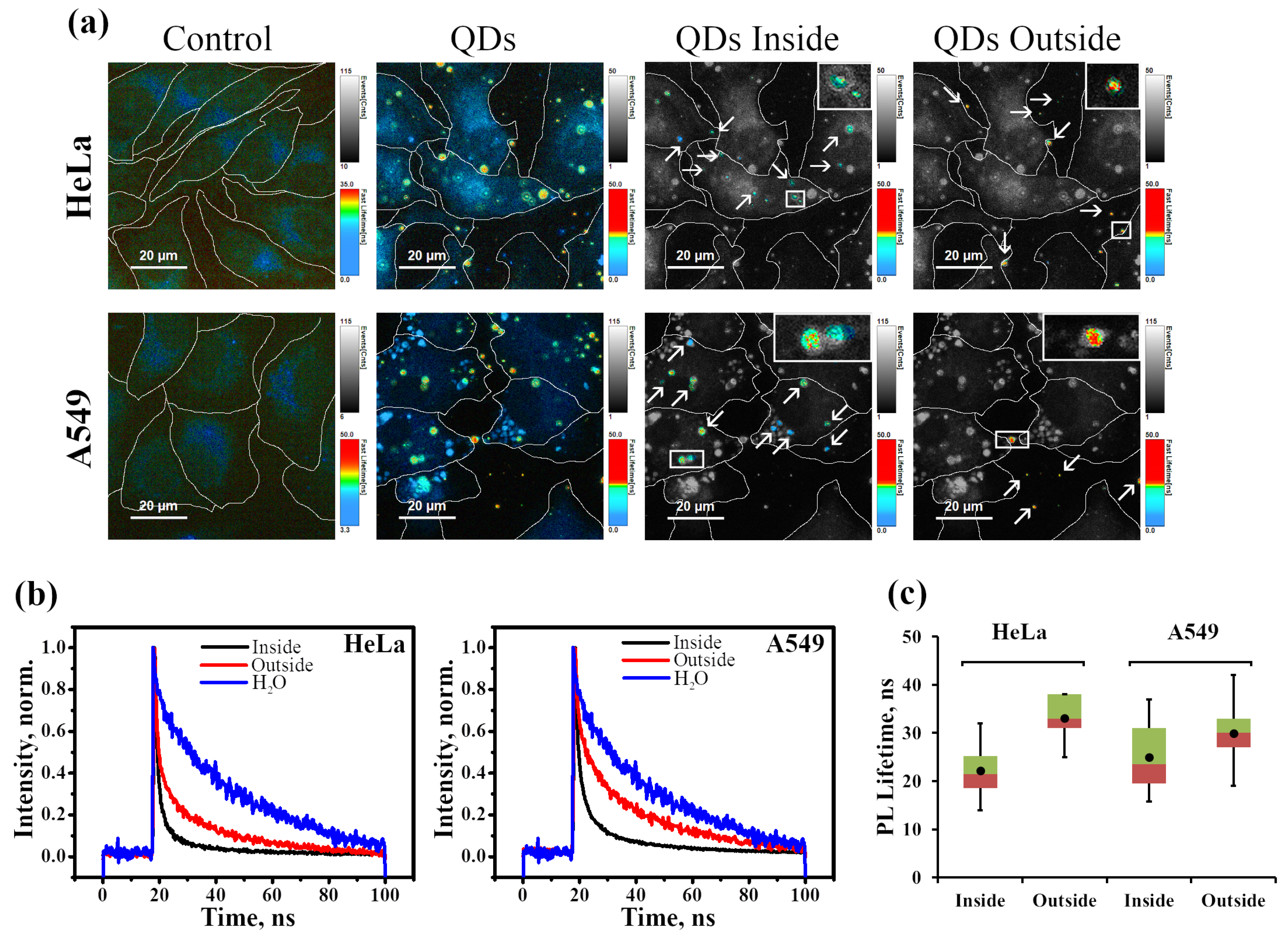
2.2.2. Analysis of Photophysical Properties of Cell-Associated InP-QDs
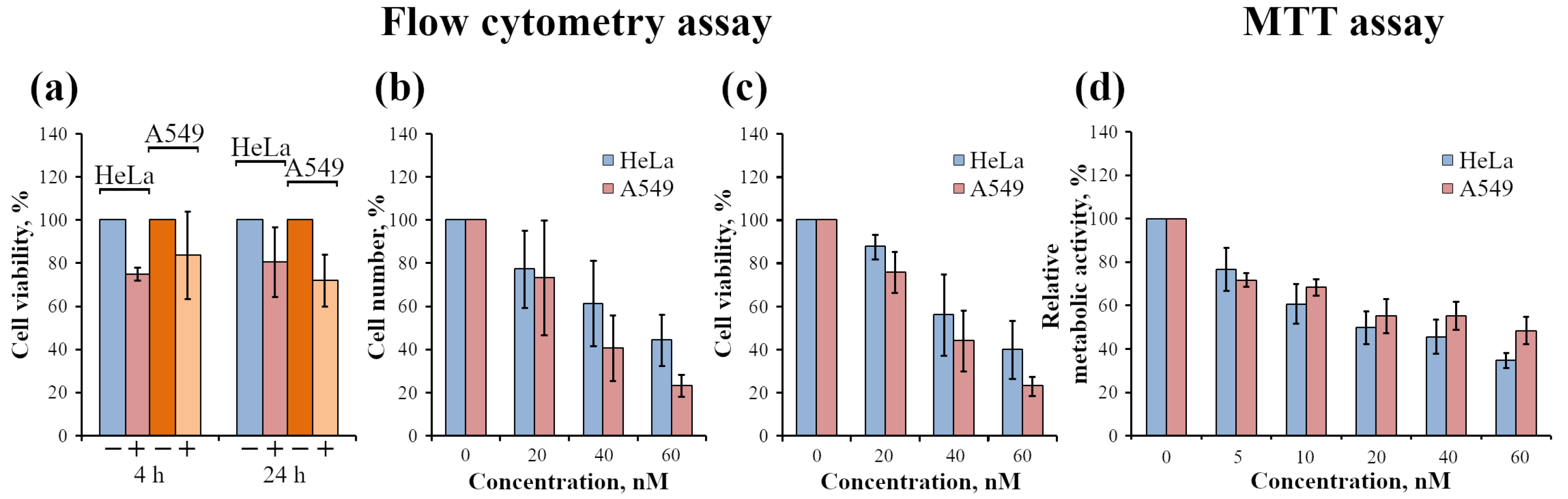
2.2.3. Analysis of InP-QD Cytotoxicity
3. Discussion
4. Materials and Methods
4.1. Analysis of the Spectral-Luminescent Properties of QDs
4.2. Estimation of Quantum Yield and Photoluminescence Lifetimes
4.3. In Vitro Models
4.4. Cell Lines
4.5. Confocal Microscopy
4.6. Analysis of Cell-Associated Fluorescence by Flow Cytometry
4.7. Estimation of Cell Survival
4.8. Evaluation of Cell Metabolic Activity by MTT Test
4.9. Statistical Data Processing
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DIC | Differential Interference Contrast |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EGF | Epidermal Growth Factor |
| FBS | Fetal Bovine Serum |
| FLIM | Fluorescence-Lifetime Imaging Microscopy |
| PBS | Phosphate-Buffered Saline |
| PEG | Polyethylene glycol |
| PL | Photoluminescence |
| QDs | Quantum Dots |
References
- Rosenthal, S.J.; Chang, J.C.; Kovtun, O.; McBride, J.R.; Tomlinson, I.D. Biocompatible quantum dots for biological applications. Chem. Biol. 2011, 18, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Turrillas, F.A.; Abad-Fuentes, A. Applications of quantum dots as probes in immunosensing of small-sized analytes. Biosens. Bioelectron. 2013, 41, 12–29. [Google Scholar] [CrossRef] [PubMed]
- McClelland, K.P.; Clemons, T.D.; Stupp, S.I.; Weiss, E.A. Semiconductor Quantum Dots Are Efficient and Recyclable Photocatalysts for Aqueous PET-RAFT Polymerization. ACS Macro Lett. 2019, 9, 7–13. [Google Scholar] [CrossRef]
- Zhu, Y.; Egap, E. PET-RAFT polymerization catalyzed by cadmium selenide quantum dots (QDs): Grafting-from QDs photocatalysts to make polymer nanocomposites. Polym. Chem. 2020, 11, 1018–1024. [Google Scholar] [CrossRef]
- Zhu, Y.; Jin, T.; Lian, T.; Egap, E. Enhancing the efficiency of semiconducting quantum dot photocatalyzed atom transfer radical polymerization by ligand shell engineering. J. Chem. Phys. 2021, 154, 204903. [Google Scholar] [CrossRef]
- López, E.; Figueroa, S.; Oset-Gasque, M.J.; González, M.P. Apoptosis and necrosis: Two distinct events induced by cadmium in cortical neurons in culture. Br. J. Pharmacol. 2003, 138, 901–911. [Google Scholar] [CrossRef]
