A Mixture of Chemicals Found in Human Amniotic Fluid Disrupts Brain Gene Expression and Behavior in Xenopus laevis
Abstract
1. Introduction
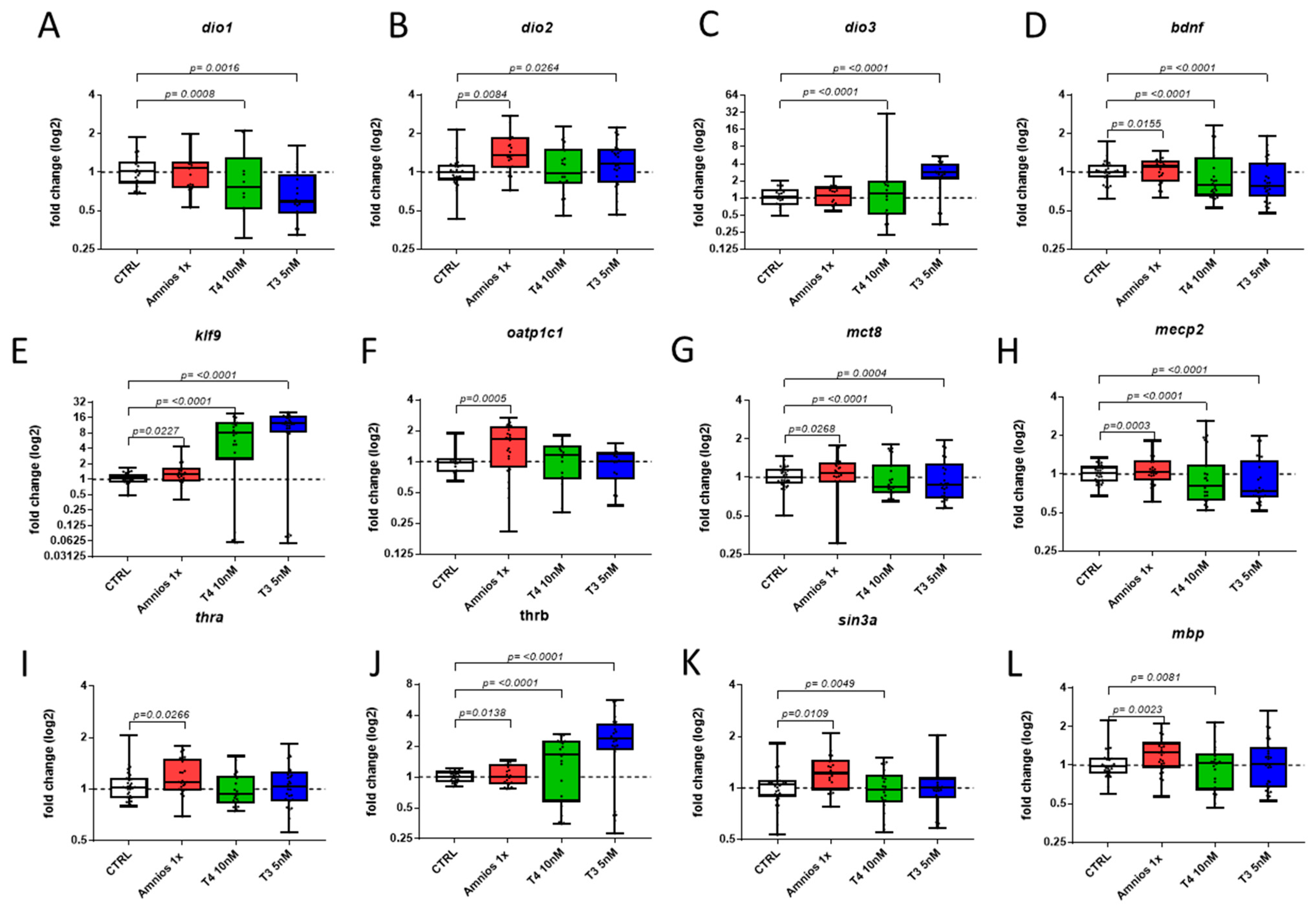
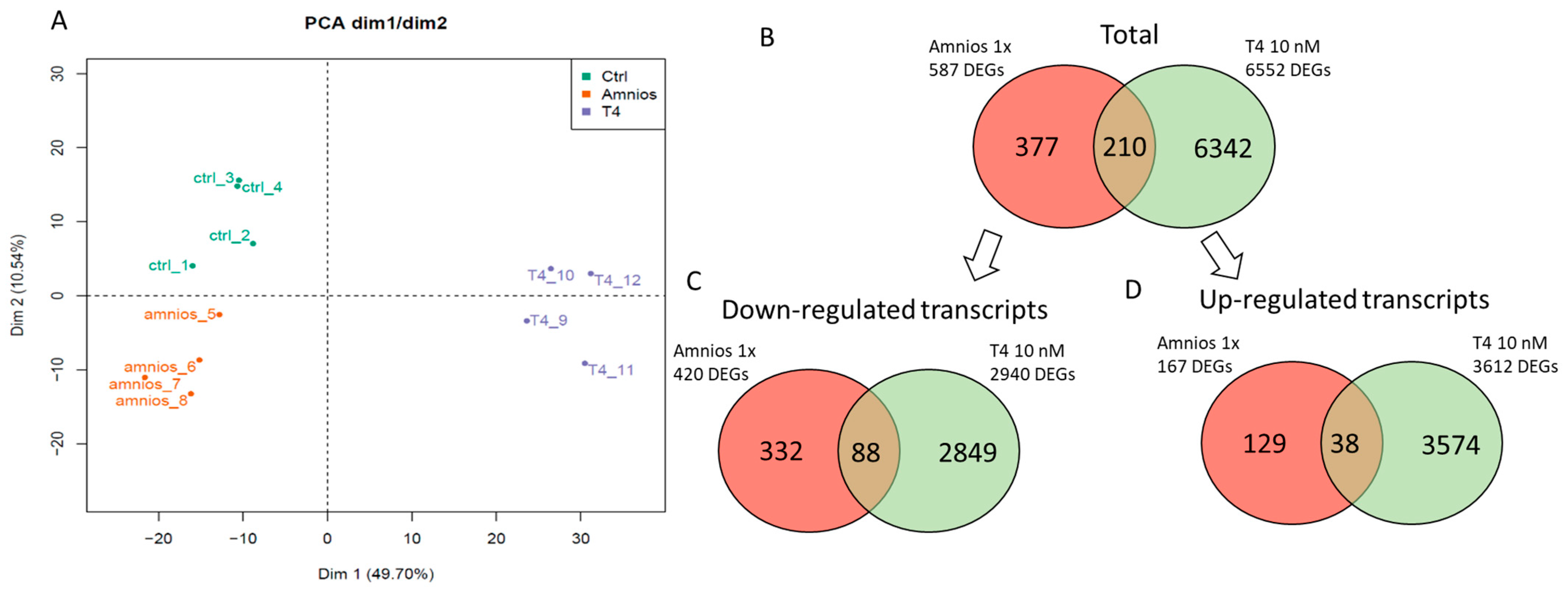
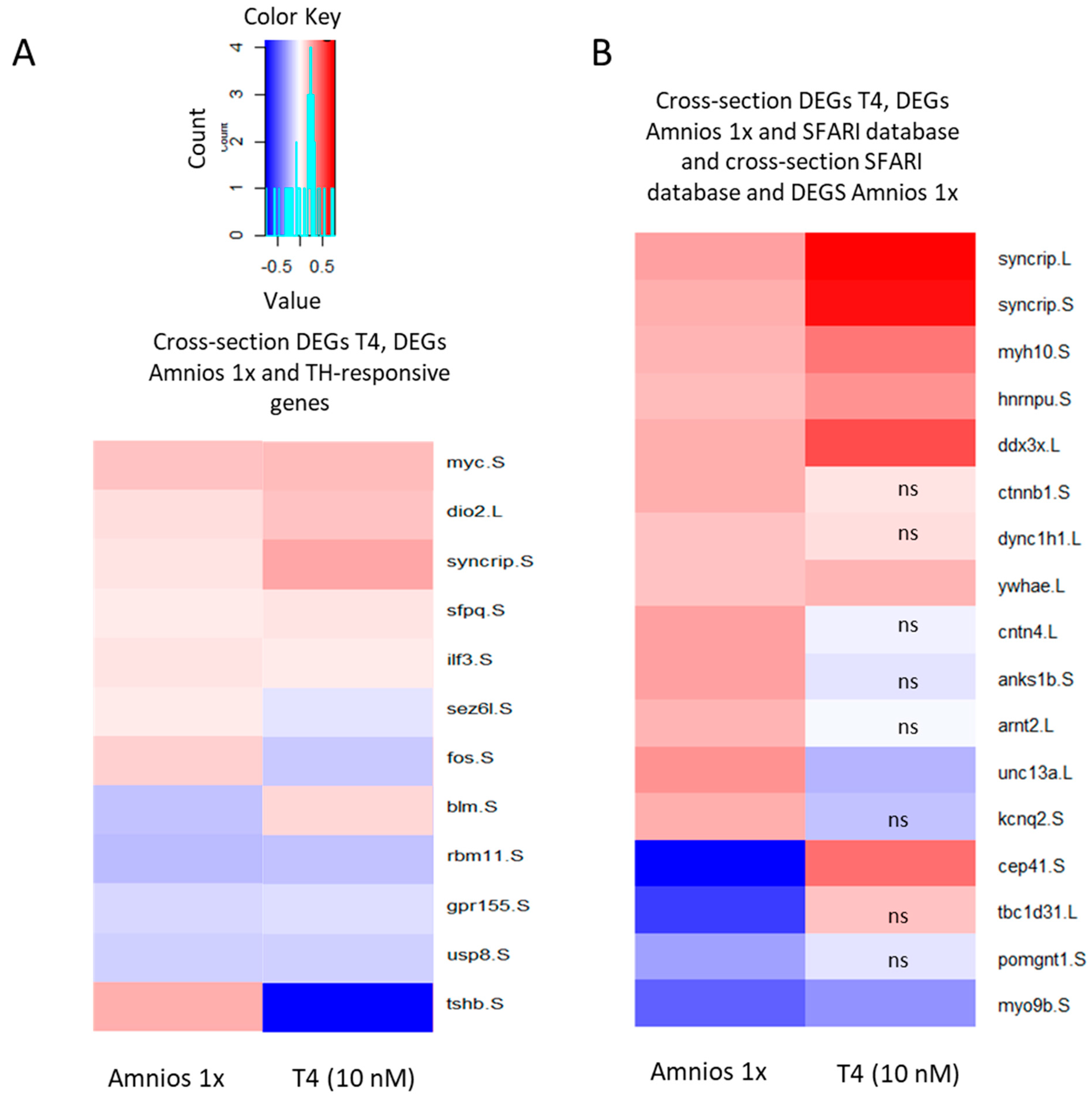
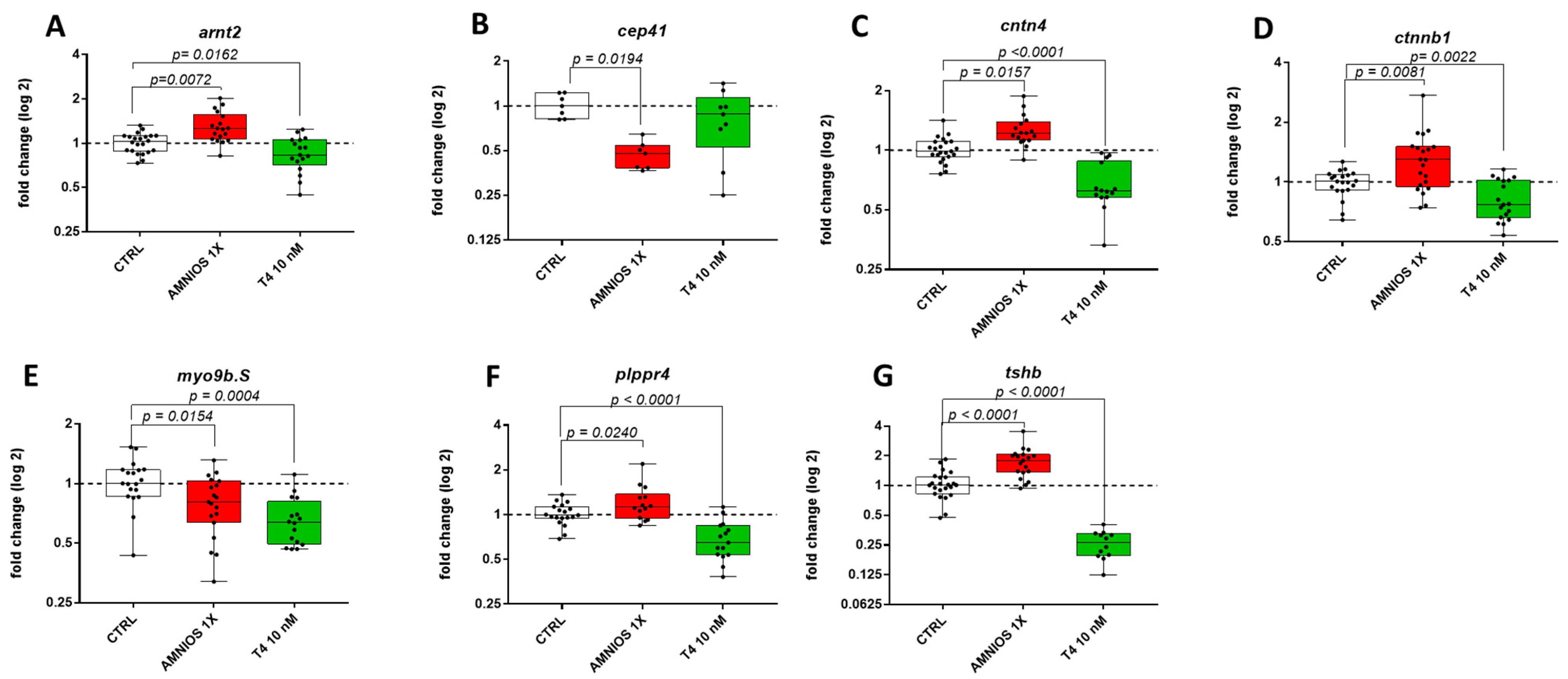
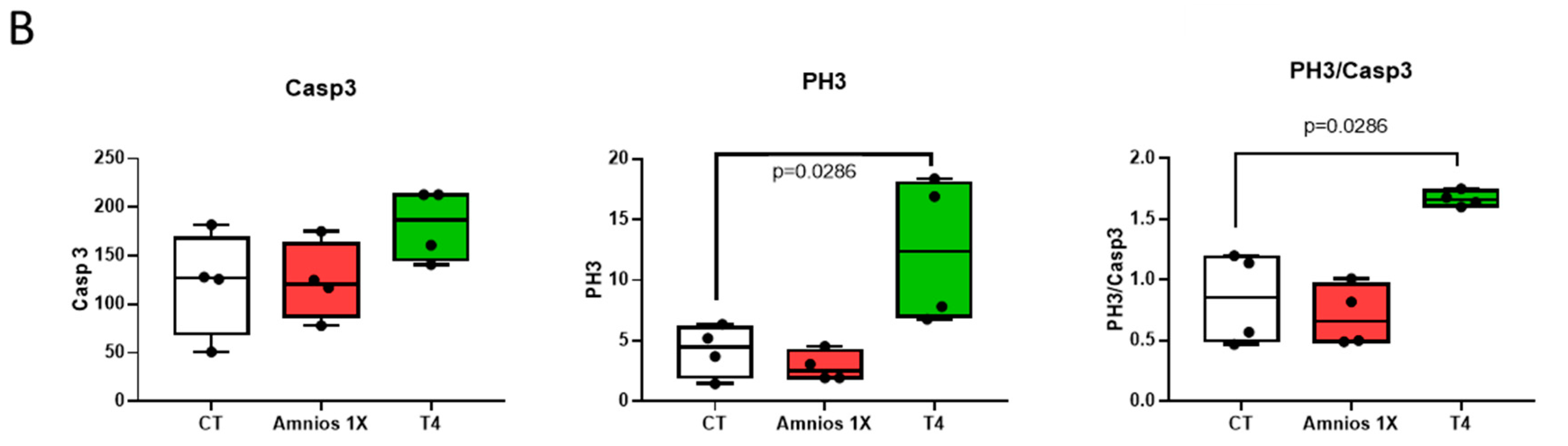
2. Results
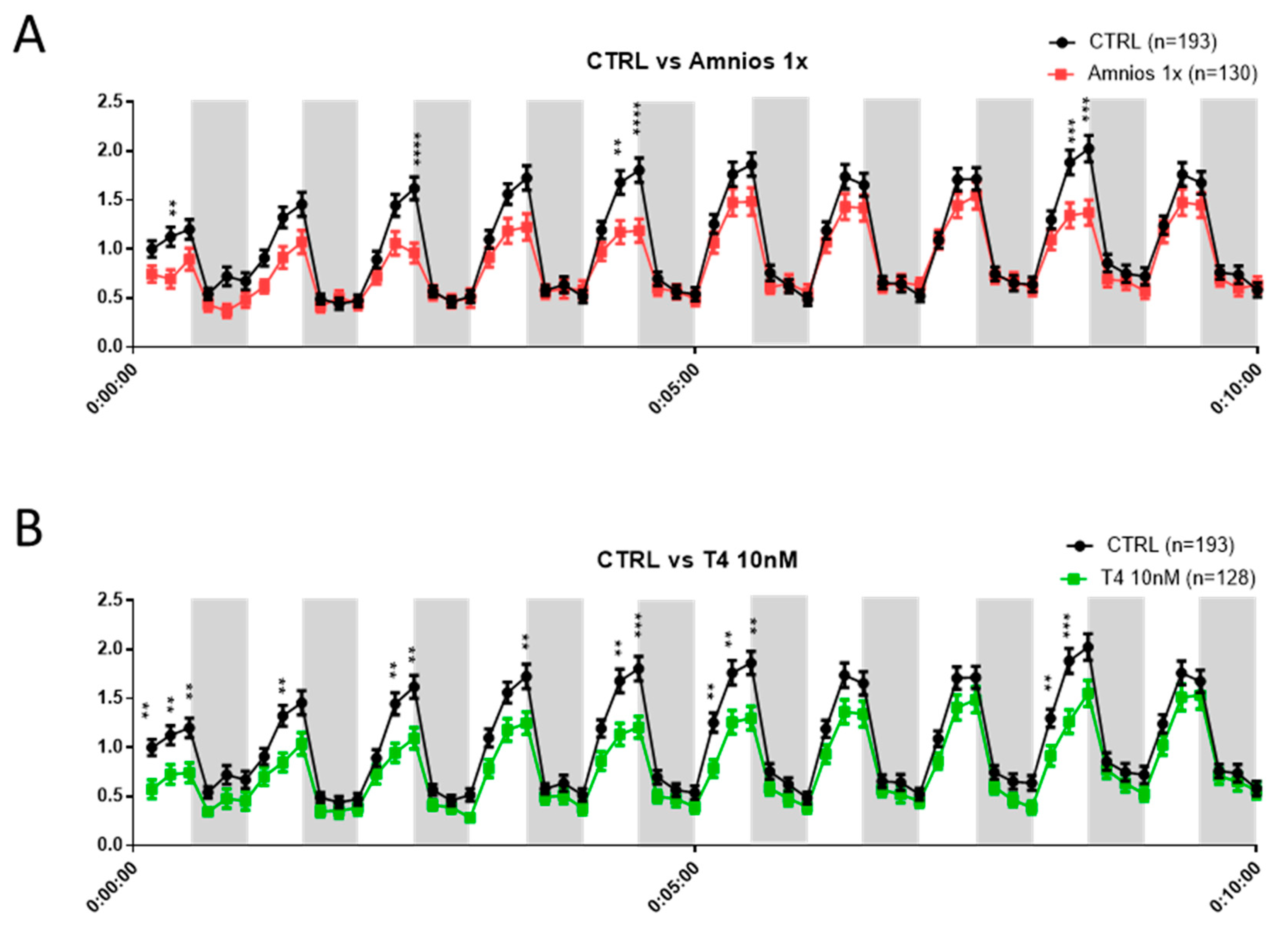
Exposure to an Amniotic Mixture Alters the Tadpole Behavior
3. Discussion
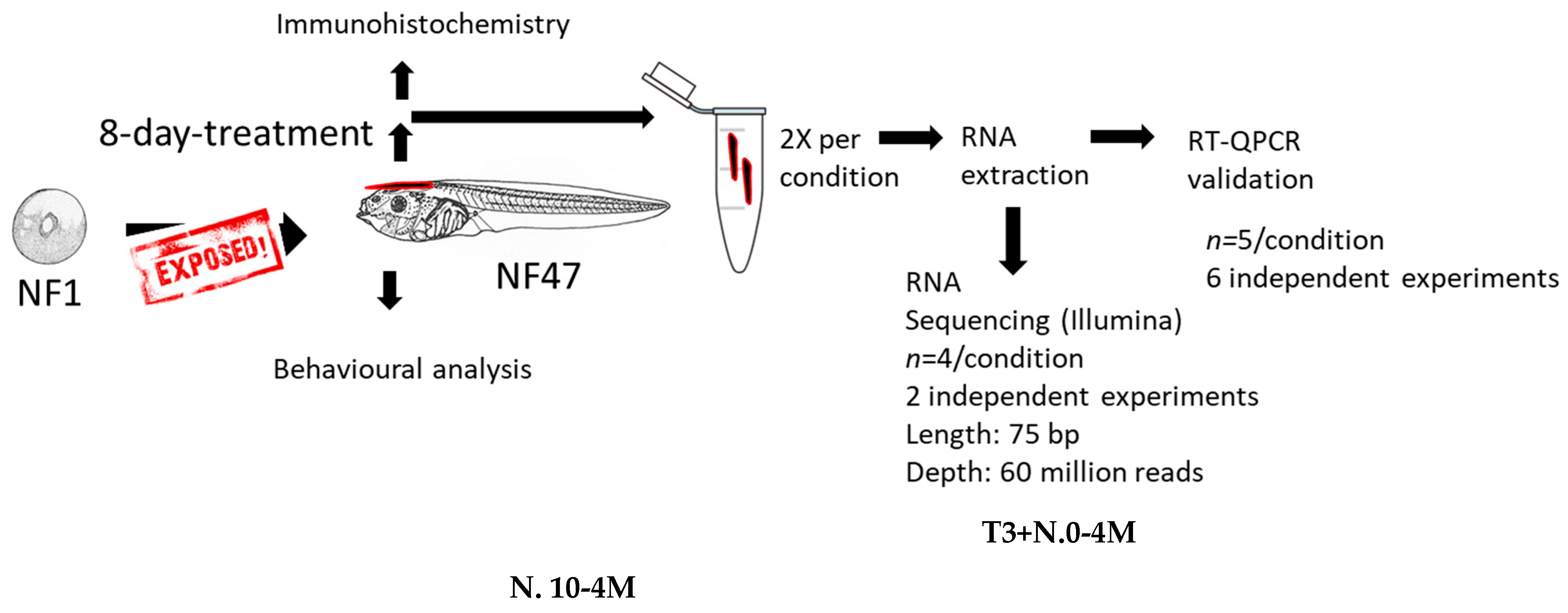
4. Material and Methods
4.1. Chemical Exposure
4.2. RNA Extraction
4.3. RT-qPCR
4.4. RNA-Sequencing
4.5. DAVID
4.6. Mobility
4.7. Immunohistochemistry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Railton, T.C. Sporadic Cretinism Treated by Administration of the Thyroid Gland. Br. Med. J. 1894, 1, 1180–1181. [Google Scholar] [CrossRef] [PubMed]
- Smith, T. Case of sporadic cretinism treated with thyroid gland. Br. Med. J. 1894, 1, 1178–1180. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J. Thyroid hormone receptors in brain development and function. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 249–259. [Google Scholar] [CrossRef]
- Nunez, J.; Celi, F.S.; Ng, L.; Forrest, D. Multigenic control of thyroid hormone functions in the nervous system. Mol. Cell. Endocrinol. 2008, 287, 1–12. [Google Scholar] [CrossRef]
- Pharoah, P.O.; Ellis, S.M.; Ekins, R.P.; Williams, E.S. Maternal thyroid function, iodine deficiency and fetal development. Clin. Endocrinol. 1976, 5, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Pop, V.J.; Kuijpens, J.L.; van Baar, A.L.; Verkerk, G.; van Son, M.M.; de Vijlder, J.J.; Vulsma, T.; Wiersinga, W.M.; Drexhage, H.A.; Vader, H.L. Low maternal free thyroxine concentrations during early pregnancy are associated with impaired psychomotor development in infancy. Clin. Endocrinol. 