- Mo, D.; Hu, L.; Zeng, G.; Chen, G.; Wan, J.; Yu, Z.; Huang, Z.; He, K.; Zhang, C.; Cheng, M. Cadmium-containing quantum dots: Properties, applications, and toxicity. Appl. Microbiol. Biotechnol. 2017, 101, 2713–2733. [Google Scholar] [CrossRef]
- Kirchner, C.; Liedl, T.; Kudera, S.; Pellegrino, T.; Muñoz Javier, A.; Gaub, H.E.; Stölzle, S.; Fertig, N.; Parak, W.J. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005, 5, 331–338. [Google Scholar] [CrossRef]
- Smith, A.M.; Duan, H.; Mohs, A.M.; Nie, S. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv. Drug Deliver. Rev. 2008, 60, 1226–1240. [Google Scholar] [CrossRef]
- Prasad, B.R.; Nikolskaya, N.; Connolly, D.; Smith, T.J.; Byrne, S.J.; Gerard, V.A.; Gunko, Y.K.; Rochev, Y. Long-term exposure of CdTe quantum dots on PC12 cellular activity and the determination of optimum non-toxic concentrations for biological use. J. Nanobiotechnol. 2010, 8, 7. [Google Scholar] [CrossRef]
- Chibli, H.; Carlini, L.; Park, S.; Dimitrijevic, N.M.; Nadeau, J.L. Cytotoxicity of InP/ZnS quantum dots related to reactive oxygen species generation. Nanoscale 2011, 3, 2552. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Chen, Z.; Gao, S.; Goh, B.L.; Samsudin, I.B.; Lwe, K.W.; Wu, Y.; Wu, C.; Su, X. Recent advances in non-toxic quantum dots and their biomedical applications. Prog. Nat. Sci. Mater. Int. 2019, 29, 628–640. [Google Scholar] [CrossRef]
- Young, K.T.; Wang, Y.; Roy, I.; Rui, H.; Swihart, M.T.; Law, W.C.; Kwak, S.K.; Ye, L.; Liu, J.; Mahajan, S.D.; et al. Preparation of quantum dot/drug nanoparticles formulations for traceable targeted delivery and therapy. Theranostics 2012, 2, 681–694. [Google Scholar] [CrossRef]
- Brkić, S. Applicability of Quantum Dots in Biomedical Science. In Ionizing Radiation Effects and Applications; IntechOpen: London, UK, 2018; pp. 21–39. [Google Scholar]
- Granada-Ramírez, D.; Arias-Cerón, J.S.; Rodríguez-Fragoso, P.; Vazquez, F.; Luna-Arias, J.P.; Herrera-Perez, J.L.; Mendoza-Alvarez, J. Quantum dots for biomedical applications. In Nanobiomaterials Nanostructured Materials for Biomedical Applications; Elsevier: Amsterdam, The Netherlands, 2018; pp. 411–436. [Google Scholar]
- Dong, X.-P.; Wang, X.; Xu, H. TRP channels of intracellular membranes. J. Neurochem. 2010, 113, 313–328. [Google Scholar] [CrossRef]
- Salova, A.V.; Belyaeva, T.N.; Leontieva, E.A.; Zlobina, M.V.; Kharchenko, M.V.; Kornilova, E.S. Quantum dots implementation as a label for analysis of early stages of EGF receptor endocytosis: A comparative study on cultured cells. Oncotarget 2016, 7, 6029–6047. [Google Scholar] [CrossRef] [PubMed]
- Salova, A.V.; Belyaeva, T.N.; Leontieva, E.A.; Kornilova, E.S. EGF receptor lysosomal degradation is delayed in the cells stimulated with EGF-Quantum dot bioconjugate but earlier key events of endocytic degradative pathway are similar to that of native EGF. Oncotarget 2017, 8, 44335–44350. [Google Scholar] [CrossRef]
- Yu, W.W.; Qu, L.; Guo, W.; Peng, X. Experimental Determination of the Extinction Coefficient of CdTe, CdSe, and CdS. Nanocryst. Chem. Mater. 2003, 15, 2854–2860. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, P.; Zhao, X.; Qua, L.; Lai, X. InP and Sn:InP based quantum dot sensitized solar cells. J. Mater. Chem. A 2015, 3, 21922. [Google Scholar] [CrossRef]
- Sannaikar, M.S.; Inamdar, L.S.; Pujar, G.H.; Wari, M.N.; Balasinor, N.H.; Inamdar, S.R. Comprehensive study of interaction between biocompatible PEG-InP/ZnS QDs and bovine serum albumin. Luminescence 2017, 33, 495–504. [Google Scholar] [CrossRef]
- De, C.K.; Routh, T.; Roy, D.; Mandal, S.; Mandal, P.K. Highly Photoluminescent InP Based Core Alloy Shell QDs from Air-Stable Precursors: Excitation Wavelength Dependent Photoluminescence Quantum Yield, Photoluminescence Decay Dynamics, and Single Particle Blinking Dynamics. J. Phys. Chem. 2018, 122, 964–973. [Google Scholar] [CrossRef]
- Crisp, R.W.; Kirkwood, N.; Grimaldi, G.; Kinge, S.; Siebbeles, L.D.A.; Houtepen, A.J. Highly Photoconductive InP Quantum Dots Films and Solar Cells (2018). ACS Appl. Energy Mater. 2018, 1, 6569–6576. [Google Scholar] [CrossRef] [PubMed]
- Damalakiene, L.; Karabanovas, V.; Bagdonas, S.; Rotomskis, R. Fluorescence-Lifetime Imaging Microscopy for Visualization of Quantum Dots’ Endocytic Pathway. Int. J. Mol. Sci. 2016, 17, 473. [Google Scholar] [CrossRef]