1999, 50, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Levie, D.; Derakhshan, A.; Shu, H.; Broeren, M.; de Poortere, R.; Peeters, R.; Bornehag, C.-G.; Demeneix, B.; Korevaar, T. The association of maternal iodine status in early pregnancy with thyroid function in the SELMA study. Thyroid 2019, 29, 1660–1668. [Google Scholar] [CrossRef]
- Obregon, M.J.; Calvo, R.M.; Escobar Del Rey, F.; Morreale de Escobar, G. Ontogenesis of thyroid function and interactions with maternal function. Endocr. Dev. 2007, 10, 86–98. [Google Scholar] [CrossRef]
- Korevaar, T.I.M.; Muetzel, R.; Medici, M.; Chaker, L.; Jaddoe, V.W.V.; De Rijke, Y.B.; Steegers, E.P.; Visser, T.J.; White, T.; Peeters, R.P. Association of maternal thyroid function during early pregnancy with off spring IQ and brain morphology in childhood: A population-based prospective cohort study. Artic. Lancet Diabetes Endocrinol. 2016, 4, 35–43. [Google Scholar] [CrossRef]
- Honda, J.; Ogawa, K.; Taniguchi, K. [Immunohistochemical and morphometric studies on the development of the thyroid, parathyroid and ultimobranchial body in Xenopus laevis Daudin]. Jikken Dobutsu 1993, 42, 23–32. [Google Scholar] [CrossRef]
- Tata, J.R. Early metamorphic competence of Xenopus larvae. Dev. Biol. 1968, 18, 415–440. [Google Scholar] [CrossRef]
- Baker, B.S.; Tata, J.R. Accumulation of proto-oncogene c-erb-A related transcripts during Xenopus development: Association with early acquisition of response to thyroid hormone and estrogen. EMBO J. 1990, 9, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.B.; Wong, J.; Puzianowska-Kuznicka, M.; Stolow, M.A. Tadpole competence and tissue-specific temporal regulation of amphibian metamorphosis: Roles of thyroid hormone and its receptors. BioEssays News Rev. Mol. Cell. Dev. Biol. 1996, 18, 391–399. [Google Scholar] [CrossRef]