- Matiushkina, A.; Litvinov, I.; Bazhenova, A.; Belyaeva, T.; Dubavik, A.; Veniaminov, A.; Maslov, V.; Kornilova, E.; Orlova, A. Time- and Spectrally-Resolved Photoluminescence Study of Alloyed CdxZn1-xSeyS1-y/ZnS Quantum Dots and Their Nanocomposites with SPIONs in Living Cells. Int. J. Mol. Sci. 2022, 23, 4061. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.K.; Belyaeva, T.N.; Salova, A.V.; Aksenov, N.D.; Leontieva, E.A.; Shkalova, L.V.; Kornilova, E.S. Quantum Dots based on Indium Phosphide (InP): The Effect of Chemical Modifications of the Organic Shell on Interaction with Cultured Cells of Various Origins. Cell Tissue Biol. 2018, 12, 135–145. [Google Scholar] [CrossRef]
- Abraham, B.G.; Tkachenko, N.V.; Santala, V.; Lemmetyinen, H.; Karp, M. Bidirectional Fluorescence Resonance Energy Transfer (FRET) in Mutated and Chemically Modified Yellow Fluorescent Protein (YFP). Bioconjug. Chem. 2011, 22, 227–234. [Google Scholar] [CrossRef]
- Qu, S.; Sun, F.; Qiao, Z.; Li, J.; Shang, L. In Situ Investigation on the Protein Corona Formation of Quantum Dots by Using Fluorescence Resonance Energy Transfer. Small 2020, 16, 1907633. [Google Scholar] [CrossRef]
- Mindell, J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef]
- Collins, M.P.; Forgac, M. Regulation and function of V-ATPases in physiology and disease. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183341. [Google Scholar] [CrossRef]
- Zhang, L.W.; Monteiro-Riviere, N.A. Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol. Sci. 2009, 110, 138–155. [Google Scholar] [CrossRef]
- Karabanovas, V.; Zitkus, Z.; Kuciauskas, D.; Rotomskis, R.; Valius, M. Surface Properties of Quantum Dots Define Their Cellular Endocytic Routes, Mitogenic Stimulation and Suppression of Cell Migration. J. Biomed. Nanotechnol. 2014, 10, 775–786. [Google Scholar] [CrossRef]
- Kamentseva, R.; Kosheverova, V.; Kharchenko, M.; Zlobina, M.; Salova, A.; Belyaeva, T.; Nikolsky, N.; Kornilova, E. Functional cycle of EEA1-positive early endosome: Direct evidence for pre-existing compartment of degradative pathway. PLoS ONE 2020, 15, e0232532. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.T.; Zolnik, B.S.; McLeland, C.B.; Clogston, J.; Zheng, J.; McNeil, S.E. Induction of autophagy in porcine kidney cells by quantum dots: A common cellular response to nanomaterials? Toxicol. Sci. 2008, 106, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, V.; Chibli, H.; Fiammengo, R.; Galeone, A.; Malvindi, M.A.; Vecchio, G.; Cingolani, R.; Nadeau, J.L.; Pompa, P.P. InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: In vitro and in vivo toxicity assessment. Nanoscale 2013, 5, 307–317. [Google Scholar] [CrossRef]
- Tang, S.; Allagadda, V.; Chibli, H.; Nadeau, J.L.; Mayer, G.D. Comparison of cytotoxicity and expression of metal regulatory genes in zebrafish (Danio rerio) liver cells exposed to cadmium sulfate, zinc sulfate and quantum dots. Metallomics 2013, 5, 1411. [Google Scholar] [CrossRef] [PubMed]
- Soenen, S.J.; Manshian, B.B.; Aubert, T.; Himmelreich, U.; Demeester, J.; De Smedt, S.C.; Hens, Z.; Braeckmans, K. Cytotoxicity of cadmium-free quantum dots and their use in cell bioimaging. Chem. Res. Toxicol. 2014, 27, 1050–1059. [Google Scholar] [CrossRef]
- Liu, J.; Hu, R.; Liu, J.; Zhang, B.; Wang, Y.; Liu, X.; Law, W.-C.; Liu, L.; Ye, L.; Yong, K.-T. Cytotoxicity assessment of functionalized CdSe, CdTe and InP quantum dots in two human cancer cell models. Mater. Sci. Eng. C 2015, 57, 222–231. [Google Scholar] [CrossRef]
- Lin, G.; Ouyang, Q.; Hu, R.; Ding, Z.; Tian, J.; Yin, F.; Xu, G.; Chen, Q.; Wang, X.; Yong, K.T. In vivo toxicity assessment of non-cadmium quantum dots in BALB/c mice. Nanomed. Nanotechnol. 2015, 11, 341–350. [Google Scholar] [CrossRef]
- Williams, Y.; Sukhanova, A.; Conroy, J. Surface Density of Charged Functional Groups on Quantum Dots Determines Their Intracellular Compartmentalization and Biocompatibility. Asp. Nanotechnol. 2017, 1, 20–31. [Google Scholar]
- Gao, X.; Chan, W.C.W.; Nie, S. Quantum-dot nanocrystals for ultrasensitive biological labeling and multicolor optical encoding. J. Biomed. Opt. 2002, 7, 532. [Google Scholar] [CrossRef]
- Durisic, N.; Godin, A.G.; Walters, D.; Grütter, P.; Wiseman, P.W.; Heyes, C.D. Probing the “dark” fraction of core-shell quantum dots by ensemble and single particle pH-dependent spectroscopy. ACS Nano 2011, 5, 9062–9073. [Google Scholar] [CrossRef]
- Aldana, J.; Lavelle, N.; Wang, Y.J.; Peng, X.G. Size-Dependent Dissociation pH of Thiolate Ligands from Cadmium Chalcogenide Nanocrystals. J. Am. Chem. Soc. 2005, 127, 2496–2504. [Google Scholar] [CrossRef]
- Stroyuk, A.L.; Kryukov, A.I.; Kuchmii, S.Y.; Pokhodenko, V.D. Quantum size effects in photonics of semiconductor nanoparticles. Theor. Exp. Chem. 2005, 41, 67–91. [Google Scholar] [CrossRef]