- Fini, J.B.; Le Mével, S.; Palmier, K.; Darras, V.M.; Punzon, I.; Richardson, S.J.; Clerget-Froidevaux, M.S.; Demeneix, B.A. Thyroid hormone signaling in the Xenopus laevis embryo is functional and susceptible to endocrine disruption. Endocrinology 2012, 153, 5068–5081. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, T.J.; Zota, A.R.; Schwartz, J.M. Environmental Chemicals in Pregnant Women in the United States: NHANES 2003–2004. Environ. Health Perspect. 2011, 119, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Saaristo, M.; Brodin, T.; Balshine, S.; Bertram, M.G.; Brooks, B.W.; Ehlman, S.M.; McCallum, E.S.; Sih, A.; Sundin, J.; Wong, B.B.M.; et al. Direct and indirect effects of chemical contaminants on the behaviour, ecology and evolution of wildlife. Proc. Biol. Sci. 2018, 285, 20181297. [Google Scholar] [CrossRef] [PubMed]
- Boas, M.; Feldt-rasmussen, U.; Main, K.M. Molecular and Cellular Endocrinology Thyroid effects of endocrine disrupting chemicals. Mol. Cell. Endocrinol. 2012, 355, 240–248. [Google Scholar] [CrossRef]
- Crofton, K.M. Thyroid disrupting chemicals: Mechanisms and mixtures. Int. J. Androl. 2008, 31, 209–223. [Google Scholar] [CrossRef]
- Fini, J.; Mughal, B.B.; Mével, S.L.; Leemans, M.; Lettmann, M.; Spirhanzlova, P.; Affaticati, P.; Jenett, A. Human amniotic fluid contaminants alter thyroid hormone signalling and early brain development in Xenopus embryos. Sci. Rep. 2017, 7, 43786. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef]
- Kavčič, N.; Pegan, K.; Turk, B. Lysosomes in programmed cell death pathways: From initiators to amplifiers. Biol. Chem. 2017, 398, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Chatonnet, F.; Flamant, F.; Morte, B. A temporary compendium of thyroid hormone target genes in brain. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Rosina, E.; Battan, B.; Siracusano, M.; Di Criscio, L.; Hollis, F.; Pacini, L.; Curatolo, P.; Bagni, C. Disruption of mTOR and MAPK pathways correlates with severity in idiopathic autism. Transl. Psychiatry 2019, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-H.; Lee, Y.-S. Cell type-specific roles of RAS-MAPK signaling in learning and memory: Implications in neurodevelopmental disorders. Neurobiol. Learn. Mem. 2016, 135, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Vithayathil, J.; Pucilowska, J.; Landreth, G.E. ERK/MAPK signaling and autism spectrum disorders. In Progress in Brain Research; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 241, pp. 63–112. [Google Scholar]
- Faridar, A.; Jones-Davis, D.; Rider, E.; Li, J.; Gobius, I.; Morcom, L.; Richards, L.J.; Sen, S.; Sherr, E.H. Mapk/Erk activation in an animal model of social deficits shows a possible link to autism. Mol. Autism 2014, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.K.; Cline, H.T. Thyroid Hormone Acts Locally to Increase Neurogenesis, Neuronal Differentiation, and Dendritic Arbor Elaboration in the Tadpole Visual System. J. Neurosci. 2016, 36, 10356–10375. [Google Scholar] [CrossRef]
- Straka, H.; Simmers, J. Xenopus laevis: An ideal experimental model for studying the developmental dynamics of neural network assembly and sensory-motor computations. Dev. Neurobiol. 2012, 72, 649–663. [Google Scholar] [CrossRef]
- Leloup, J.; Buscaglia, M. La triiodothyronine, hormone de la metamorphose des Amphibiens. C. R. Acad. Sci. Paris D 1977, 284, 2261–2263. [Google Scholar]
- Regard, E. Cytophysiology of the Amphibian Thyroid Gland through Larval Development and Metamorphosis. Int. Rev. Cytol. 1978, 52, 81–118. [Google Scholar] [CrossRef]
- Prati, M.; Calvo, R.; Morreale De Escobar, G. L-thyroxine and 3,5,3′-triiodothyronine concentrations in the chicken egg and in the embryo before and after the onset of thyroid function. Endocrinology 1992, 130, 2651–2659. [Google Scholar] [CrossRef]