- Liu, Y.-S.; Sun, Y.; Vernier, P.T.; Liang, C.-H.; Chong, S.Y.C.; Gundersen, M.A. pH-Sensitive Photoluminescence of CdSe/ZnSe/ZnS Quantum Dots in Human Ovarian Cancer Cells. J. Phys. Chem. C 2007, 111, 2872–2878. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, I.K.; Belyaeva, T.N.; Leontieva, E.A.; Orlova, A.O.; Kornilova, E.S. Changes in the Fluorescence Characteristics of Quantum Dots Based on InP/ZnS during the Interaction with Cells. Cell Tissue Biol. 2021, 15, 90–97. [Google Scholar] [CrossRef]
- Kritchenkov, I.S.; Solomatina, A.I.; Kozina, D.O.; Porsev, V.V.; Sokolov, V.V.; Shirmanova, M.V.; Lukina, M.M.; Komarova, A.D.; Shcheslavskiy, V.I.; Belyaeva, T.N.; et al. Biocompatible Ir(III) Complexes as Oxygen Sensors for Phosphorescence Lifetime Imaging. Molecules 2021, 26, 2898. [Google Scholar] [CrossRef]
- Chen, T.; Li, L.; Xu, G.; Wang, X.; Wang, J.; Chen, Y.; Lin, G. Cytotoxicity of InP/ZnS quantum dots with different surface functional groups toward two lung-derived cell lines. Front. Pharmacol. 2018, 9, 763. [Google Scholar] [CrossRef]
- Ayupova, D.; Dobhal, G.; Laufersky, G.; Nann, T.; Goreham, R. An In Vitro Investigation of Cytotoxic Effects of InP/Zns Quantum Dots with Different Surface Chemistries. Nanomaterials 2019, 9, 135. [Google Scholar] [CrossRef]
- Penjweini, R.; Roarke, B.; Alspaugh, G.; Gevorgyan, A.; Andreoni, A.; Pasut, A.; Sackett, D.L.; Knutson, J.R. Single cell-based fluorescence lifetime imaging of intracellular oxygenation and metabolism. Redox Biol. 2020, 34, 101549. [Google Scholar] [CrossRef]
- Shang, L.; Yang, L.; Wang, H.; Nienhaus, G.U. In Situ Monitoring of the Intracellular Stability of Nanoparticles by Using Fluorescence Lifetime Imaging. Small 2015, 12, 868–873. [Google Scholar] [CrossRef]
- Conroy, J.; Byrne, S.J.; Gunko, Y.K.; Rakovich, Y.P.; Donegan, J.F.; Davies, A.; Kelleher, D.; Volkov, Y. CdTe Nanoparticles Display Tropism to Core Histones and Histone-Rich Cell Organelles. Small 2008, 4, 2006–2015. [Google Scholar] [CrossRef]
- Schlegel, G.; Bohnenberger, J.; Potapova, I.; Mews, A. Fluorescence Decay Time of Single Semiconductor Nanocrystals. Phys. Rev. Lett. 2002, 88, 137401. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.A. Photoluminescence of Solutions: With Applications to Photochemistry and Analytical Chemistry; Elsevier Publishing Co.: Parker, CA, USA, 1968. [Google Scholar]
- Lakowicz, E.; Joseph, R. Principles of Fluorescence Spectroscopy; Springer Science + Business Media: Boston, MA, USA, 2013. [Google Scholar]