- McNabb, F.M.A.; Darras, V.M. Thyroids. In Sturkie’s Avian Physiology, 6th ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 535–547. ISBN 978-0-12-407160-5. [Google Scholar]
- Ruuskanen, S.; Darras, V.M.; Visser, M.E.; Groothuis, T.G.G. Effects of experimentally manipulated yolk thyroid hormone levels on offspring development in a wild bird species. Horm. Behav. 2016, 81, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Flickinger, R.A. Sequential appearance of monoiodotyrosine, diiodotyrosine, and thyroxine in the developing frog embryo. Gen. Comp. Endocrinol. 1964, 4, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Denef, C. Paracrinicity: The story of 30 years of cellular pituitary crosstalk. J. Neuroendocrinol. 2008, 20, 1–70. [Google Scholar] [CrossRef] [PubMed]
- Licht, P.; Denver, R.J. Regulation of pituitary thyrotropin secretion. Prog. Clin. Biol. Res. 1990, 342, 427–432. [Google Scholar] [PubMed]
- Büyükgebiz, A. Newborn screening for congenital hypothyroidism. JCRPE J. Clin. Res. Pediatr. Endocrinol. 2013, 5, 8–12. [Google Scholar] [CrossRef]
- Tonyushkina, K.N.; Shen, M.C.; Ortiz-Toro, T.; Karlstrom, R.O. Embryonic exposure to excess thyroid hormone causes thyrotrope cell death. J. Clin. Investig. 2014, 124, 321–327. [Google Scholar] [CrossRef]
- Zucchi, R.; Rutigliano, G.; Saponaro, F. Novel thyroid hormones. Endocrine 2019, 66, 95–104. [Google Scholar] [CrossRef]
- Tindall, A.J.; Morris, I.D.; Pownall, M.E.; Isaacs, H.V. Expression of enzymes involved in thyroid hormone metabolism during the early development of Xenopus tropicalis. Biol. Cell 2007, 99, 151–163. [Google Scholar] [CrossRef]
- Dubois, G.M.; Sebillot, A.; Kuiper, G.G.J.M.; Verhoelst, C.H.J.; Darras, V.M.; Visser, T.J.; Demeneix, B.A. Deiodinase activity is present in Xenopus laevis during early embryogenesis. Endocrinology 2006, 147, 4941–4949. [Google Scholar] [CrossRef]
- Spirhanzlova, P.; Leemans, M.; Demeneix, B.A.; Fini, J.-B. Following Endocrine-Disrupting Effects on Gene Expression in Xenopus laevis. Cold Spring Harb. Protoc. 2019, 2019, prot098301. [Google Scholar] [CrossRef]

| n° | Family | Molecule | Concentration 1X (Actual Concentration Found in Amniotic Fluid) |
|---|---|---|---|
| 1 | Phenol | Bisphenol A | 0.2 × 10−8 M |
| 2 | Phenol | Triclosan | 0.7 × 10−7 M |
| 3 | Phenol | Benzophenone-3 | 0.86 × 10−7 M |
| 4 | Phthalate | Dibutyl phthalate | 0.24 × 10−6 M |
| 5 | Phthalate | Di-2-ethylhexyl phthalate | 0.1 × 10−6 M |
| 6 | Organochlorine pesticide | Hexachlorobenzene | 0.8 × 10−11 M |
| 7 | Organochlorine pesticide | Dichlorodiphenyldichloroethylene | 0.66 × 10−9 M |
| 8 | Perfluorinated compound | Perfluorooctanoic acid | 0.43 × 10−8 M |
| 9 | Perfluorinated compound | Perfluorooctanesulfonic acid | 0.8 × 10−8 M |
| 10 | Poly aromatic hydroxylated compound | 2-napthol | 0.5 × 10−8 M |
| 11 | Polychlorinated compound | Sodium perchlorate monohydrate | 0.3 × 10−8 M |
| 12 | Polybrominated compound | Decabromodiphenyl ether | 0.63 × 10−9 M |
| 13 | Polychlorinated compound | PCB-153 | 0.2 × 10−8 M |
| 14 | Heavy metal | Methyl mercury(III) chloride | 0.5 × 10−7 M |
| 15 | Heavy metal | Lead (II) chloride | 0.21 × 10−8 M |
| Exposure | Gene | Pathway | |
|---|---|---|---|
| Mixture 1x | cacnb1.S (calcium channel, voltage-dependent, beta 1 subunit S) nfkb2.S (nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) S) myd88.S (myeloid differentiation primary response 88 S) irak4.L (interleukin 1 receptor associated kinase 4 L) fos.S (FBJ murine osteosarcoma viral oncogene S) myc.S (v-myc avian myelocytomatosis viral oncogene S) hspa8.L (heat shock protein family A (Hsp70) member 8) | MAPK signaling pathway | |