| , ns | , ns | , ns | <>, ns | ||||
|---|---|---|---|---|---|---|---|
| 0.77 ± 0.012 | 51 ± 2 | 0.192 ± 0.02 | 10 ± 1 | 0.04 ± 0.01 | 1.5 ± 1.1 | 49 ± 2 | |
| PBS, pH 7.4 | 0.82 ± 0.01 | 44 ± 0.27 | 0.14 ± 0.01 | 7.6 ± 1.1 | 0.03 ± 0.01 | 1 ± 0.65 | 42 ± 1 |
| PBS, pH 4.0 | 0.79 ± 0.04 | 38 ± 1.1 | 0.19 ± 0.02 | 6.3 ± 0.91 | 0.02 ± 0.03 | 0.05 ± 0.04 | 37 ± 1 |
| , ns | , ns | , ns | <>, ns | ||||
|---|---|---|---|---|---|---|---|
| 0.77 ± 0.012 | 51 ± 2 | 0.192 ± 0.02 | 10 ± 1 | 0.04 ± 0.01 | 1.5 ± 1.1 | 49 ± 2 | |
| Inside HeLa cells | 0.2 ± 0.02 | 37 ± 6 | 0.4 ± 0.1 | 7 ± 2 | 0.4 ± 0.1 | 2 ± 0.3 | 26 ± 5 |
| Outside HeLa cells | 0.45 ± 0.001 | 44 ± 4 | 0.39 ± 0.002 | 9 ± 1 | 0.16 ± 0.004 | 2 ± 0.4 | 38 ± 4 |
| Inside A549 cells | 0.25 ± 0.002 | 29 ± 3 | 0.44 ± 0.003 | 5 ± 0.7 | 0.31 ± 0.005 | 1.4 ± 0.2 | 22 ± 2 |
| Outside A549 cells | 0.57 ± 0.005 | 35 ± 2 | 0.38 ± 0.001 | 6 ± 0.5 | 0.1 ± 0.005 | 2 ± 1 | 32 ± 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Litvinov, I.; Salova, A.; Aksenov, N.; Kornilova, E.; Belyaeva, T. Microenvironmental Impact on InP/ZnS-Based Quantum Dots in In Vitro Models and in Living Cells: Spectrally- and Time-Resolved Luminescence Analysis. Int. J. Mol. Sci. 2023, 24, 2699. https://doi.org/10.3390/ijms24032699
Litvinov I, Salova A, Aksenov N, Kornilova E, Belyaeva T. Microenvironmental Impact on InP/ZnS-Based Quantum Dots in In Vitro Models and in Living Cells: Spectrally- and Time-Resolved Luminescence Analysis. International Journal of Molecular Sciences. 2023; 24(3):2699. https://doi.org/10.3390/ijms24032699
Chicago/Turabian StyleLitvinov, Ilia, Anna Salova, Nikolay Aksenov, Elena Kornilova, and Tatiana Belyaeva. 2023. "Microenvironmental Impact on InP/ZnS-Based Quantum Dots in In Vitro Models and in Living Cells: Spectrally- and Time-Resolved Luminescence Analysis" International Journal of Molecular Sciences 24, no. 3: 2699. https://doi.org/10.3390/ijms24032699
APA StyleLitvinov, I., Salova, A., Aksenov, N., Kornilova, E., & Belyaeva, T. (2023). Microenvironmental Impact on InP/ZnS-Based Quantum Dots in In Vitro Models and in Living Cells: Spectrally- and Time-Resolved Luminescence Analysis. International Journal of Molecular Sciences, 24(3), 2699. https://doi.org/10.3390/ijms24032699