| Mixture 1x | ctns.L (cystinosin, lysosomal cystine transporter glb1l.L (galactosidase beta 1 like L) naga.L (N-acetylgalactosaminidase, alpha- L) mfsd8.L (major facilitator superfamily domain containing 8 L) galc.L (galactosylceramidase L) arsa.1.S (arylsulfatase A, gene 1 S) ap3s2.S (adaptor related protein complex 3 sigma 2 subunit S) gm2a.L (GM2 ganglioside activator L) | Lysosome | |
| T4 (10 nM) | gria1.L/S (glutamate receptor, ionotropic, AMPA 1 L/S) kiss1.S (kisspeptin S) tshb.L.S (thyroid stimulating hormone, beta L/S) gabrd.L (gamma-aminobutyric acid (GABA) A receptor, delta L) gnrh2.L (gonadotropin releasing hormone 2 L) htr2C.L (5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled L) glrb.L/S (glycine receptor beta L/S) gabbr1.S (gamma-aminobutyric acid (GABA) B receptor, 1 S) pth2R.L (parathyroid hormone 2 receptor L) pyy.S (peptide YY S) pth2R.L (nociceptin receptor-like S) sstr5.S (somatostatin receptor 5 S) nmur1.L (neuromedin U receptor 1 L) grpr.L (gastrin releasing peptide receptor L) p2rx5.L (purinergic receptor P2X, ligand gated ion channel, 5 L) gpr83.2.L (G protein-coupled receptor 83 L) s1pr5. L (sphingosine-1-phosphate receptor 5 L) avpr1a.L (arginine vasopressin receptor 1A L) nts.L (neurotensin L) drd1.S (dopamine receptor D1 S) chrna7.S (cholinergic receptor, nicotinic alpha 7 S) galr3.L/S (galanin receptor 3 L/S) adm.S (adrenomedullin S) thrb.L (thyroid hormone receptor, beta L) | vip.S (vasoactive intestinal peptide S) htr5.L (5-hydroxytryptamine (serotonin) receptor 5A, G protein-coupled L) mc5r.L (melanocortin 5 receptor L) tacr3.L (tachykinin receptor 3 L) calcr.S (calcitonin receptor S) lpar1.L (lysophosphatidic acid receptor 1 L) drd2.L/S (dopamine receptor D2 L/S) grik2 (glutamate receptor, ionotropic, kainate 2) crhr2.S (corticotropin releasing hormone receptor 2 L/S) cga.L/S (glycoprotein hormones, alpha polypeptide S) ednrb2.S (endothelin receptor B subtype 2 S) lpar1.S (lysophosphatidic acid receptor 1 S) trh.L (thyrotropin-releasing hormone L) adcyap1.L (adenylate cyclase activating polypeptide 1 (pituitary) L) ghr.L (growth hormone receptor L) aplnr.L (apelin receptor L) pdyn.L/S (prodynorphin L) s1pr1.L (sphingosine-1-phosphate receptor 1 L) penk.L (proenkephalin L) prl.1.S (prolactin, gene 1) tac1.L/S (tachykinin precursor 1 L/S) | Neuroactive ligand-receptor interaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leemans, M.; Spirhanzlova, P.; Couderq, S.; Le Mével, S.; Grimaldi, A.; Duvernois-Berthet, E.; Demeneix, B.; Fini, J.-B. A Mixture of Chemicals Found in Human Amniotic Fluid Disrupts Brain Gene Expression and Behavior in Xenopus laevis. Int. J. Mol. Sci. 2023, 24, 2588. https://doi.org/10.3390/ijms24032588
Leemans M, Spirhanzlova P, Couderq S, Le Mével S, Grimaldi A, Duvernois-Berthet E, Demeneix B, Fini J-B. A Mixture of Chemicals Found in Human Amniotic Fluid Disrupts Brain Gene Expression and Behavior in Xenopus laevis. International Journal of Molecular Sciences. 2023; 24(3):2588. https://doi.org/10.3390/ijms24032588
Chicago/Turabian StyleLeemans, Michelle, Petra Spirhanzlova, Stephan Couderq, Sébastien Le Mével, Alexis Grimaldi, Evelyne Duvernois-Berthet, Barbara Demeneix, and Jean-Baptiste Fini. 2023. "A Mixture of Chemicals Found in Human Amniotic Fluid Disrupts Brain Gene Expression and Behavior in Xenopus laevis" International Journal of Molecular Sciences 24, no. 3: 2588. https://doi.org/10.3390/ijms24032588
APA StyleLeemans, M., Spirhanzlova, P., Couderq, S., Le Mével, S., Grimaldi, A., Duvernois-Berthet, E., Demeneix, B., & Fini, J.-B. (2023). A Mixture of Chemicals Found in Human Amniotic Fluid Disrupts Brain Gene Expression and Behavior in Xenopus laevis. International Journal of Molecular Sciences, 24(3), 2588. https://doi.org/10.3390/ijms24032588





